Peripheral Inflammatory Cytokine Signature Mirrors Motor Deficits in Mucolipidosis IV
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Blood Sample Collection and Processing
2.3. Animal Studies and Sample Collection
2.4. Cytokine Luminex Immunoassays
2.5. Statistical and Multivariate Analyses
3. Results
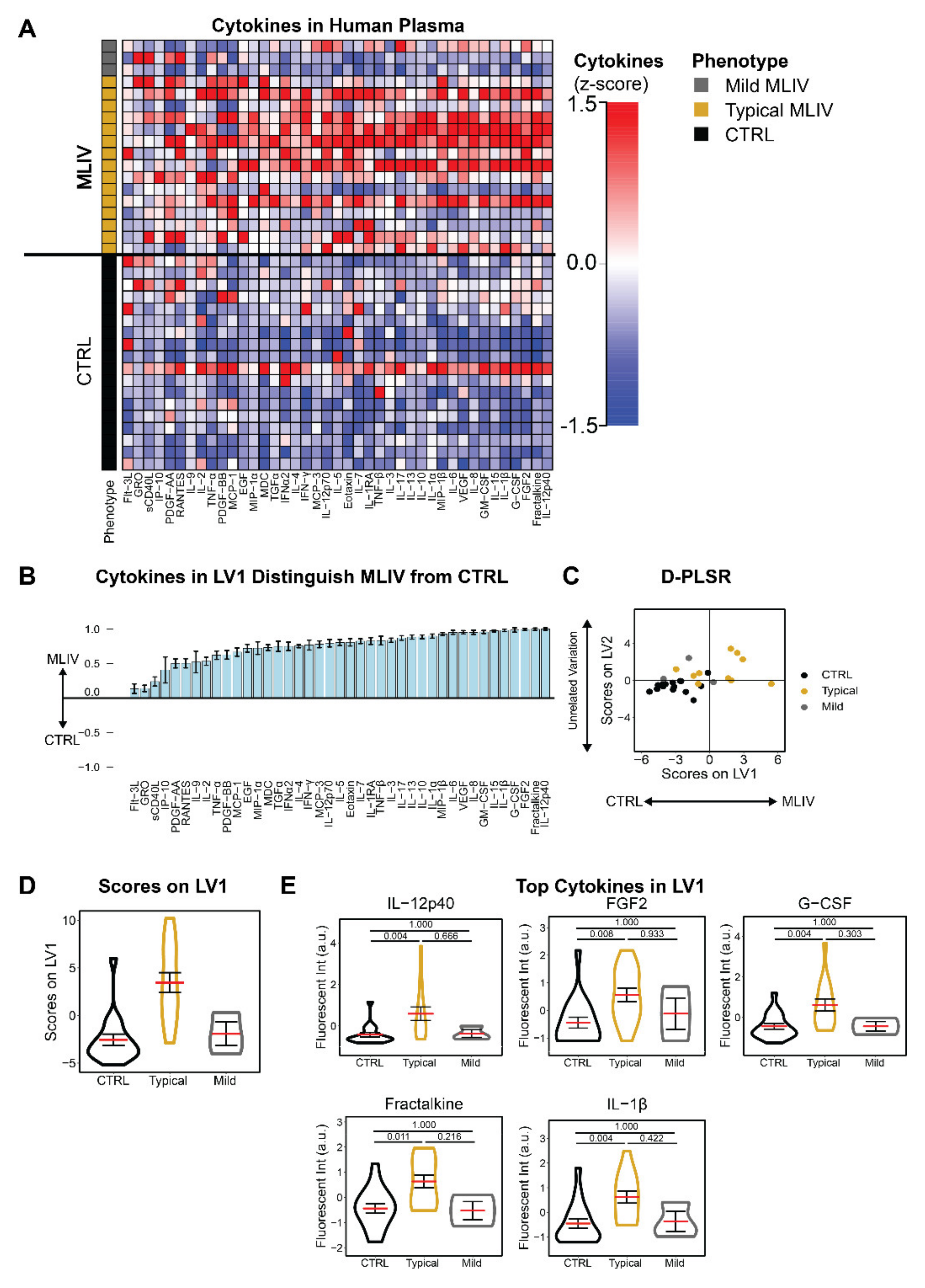
3.1. Mucolipidosis Type IV Patients Exhibit Pro-Inflammatory Blood Signatures Compared to Familial Controls
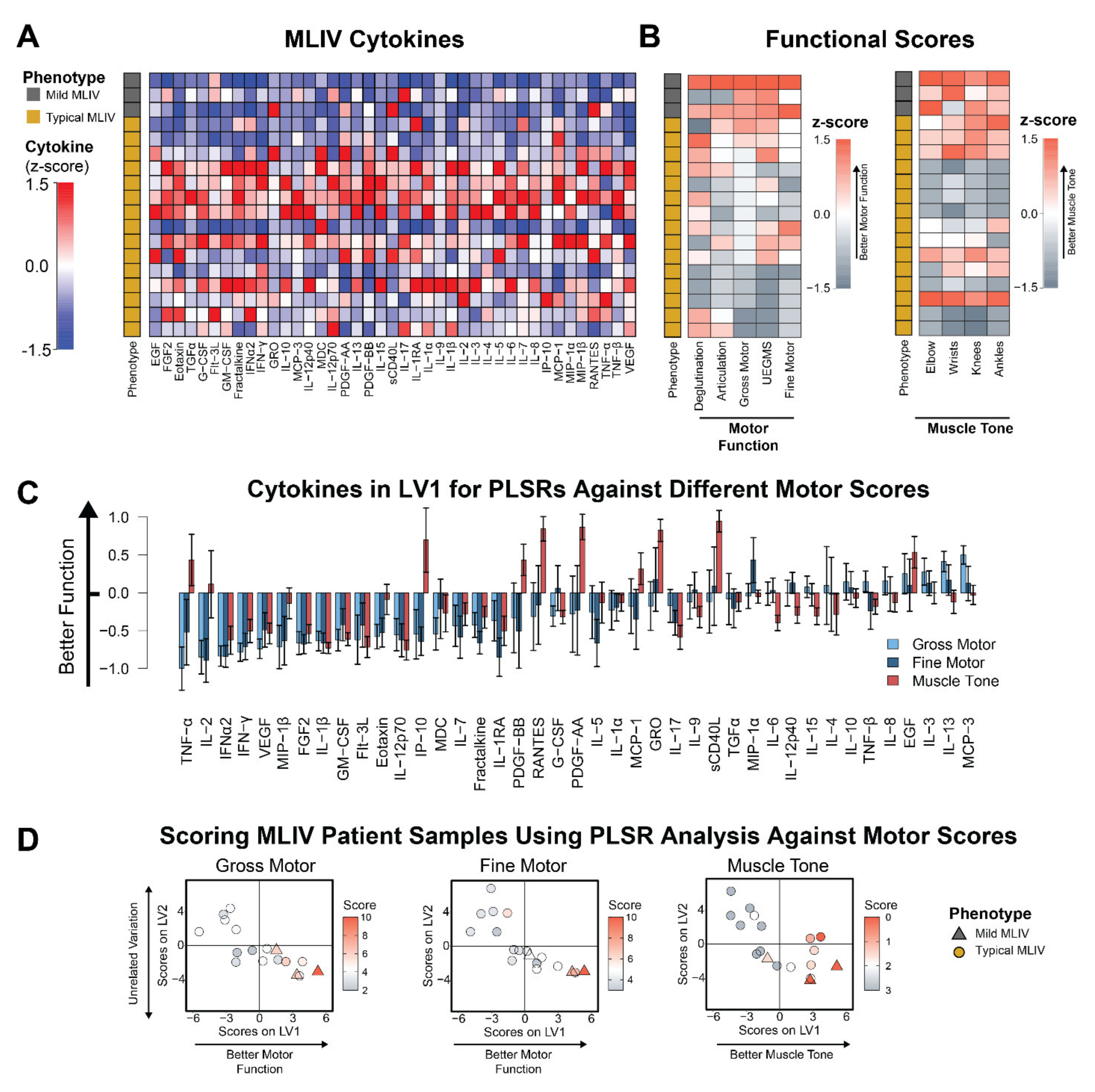
3.2. Plasma Cytokines Are Increased in Patients with Poor Motor Function and Hypertonicity
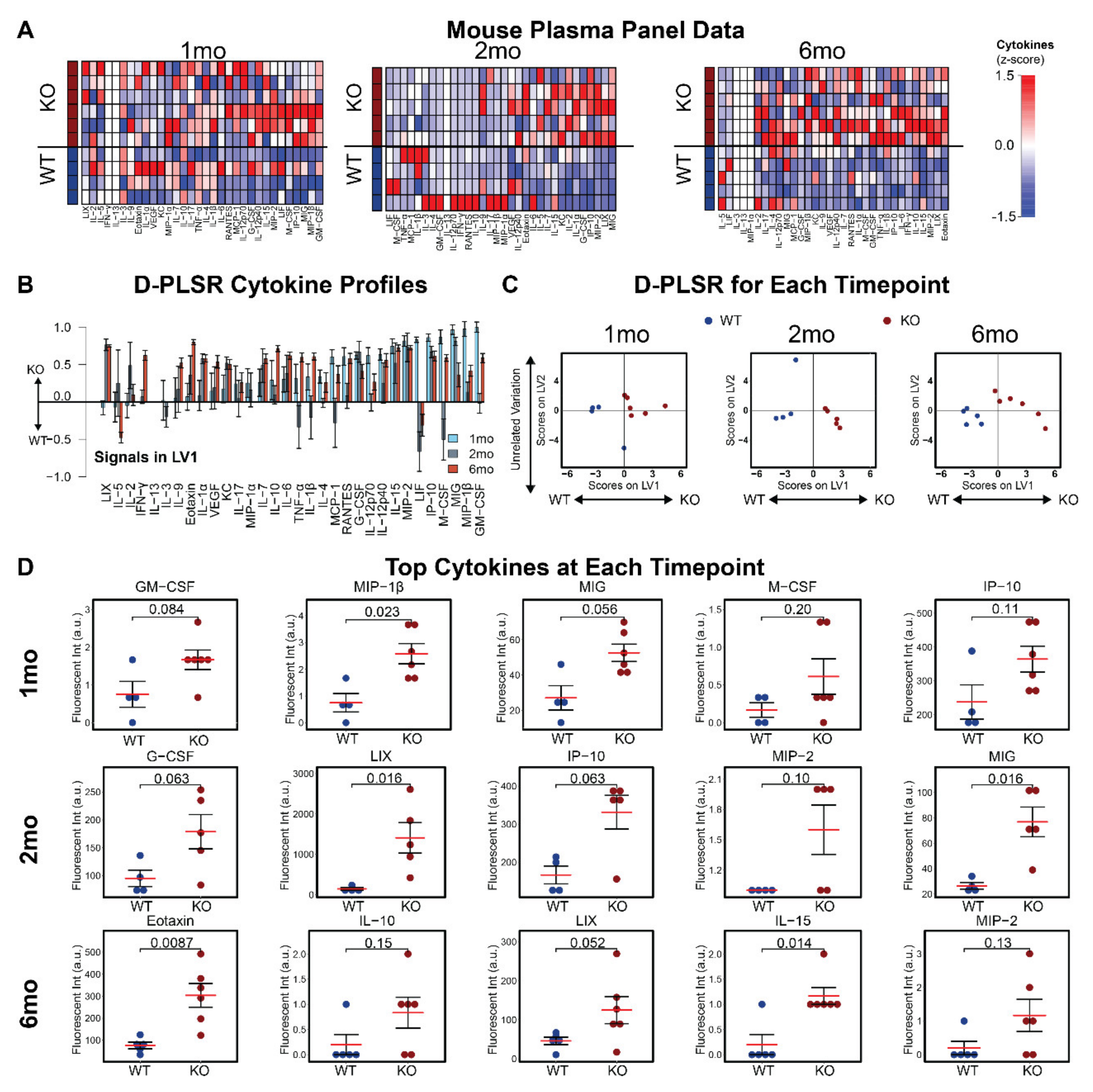
3.3. Plasma Cytokines Are Increased in Mcoln1−/− Mice
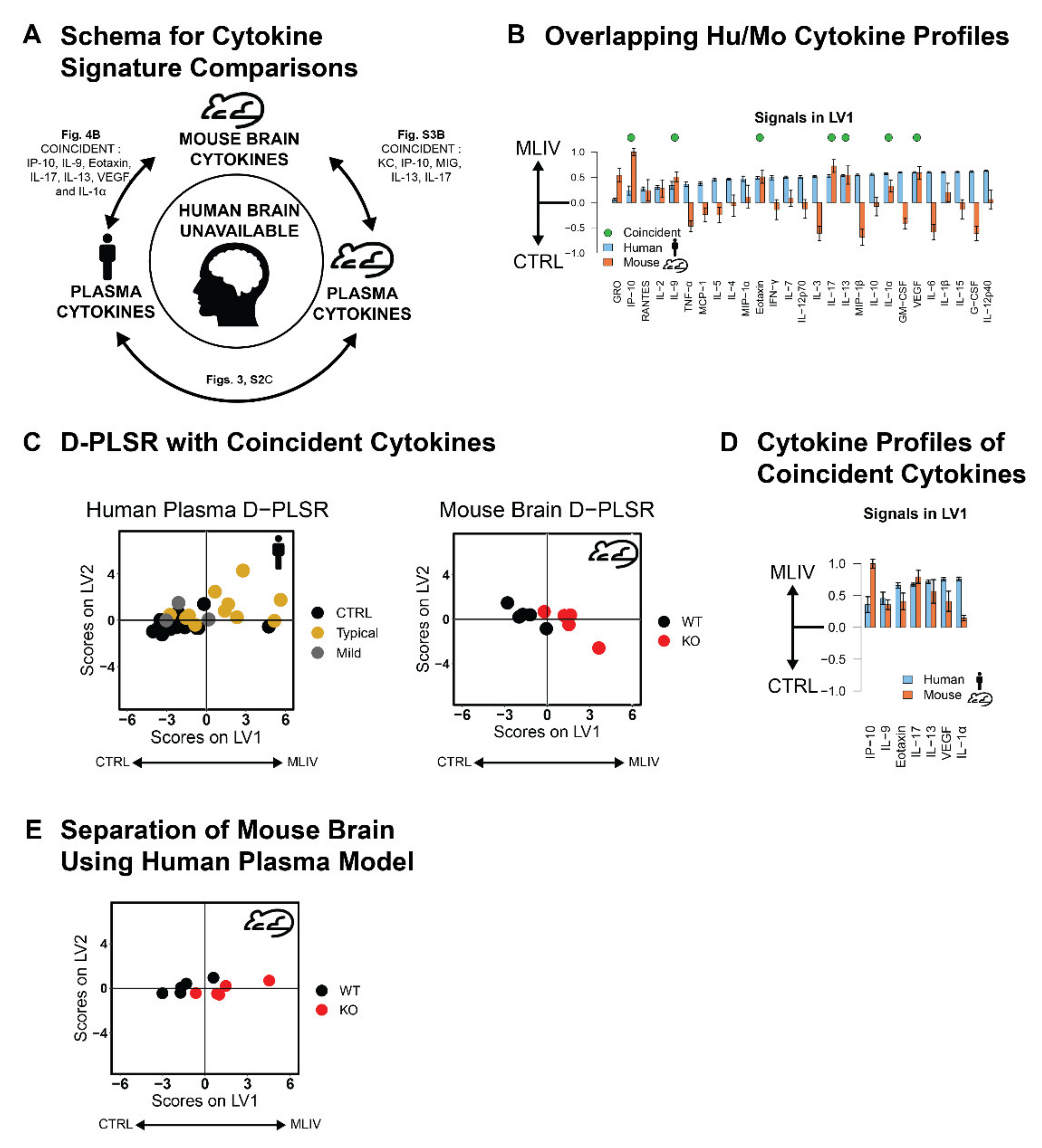
3.4. Plasma Cytokine Signatures Overlap with Brain Cytokine Signature in Mcoln1−/− Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Altarescu, G.; Sun, M.; Moore, D.F.; Smith, J.A.; Wiggs, E.A.; Solomon, B.I.; Patronas, N.J.; Frei, K.P.; Gupta, S.; Kaneski, C.R.; et al. The Neurogenetics of Mucolipidosis Type IV. Neurology 2002, 59, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Chitayat, D.; Meunier, C.M.; Hodgkinson, K.A.; Silver, K.; Flanders, M.; Anderson, I.J.; Little, J.M.; Whiteman, D.A.H.; Carpenter, S. Mucolipidosis type IV: Clinical Manifestations and Natural History. Am. J. Med. Genet. 1991, 41, 313–318. [Google Scholar] [CrossRef]
- Grishchuk, Y.; Sri, S.; Rudinskiy, N.; Ma, W.; Stember, K.G.; Cottle, M.W.; Sapp, E.; Difiglia, M.; Muzikansky, A.; Betensky, R.A.; et al. Behavioral Deficits, Early Gliosis, Dysmyelination and Synaptic Dysfunction in a Mouse Model of Mucolipidosis IV. Acta Neuropathol. Commun. 2014, 2, 133. [Google Scholar] [CrossRef] [PubMed]
- Grishchuk, Y.; Stember, K.G.; Matsunaga, A.; Olivares, A.M.; Cruz, N.M.; King, V.E.; Humphrey, D.M.; Wang, S.L.; Muzikansky, A.; Betensky, R.A.; et al. Retinal Dystrophy and Optic Nerve Pathology in the Mouse Model of Mucolipidosis IV. Am. J. Pathol. 2015, 186, 199–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandra, M.; Zhou, H.; Li, Q.; Muallem, S.; Hofmann, S.L.; Soyombo, A.A. A Role for the Ca2+ Channel TRPML1 in Gastric Acid Secretion, Based on Analysis of Knockout Mice. Gastroenterology 2011, 140, 857–867.e1. [Google Scholar] [CrossRef] [Green Version]
- Frei, K.P.; Patronas, N.J.; Crutchfield, K.E.; Altarescu, G.; Schiffmann, R. Mucolipidosis Type IV: Characteristic MRI Findings. Neurology 1998, 51, 565–569. [Google Scholar] [CrossRef]
- Schiffmann, R.; Mayfield, J.; Swift, C.; Nestrasil, I. Quantitative Neuroimaging in Mucolipidosis Type IV. Mol. Genet. Metab. 2013, 111, 147–151. [Google Scholar] [CrossRef] [Green Version]
- Mepyans, M.; Andrzejczuk, L.; Sosa, J.; Smith, S.; Herron, S.; DeRosa, S.; Slaugenhaupt, S.; Misko, A.; Grishchuk, Y.; Kiselyov, K. Early Evidence of Delayed Oligodendrocyte Maturation in the Mouse Model of Mucolipidosis Type IV. Dis. Model. Mech. 2020, 13, dmm044230. [Google Scholar] [CrossRef]
- Grishchuk, Y.; Peña, K.A.; Coblentz, J.; King, V.E.; Humphrey, D.M.; Wang, S.L.; Kiselyov, K.I.; Slaugenhaupt, S.A. Impaired Myelination and Reduced Brain Ferric Iron in the Mouse Model of Mucolipidosis IV. Dis. Model. Mech. 2015, 8, 1591–1601. [Google Scholar]
- Weinstock, L.D.; Furness, A.M.; Herron, S.S.; Smith, S.S.; Sankar, S.B.; DeRosa, S.G.; Gao, D.; Mepyans, M.E.; Rosato, A.S.; Medina, D.L.; et al. Fingolimod Phosphate Inhibits Astrocyte Inflammatory Activity in Mucolipidosis IV. Hum. Mol. Genet. 2018, 27, 2725–2738. [Google Scholar] [CrossRef]
- Shim, H.G.; Jang, S.-S.; Kim, S.H.; Hwang, E.M.; Min, J.O.; Kim, H.Y.; Kim, Y.S.; Ryu, C.; Chung, G.; Kim, Y.; et al. TNF-α Increases the Intrinsic Excitability of Cerebellar Purkinje Cells through Elevating Glutamate Release in Bergmann Glia. Sci. Rep. 2018, 8, 11589. [Google Scholar] [CrossRef]
- Gruol, D.L.; Nelson, T.E. Purkinje Neuron Physiology Is Altered by the Inflammatory Factor Interleukin-6. Cerebellum 2005, 4, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Bosch, M.E.; Kielian, T. Neuroinflammatory Paradigms in Lysosomal Storage Diseases. Front. Neurosci. 2015, 9, 417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cintas, H.L.; Parks, R.; Don, S.; Gerber, L. Brief Assessment of Motor Function: Content Validity and Reliability of the Upper Extremity Gross Motor Scale. Phys. Occup. Ther. Pediatr. 2011, 31, 440–450. [Google Scholar] [CrossRef]
- Cintas, H.L.; Siegel, K.L.; Furst, G.P.; Gerber, L.H. Brief Assessment of Motor Function: Reliability and Concurrent Validity of the Gross Motor Scale. Am. J. Phys. Med. Rehabil. 2003, 82, 33–41. [Google Scholar] [CrossRef]
- Sonies, B.C.; Cintas, H.L.; Parks, R.; Miller, J.; Caggiano, C.; Gottshall, S.G.; Gerber, L. Brief Assessment of Motor Function: Content Validity and Reliability of the Oral Motor Scales. Am. J. Phys. Med. Rehabil. 2009, 88, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Parks, R.; Cintas, H.L.; Chaffin, M.C.; Gerber, L. Brief Assessment of Motor Function: Content Validity and Reliability of the Fine Motor Scale. Pediatr. Phys. Ther. 2007, 19, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Fosang, A.L.; Galea, M.; McCoy, A.T.; Reddihough, D.S.; Story, I. Measures of Muscle and Joint Performance in the Lower Limb of Children with Cerebral Palsy. Dev. Med. Child Neurol. 2003, 45, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Vogel, D.Y.S.; Kooij, G.; Heijnen, P.D.A.M.; Breur, M.; Peferoen, L.A.N.; van der Valk, P.; de Vries, H.E.; Amor, S.; Dijkstra, C.D. GM-CSF Promotes Migration of Human Monocytes Across the Blood Brain Barrier. Eur. J. Immunol. 2015, 45, 1808–1819. [Google Scholar] [CrossRef]
- Carter, S.L.; Müller, M.; Manders, P.M.; Campbell, I.L. Induction of the Genes for Cxcl9 and Cxcl10 Is Dependent on IFN-Gamma but Shows Differential Cellular Expression in Experimental Autoimmune Encephalomyelitis and by Astrocytes and Microglia in Vitro. Glia 2007, 55, 1728–1739. [Google Scholar] [CrossRef]
- Jana, M.; Dasgupta, S.; Pal, U.; Pahan, K. IL-12 p40 Homodimer, the So-Called Biologically Inactive Molecule, Induces Nitric Oxide Synthase in Microglia via IL-12R Beta 1. Glia 2009, 57, 1553–1565. [Google Scholar] [CrossRef] [Green Version]
- Hanisch, U.-K.; Lyons, S.A.; Prinz, M.; Nolte, C.; Weber, J.R.; Kettenmann, H.; Kirchhoff, F. Mouse Brain Microglia Express Interleukin-15 and Its Multimeric Receptor Complex Functionally Coupled to Janus Kinase Activity. J. Biol. Chem. 1997, 272, 28853–28860. [Google Scholar] [CrossRef] [Green Version]
- Vardi, A.; Pri-Or, A.; Wigoda, N.; Grishchuk, Y.; Futerman, A.H. Proteomics Analysis of a Human Brain Sample from a Mucolipidosis Type IV Patient Reveals Pathophysiological Pathways. Orphanet J. Rare Dis. 2021, 16, 1–13. [Google Scholar] [CrossRef]
- De Rosa, S.; Salani, M.; Smith, S.; Sangster, M.; Miller-Browne, V.; Wassmer, S.; Xiao, R.; Vandenberghe, L.; Slaugenhaupt, S.; Misko, A.; et al. MCOLN1 Gene Therapy Corrects Neurologic Dysfunction in the Mouse Model of Mucolipidosis IV. Hum. Mol. Genet. 2021, 30, 908–922. [Google Scholar] [CrossRef] [PubMed]
- Boudewyn, L.C.; Sikora, J.; Kuchar, L.; Ledvinova, J.; Grishchuk, Y.; Wang, S.L.; Dobrenis, K.; Walkley, S.U. N-Butyldeoxynojirimycin Delays Motor Deficits, Cerebellar Microgliosis, and Purkinje Cell Loss in a Mouse Model of Mucolipidosis Type IV. Neurobiol. Dis. 2017, 105, 257–270. [Google Scholar] [CrossRef]
- Venugopal, B.; Browning, M.F.; Curcio-Morelli, C.; Varro, A.; Michaud, N.; Nanthakumar, N.; Walkley, S.U.; Pickel, J.; Slaugenhaupt, S.A. Neurologic, Gastric, and Opthalmologic Pathologies in a Murine Model of Mucolipidosis Type IV. Am. J. Hum. Genet. 2007, 81, 1070–1083. [Google Scholar] [CrossRef] [Green Version]
- Vitner, E.B.; Farfel-Becker, T.; Ferreira, N.S.; Leshkowitz, D.; Sharma, P.; Lang, K.; Futerman, A.H. Induction of the Type I Interferon Response in Neurological Forms of Gaucher Disease. J. Neuroinflamm. 2016, 13, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, L.B.; Winslow, A.R.; Proctor, E.A.; McGuone, D.; Mordes, D.A.; Frosch, M.P.; Hyman, B.T.; Lauffenburger, D.A.; Haigis, K.M. Identification of Neurotoxic Cytokines by Profiling Alzheimer’s Disease Tissues and Neuron Culture Viability Screening. Sci. Rep. 2015, 5, 16622. [Google Scholar] [CrossRef]
- Di Paola, S.; Rosato, A.S.; Medina, D.L. TRPML1: The Ca(2+)Retaker of the Lysosome. Cell Calcium 2018, 69, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Xu, M.; Wu, Y.; Syeda, A.K.R.; Dong, X.-P. Multiple Facets of TRPML1 in Autophagy. Cell Calcium 2020, 88, 102196. [Google Scholar] [CrossRef]
- Dayam, R.M.; Saric, A.; Shilliday, R.E.; Botelho, R.J. The Phosphoinositide-Gated Lysosomal Ca(2+) Channel, TRPML1, Is Required for Phagosome Maturation. Traffic 2015, 16, 1010–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bretou, M.; Sáez, P.J.; Sanséau, D.; Maurin, M.; Lankar, D.; Chabaud, M.; Spampanato, C.; Malbec, O.; Barbier, L.; Muallem, S.; et al. Lysosome Signaling Controls the Migration of Dendritic Cells. Sci. Immunol. 2017, 2, eaak9573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodridge, J.P.; Jacobs, B.; Saetersmoen, M.L.; Clement, D.; Hammer, Q.; Clancy, T.; Skarpen, E.; Brech, A.; Landskron, J.; Grimm, C.; et al. Remodeling of Secretory Lysosomes during Education Tunes Functional Potential in NK Cells. Nat. Commun. 2019, 10, 514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clement, D.; Goodridge, J.P.; Grimm, C.; Patel, S.; Malmberg, K.-J. TRP Channels as Interior Designers: Remodeling the Endolysosomal Compartment in Natural Killer Cells. Front. Immunol. 2020, 11, 753. [Google Scholar] [CrossRef] [PubMed]
- Santoni, G.; Morelli, M.B.; Amantini, C.; Nabissi, M.; Santoni, M.; Santoni, A. Involvement of the TRPML Mucolipin Channels in Viral Infections and Anti-Viral Innate Immune Responses. Front. Immunol. 2020, 11, 739. [Google Scholar] [CrossRef]
- Medina, D.L.; Di Paola, S.; Peluso, I.; Armani, A.; De Stefani, D.; Venditti, R.; Montefusco, S.; Scotto-Rosato, A.; Prezioso, C.; Forrester, A.; et al. Lysosomal Calcium Signalling Regulates Autophagy through Calcineurin and TFEB. Nat. Cell Biol. 2015, 17, 288–299. [Google Scholar] [CrossRef] [Green Version]
- Palmieri, M.; Impey, S.; Kang, H.; di Ronza, A.; Pelz, C.; Sardiello, M.; Ballabio, A. Characterization of the CLEAR Network Reveals an Integrated Control of Cellular Clearance Pathways. Hum. Mol. Genet. 2011, 20, 3852–3866. [Google Scholar] [CrossRef] [Green Version]
- Settembre, C.; Medina, D.L. TFEB and the CLEAR Network. Methods Cell Biol. 2015, 126, 45–62. [Google Scholar] [CrossRef]
- Brady, O.A.; Martina, J.; Puertollano, R. Emerging Roles for TFEB in the Immune Response and Inflammation. Autophagy 2017, 14, 181–189. [Google Scholar] [CrossRef]
- Cougnoux, A.; Drummond, R.A.; Fellmeth, M.; Navid, F.; Collar, A.; Iben, J.; Kulkarni, A.B.; Pickel, J.; Schiffmann, R.; Wassif, C.A.; et al. Unique Molecular Signature in Mucolipidosis Type IV Microglia. J. Neuroinflamm. 2019, 16, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Curcio-Morelli, C.; Zhang, P.; Venugopal, B.; Charles, F.A.; Browning, M.F.; Cantiello, H.F.; Slaugenhaupt, S.A. Functional Multimerization of Mucolipin Channel Proteins. J. Cell. Physiol. 2009, 222, 328–335. [Google Scholar] [CrossRef]
- Venkatachalam, K.; Hofmann, T.; Montell, C. Lysosomal Localization of TRPML3 Depends on TRPML2 and the Mucolipidosis-Associated Protein TRPML1. J. Biol. Chem. 2006, 281, 17517–17527. [Google Scholar] [CrossRef] [Green Version]
- Cuajungco, M.P.; Da Silva, J.F.M.; Habibi, A.; Valadez, J.A. The Mucolipin-2 (TRPML2) Ion Channel: A Tissue-Specific Protein Crucial to Normal Cell Function. Pflügers Arch.-Eur. J. Physiol. 2015, 468, 177–192. [Google Scholar] [CrossRef] [Green Version]
- Samie, M.A.; Grimm, C.; Evans, J.A.; Curcio-Morelli, C.; Heller, S.; Slaugenhaupt, S.A.; Cuajungco, M.P. The Tissue-Specific Expression of TRPML2 (MCOLN-2) Gene Is Influenced by the Presence of TRPML1. Pflügers Arch.-Eur. J. Physiol. 2009, 459, 79–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Dayalu, R.; Matthews, S.A.; Scharenberg, A.M. TRPML Cation Channels Regulate the Specialized Lysosomal Compartment of Vertebrate B-Lymphocytes. Eur. J. Cell Biol. 2006, 85, 1253–1264. [Google Scholar] [CrossRef]
- Watts, C. The Endosome–Lysosome Pathway and Information Generation in the Immune System. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2012, 1824, 14–21. [Google Scholar] [CrossRef] [Green Version]
- Reinheckel, T. On the Road to Inflammation: Linking Lysosome Disruption, Lysosomal Protease Release and Necrotic Death of Immune Cells. Cell Cycle 2013, 12, 1994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Galluzzi, L.; Zitvogel, L.; Kroemer, G. Autophagy and Cellular Immune Responses. Immunity 2013, 39, 211–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabowski, G.A. Overview of Inflammation in Neurometabolic Diseases. Semin. Pediatr. Neurol. 2017, 24, 207–213. [Google Scholar] [CrossRef]
- Rozenfeld, P.; Feriozzi, S. Contribution of Inflammatory Pathways to Fabry Disease Pathogenesis. Mol. Genet. Metab. 2017, 122, 19–27. [Google Scholar] [CrossRef]
- Vitner, E.B.; Futerman, A.; Platt, N. Innate Immune Responses in the Brain of Sphingolipid Lysosomal Storage Diseases. Biol. Chem. 2015, 396, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Rigante, D.; Cipolla, C.; Basile, U.; Gulli, F.; Savastano, M.C. Overview of Immune Abnormalities in Lysosomal Storage Disorders. Immunol. Lett. 2017, 188, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.-H.; Wu, Y.-R.; Chen, Y.-C.; Chen, C.-M. Plasma Inflammatory Biomarkers for Huntington’s Disease Patients and Mouse Model. Brain Behav. Immun. 2015, 44, 121–127. [Google Scholar] [CrossRef] [PubMed]
| Patient | Age (Years) | Mode of Diagnosis | Phenotype | MCOLN1 Genotype | |
|---|---|---|---|---|---|
| 1 | <5 | Genetic | Typical | c.304 C > T | c.1405A > G |
| 2 | <5 | Genetic | Typical | c.406 − 2A > G | c.1336G > A |
| 3 | <5 | Genetic | Typical | c.1017_1020delACGG | c.877 G > A |
| 4 | 5–10 | Genetic | Typical | c.984 + 1G > A | c.406 − 2A > G |
| 5 | 5–10 | Genetic | Typical | c.406 − 2A > G | c.964C > T |
| 6 | 5–10 | Genetic | Typical | c.694A > C | c.785T > C |
| 7 | 11–15 | Genetic | Typical | c.406 − 2A > G | c.406 − 2A > G |
| 8 | 11–15 | Genetic | Mild | c.406 − 2A > G | c.406 − 2A > G |
| 9 | 11–15 | Genetic | Typical | c.406 − 2A > G | c.406 − 2A > G |
| 10 | 11–15 | Genetic | Typical | c.406 − 2A > G | c.406 − 2A > G |
| 11 | 11–15 | Genetic and Corneal bx | Typical | c.406 − 2A > G | Not Identified |
| 12 | 16–20 | Genetic | Typical | c.236_237ins93 | c.694A>C |
| 13 | 16–20 | Genetic | Typical | c.406 − 2A > G | c.406 − 2A > G |
| 14 | 16–20 | Genetic | Mild | c.236_237ins93 | c.1207C > T |
| 15 | 25–30 | Clinical and Gastrin | Typical | Unknown | Unknown |
| 16 | 25–30 | Genetic | Typical | c.406 − 2A > G | c.406 − 2A > G |
| 17 | 25–30 | Genetic | Typical | c406 − 2A > G | c406 − 2A > G |
| 18 | 25–30 | Genetic | Mild | c.920delT | c.1615delG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Misko, A.L.; Weinstock, L.D.; Sankar, S.B.; Furness, A.; Grishchuk, Y.; Wood, L.B. Peripheral Inflammatory Cytokine Signature Mirrors Motor Deficits in Mucolipidosis IV. Cells 2022, 11, 546. https://doi.org/10.3390/cells11030546
Misko AL, Weinstock LD, Sankar SB, Furness A, Grishchuk Y, Wood LB. Peripheral Inflammatory Cytokine Signature Mirrors Motor Deficits in Mucolipidosis IV. Cells. 2022; 11(3):546. https://doi.org/10.3390/cells11030546
Chicago/Turabian StyleMisko, Albert L., Laura D. Weinstock, Sitara B. Sankar, Amanda Furness, Yulia Grishchuk, and Levi B. Wood. 2022. "Peripheral Inflammatory Cytokine Signature Mirrors Motor Deficits in Mucolipidosis IV" Cells 11, no. 3: 546. https://doi.org/10.3390/cells11030546
APA StyleMisko, A. L., Weinstock, L. D., Sankar, S. B., Furness, A., Grishchuk, Y., & Wood, L. B. (2022). Peripheral Inflammatory Cytokine Signature Mirrors Motor Deficits in Mucolipidosis IV. Cells, 11(3), 546. https://doi.org/10.3390/cells11030546






