Genomic and Metabolomic Landscape of Right-Sided and Left-Sided Colorectal Cancer: Potential Preventive Biomarkers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Specimens
2.2. Genomic Data Acquisition
2.3. Detection of Somatic Mutations in CRC Tumor Samples
2.4. Microsatellite Instability (MSI) Detection and Tumor Mutational Burden (TMB)
2.5. Metabolomic Profiling
2.6. Polygenic Risk Analysis
3. Results
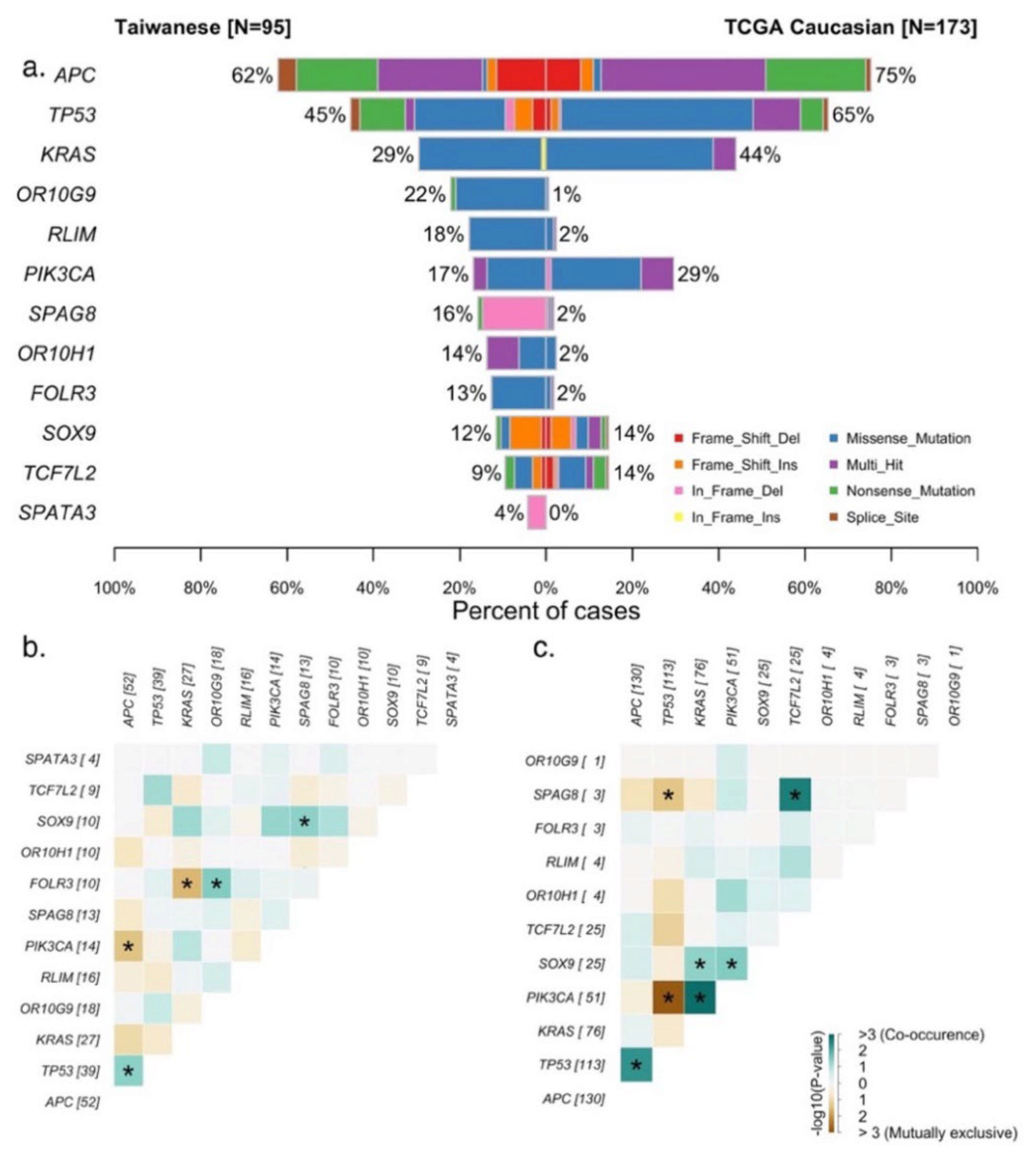
3.1. Genomic Landscape of the Taiwanese CRC Cohort
3.2. Comparison of Genetic Features between LCRC and RCRC in the Taiwanese and Caucasian Cohorts
3.3. Metabolomic Profiling of LCRC and RCRC
3.4. PRS of Taiwanese Subjects
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ministry of Health and Welfare, Taiwan, Republic of China. Cause of Death Statistics by Year (in Chinese). Available online: https://dep.mohw.gov.tw/dos/lp-5069-113.html (accessed on 18 June 2021).
- Koveitypour, Z.; Panahi, F.; Vakilian, M.; Peymani, M.; Forootan, F.S.; Esfahani, M.H.N.; Ghaedi, K. Signaling pathways involved in colorectal cancer progression. Cell Biosci. 2019, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Bufill, J.A. Colorectal Cancer: Evidence for Distinct Genetic Categories Based on Proximal or Distal Tumor Location. Ann. Intern. Med. 1990, 113, 779–788. [Google Scholar] [CrossRef]
- Hu, W.; Yang, Y.; Li, X.; Huang, M.; Xu, F.; Ge, W.; Zhang, S.; Zheng, S. Multi-omics Approach Reveals Distinct Differences in Left- and Right-Sided Colon Cancer. Mol. Cancer Res. 2017, 16, 476–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.W.; Tsai, H.L.; Huang, M.Y.; Huang, C.M.; Yeh, Y.S.; Ma, C.J.; Wang, J.Y. Different clinicopathologic features and favorable outcomes of patients with stage III left-sided colon cancer. World J. Surg. Oncol. 2015, 13, 257. [Google Scholar] [CrossRef] [Green Version]
- Cascinu, S.; Poli, D.; Zaniboni, A.; Lonardi, S.; Labianca, R.; Sobrero, A.; Rosati, G.; Di Bartolomeo, M.; Scartozzi, M.; Zagonel, V.; et al. The prognostic impact of primary tumour location in patients with stage II and stage III colon cancer receiving adjuvant therapy. A GISCAD analysis from three large randomised trials. Eur. J. Cancer 2019, 111, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ulanja, M.B.; Rishi, M.; Beutler, B.D.; Sharma, M.; Patterson, D.R.; Gullapalli, N.; Ambika, S. Colon Cancer Sidedness, Presentation, and Survival at Different Stages. J. Oncol. 2019, 2019, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Ha, G.W.; Kim, J.H.; Lee, M.R. Oncologic Effects of Primary Tumor-Sidedness on Patients with Stages 1–3 Colon Cancer: A Meta-Analysis. Ann. Surg. Oncol. 2019, 26, 1366–1375. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Lopez, N.E.; Eisenstein, S.; Schnickel, G.T.; Sicklick, J.K.; Ramamoorthy, S.L.; Clary, B.M. Synchronous metastatic colon cancer and the importance of primary tumor laterality–A National Cancer Database analysis of right- versus left-sided colon cancer. Am. J. Surg. 2019, 220, 408–414. [Google Scholar] [CrossRef]
- Ahmed, S.; Pahwa, P.; Le, D.; Chalchal, H.; Chandra-Kanthan, S.; Iqbal, N.; Fields, A. Primary Tumor Location and Survival in the General Population With Metastatic Colorectal Cancer. Clin. Color. Cancer 2018, 17, e201–e206. [Google Scholar] [CrossRef]
- Ciombor, K.K.; Bekaii-Saab, T. A Comprehensive Review of Sequencing and Combination Strategies of Targeted Agents in Metastatic Colorectal Cancer. Oncologist 2018, 23, 25–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, D.; Lueza, B.; Douillard, J.-Y.; Peeters, M.; Lenz, H.-J.; Venook, A.; Heinemann, V.; Van Cutsem, E.; Pignon, J.-P.; Tabernero, J.; et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann. Oncol. 2017, 28, 1713–1729. [Google Scholar] [CrossRef]
- You, X.-H.; Jiang, Y.-H.; Fang, Z.; Sun, F.; Li, Y.; Wang, W.; Xia, Z.-J.; Wang, X.-Z.; Ying, H.-Q. Chemotherapy plus bevacizumab as an optimal first-line therapeutic treatment for patients with right-sided metastatic colon cancer: A meta-analysis of first-line clinical trials. ESMO Open 2020, 5, e000605. [Google Scholar] [CrossRef] [Green Version]
- Law, P.J.; Timofeeva, M.; Fernandez-Rozadilla, C.; Broderick, P.; Studd, J.; Fernandez-Tajes, J.; Farrington, S.; Svinti, V.; Palles, C.; The PRACTICAL Consortium; et al. Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, A.J.; Puzio-Kuter, A.M. The Control of the Metabolic Switch in Cancers by Oncogenes and Tumor Suppressor Genes. Science 2010, 330, 1340–1344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.-Y.; Yang, J.-H.; Yeh, E.-C.; Tsai, M.-F.; Kao, H.-J.; Lo, C.-Z.; Chang, L.-P.; Lin, W.-J.; Hsieh, F.-J.; Belsare, S.; et al. Genetic profiles of 103,106 individuals in the Taiwan Biobank provide insights into the health and history of Han Chinese. NPJ Genom. Med. 2021, 6, 1–10. [Google Scholar] [CrossRef]
- Chen, C.-H.; Yang, J.-H.; Chiang, C.W.; Hsiung, C.-N.; Wu, P.-E.; Chang, L.-C.; Chu, H.-W.; Chang, J.; Song, I.W.; Yang, S.-L.; et al. Population structure of Han Chinese in the modern Taiwanese population based on 10,000 participants in the Taiwan Biobank project. Hum. Mol. Genet. 2016, 25, 5321–5331. [Google Scholar] [CrossRef] [Green Version]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.B.; Sougnez, C.; Gabriel, S.B.; Meyerson, M.L.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef]
- Cortes-Ciriano, I.; Lee, S.; Park, W.-Y.; Kim, T.-M.; Park, P.J. A molecular portrait of microsatellite instability across multiple cancers. Nat. Commun. 2017, 8, 15180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Su, M.-W.; Chang, C.-K.; Lin, C.-W.; Ling, S.-J.; Hsiung, C.-N.; Chu, H.-W.; Wu, P.-E.; Shen, C.-Y. Blood multiomics reveal insights into population clusters with low prevalence of diabetes, dyslipidemia and hypertension. PLoS ONE 2020, 15, e0229922. [Google Scholar] [CrossRef] [PubMed]
- Jacob, D.; Deborde, C.; Lefebvre, M.; Maucourt, M.; Moing, A. NMRProcFlow: A graphical and interactive tool dedicated to 1D spectra processing for NMR-based metabolomics. Metabolomics 2017, 13, 36. [Google Scholar] [CrossRef] [Green Version]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The Human Serum Metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohart, F.; Le Cao, K.-A.; Gonzalez, I.; Dejean, S.; Abadi, A.J.; Gautier, B.; Bartolo, F.; Monget, P.; Coquery, J.; Yao, F.Z.; et al. mixOmics: Omics Data Integration Project. R Package Version 6.1.1. Available online: http://www.bioconductor.org/packages/release/bioc/html/mixOmics.html (accessed on 18 June 2021).
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Software and supporting material for “Second-generation PLINK: Rising to the challenge of larger and richer datasets”. GigaSci. Database 2015. [Google Scholar] [CrossRef]
- Raut, C.P.; Pawlik, T.M.; Rodriguez-Bigas, M.A. Clinicopathologic features in colorectal cancer patients with microsatellite instability. Mutat. Res. 2004, 568, 275–282. [Google Scholar] [CrossRef] [PubMed]
- He, C.-Y.; Chen, L.-Z.; Wang, Z.-X.; Sun, L.-P.; Peng, J.-J.; Wu, M.-Q.; Wang, T.-M.; Li, Y.-Q.; Yang, X.-H.; Zhou, D.-L.; et al. Performance of common genetic variants in risk prediction for colorectal cancer in Chinese: A two-stage and multicenter study. Genomics 2021, 113, 867–873. [Google Scholar] [CrossRef]
- Ma, R.; Jing, C.; Zhang, Y.; Cao, H.; Liu, S.; Wang, Z.; Chen, D.; Zhang, J.; Wu, Y.; Wu, J.; et al. The somatic mutation landscape of Chinese Colorectal Cancer. J. Cancer 2020, 11, 1038–1046. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Yang, C.; Li, X.; Luo, W.; Roy, B.; Xiong, T.; Zhang, X.; Yang, H.; Wang, J.; Ye, Z.; et al. The landscape of somatic mutation in sporadic Chinese colorectal cancer. Oncotarget 2018, 9, 27412–27422. [Google Scholar] [CrossRef] [Green Version]
- Nagahashi, M.; Wakai, T.; Shimada, Y.; Ichikawa, H.; Kameyama, H.; Kobayashi, T.; Sakata, J.; Yagi, R.; Sato, N.; Kitagawa, Y.; et al. Genomic landscape of colorectal cancer in Japan: Clinical implications of comprehensive genomic sequencing for precision medicine. Genome Med. 2016, 8, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanes, T.; McInerney-Leo, A.M.; Law, M.H.; Cummings, S. The emerging field of polygenic risk scores and perspective for use in clinical care. Hum. Mol. Genet. 2020, 29, R165–R176. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Lei, X.; Yuan, D.; Zhu, Z.; Huang, S. Accumulation of minor alleles and risk prediction in schizophrenia. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertini, I.; Cacciatore, S.; Jensen, B.V.; Schou, J.K.; Johansen, J.S.; Kruhøffer, M.; Luchinat, C.; Nielsen, D.L.; Turano, P. Metabolomic NMR Fingerprinting to Identify and Predict Survival of Patients with Metastatic Colorectal Cancer. Cancer Res. 2012, 72, 356–364. [Google Scholar] [CrossRef] [Green Version]
- Strmiska, V.; Michalek, P.; Lackova, Z.; Guran, R.; Krizkova, S.; Vaníčková, L.; Zitka, O.; Stiborova, M.; Eckschlager, T.; Klejdus, B.; et al. Sarcosine is a prostate epigenetic modifier that elicits aberrant methylation patterns through the SAM e-Dnmts axis. Mol. Oncol. 2019, 13, 1002–1017. [Google Scholar] [CrossRef] [Green Version]
- Shen, H.-Y.; Weltha, L.; Cook, J.M.; Gesese, R.; Omi, W.; Baer, S.B.; Rose, R.M.; Reemmer, J.; Boison, D. Sarcosine Suppresses Epileptogenesis in Rats With Effects on Hippocampal DNA Methylation. Front. Mol. Neurosci. 2020, 13. [Google Scholar] [CrossRef]
- Pundir, C.S.; Deswal, R.; Kumar, P. Quantitative analysis of sarcosine with special emphasis on biosensors: A review. Biomarkers 2019, 24, 415–422. [Google Scholar] [CrossRef]
- Cervena, K.; Siskova, A.; Buchler, T.; Vodicka, P.; Vymetalkova, V. Methylation-Based Therapies for Colorectal Cancer. Cells 2020, 9, 1540. [Google Scholar] [CrossRef]



| LCRC | RCRC | Both | Total | p-Value | |
|---|---|---|---|---|---|
| n = 80 | n = 59 | n = 2 | n = 141 | ||
| Age, Mean (SD) | 63.1 (12.3) | 68.5 (10.8) | 57.5 (0.7) | 65.2 (11.9) | <0.01 |
| Gender, Male (%) | 47 (58.8%) | 30 (50.8%) | 1 (50.0%) | 78 (55.3%) | |
| BMI, Mean (SD) | 24.0 (3.9) | 23.2 (3.6) | 21.2 (0.2) | 23.6 (3.8) | |
| Grade, n (%) | <0.01 | ||||
| I Well differentiated | 1 (1.2%) | 4 (6.8%) | 2 (100%) | 7 (5.0%) | |
| II Moderate differentiated | 72 (90.0%) | 44 (74.6%) | 0 (0%) | 116 (82.3%) | |
| III Poorly differentiated | 2 (2.5%) | 9 (15.3%) | 0 (0%) | 11 (7.8%) | |
| Stage, n (%) | |||||
| I | 7 (8.8%) | 3 (5.1%) | 0 (0%) | 10 (7.1%) | |
| II | 23 (28.8%) | 21 (35.6%) | 1 (50%) | 45 (31.9%) | |
| III | 34 (42.5%) | 22 (37.3%) | 1 (50%) | 57 (40.4%) | |
| IV | 11 (13.8%) | 4 (6.8%) | 0 (0%) | 15 (10.6%) | |
| TMB, Mean (SD) | 12.9 (48.2) | 21.0 (52.8) | 2.66 (0.44) | 16.2 (49.8) | |
| MSI, MSI.H (%) | 1 (1.2%) | 11 (18.6%) | 0 (0%) | 12 (8.5%) | <0.01 |
| Taiwanese Cohort | LCRC (n = 59) | RCRC (n = 35) | ||||
|---|---|---|---|---|---|---|
| TNM stage | I + II (n = 26) | III + IV (n = 33) | overall | I + II (n = 16) | III + IV (n = 19) | overall |
| APC | 54% | 61% | 58% | 62% | 76% | 71% |
| TP53 | 38% | 55% | 47% | 38% | 53% | 43% |
| KRAS | 31% | 36% | 34% | 6% | 35% | 20% |
| PIK3CA | 23% | 18% | 20% | 6% | 12% | 9% |
| TCGA Caucasian | LCRC (n = 67) | RCRC (n = 85) | ||||
| TNM stage | I + II (n = 34) | III + IV (n = 33) | I + II (n = 48) | III + IV (n = 37) | ||
| APC | 85% | 91% | 88% | 71% | 68% | 69% |
| TP53 | 71% | 85% | 78% | 46% | 70% | 57% |
| KRAS | 44% | 24% | 34% | 52% | 54% | 53% |
| PIK3CA | 27% | 6% | 16% | 38% | 43% | 40% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, M.-W.; Chang, C.-K.; Lin, C.-W.; Chu, H.-W.; Tsai, T.-N.; Su, W.-C.; Chen, Y.-C.; Chang, T.-K.; Huang, C.-W.; Tsai, H.-L.; et al. Genomic and Metabolomic Landscape of Right-Sided and Left-Sided Colorectal Cancer: Potential Preventive Biomarkers. Cells 2022, 11, 527. https://doi.org/10.3390/cells11030527
Su M-W, Chang C-K, Lin C-W, Chu H-W, Tsai T-N, Su W-C, Chen Y-C, Chang T-K, Huang C-W, Tsai H-L, et al. Genomic and Metabolomic Landscape of Right-Sided and Left-Sided Colorectal Cancer: Potential Preventive Biomarkers. Cells. 2022; 11(3):527. https://doi.org/10.3390/cells11030527
Chicago/Turabian StyleSu, Ming-Wei, Chung-Ke Chang, Chien-Wei Lin, Hou-Wei Chu, Tsen-Ni Tsai, Wei-Chih Su, Yen-Cheng Chen, Tsung-Kun Chang, Ching-Wen Huang, Hsiang-Lin Tsai, and et al. 2022. "Genomic and Metabolomic Landscape of Right-Sided and Left-Sided Colorectal Cancer: Potential Preventive Biomarkers" Cells 11, no. 3: 527. https://doi.org/10.3390/cells11030527
APA StyleSu, M.-W., Chang, C.-K., Lin, C.-W., Chu, H.-W., Tsai, T.-N., Su, W.-C., Chen, Y.-C., Chang, T.-K., Huang, C.-W., Tsai, H.-L., Wu, C.-C., Chou, H.-C., Shiu, B.-H., & Wang, J.-Y. (2022). Genomic and Metabolomic Landscape of Right-Sided and Left-Sided Colorectal Cancer: Potential Preventive Biomarkers. Cells, 11(3), 527. https://doi.org/10.3390/cells11030527







