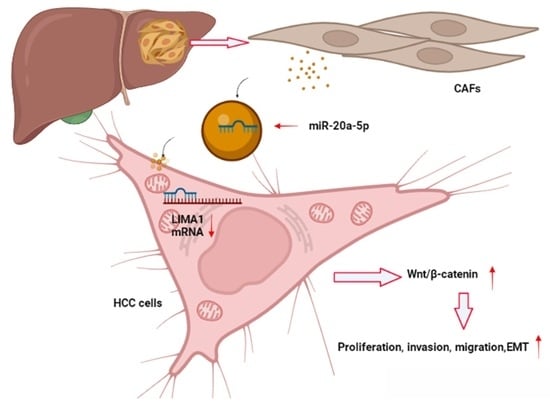
CAF-Released Exosomal miR-20a-5p Facilitates HCC Progression via the LIMA1-Mediated β-Catenin Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Specimens
2.2. Cell Culture and Transfection
2.3. RNA Isolation and Quantitative RT-PCR
2.4. Western Blotting
2.5. Coimmunoprecipitation (Co-IP)
2.6. Immunohistochemistry (IHC) and Immunofluorescence (IF)
2.7. Functional Experiments
2.8. CAF and NF Isolation
2.9. Exosome-Associated Experiments
2.10. Dual-Luciferase Reporter Assay
2.11. Mouse Models
2.12. Statistics
3. Results
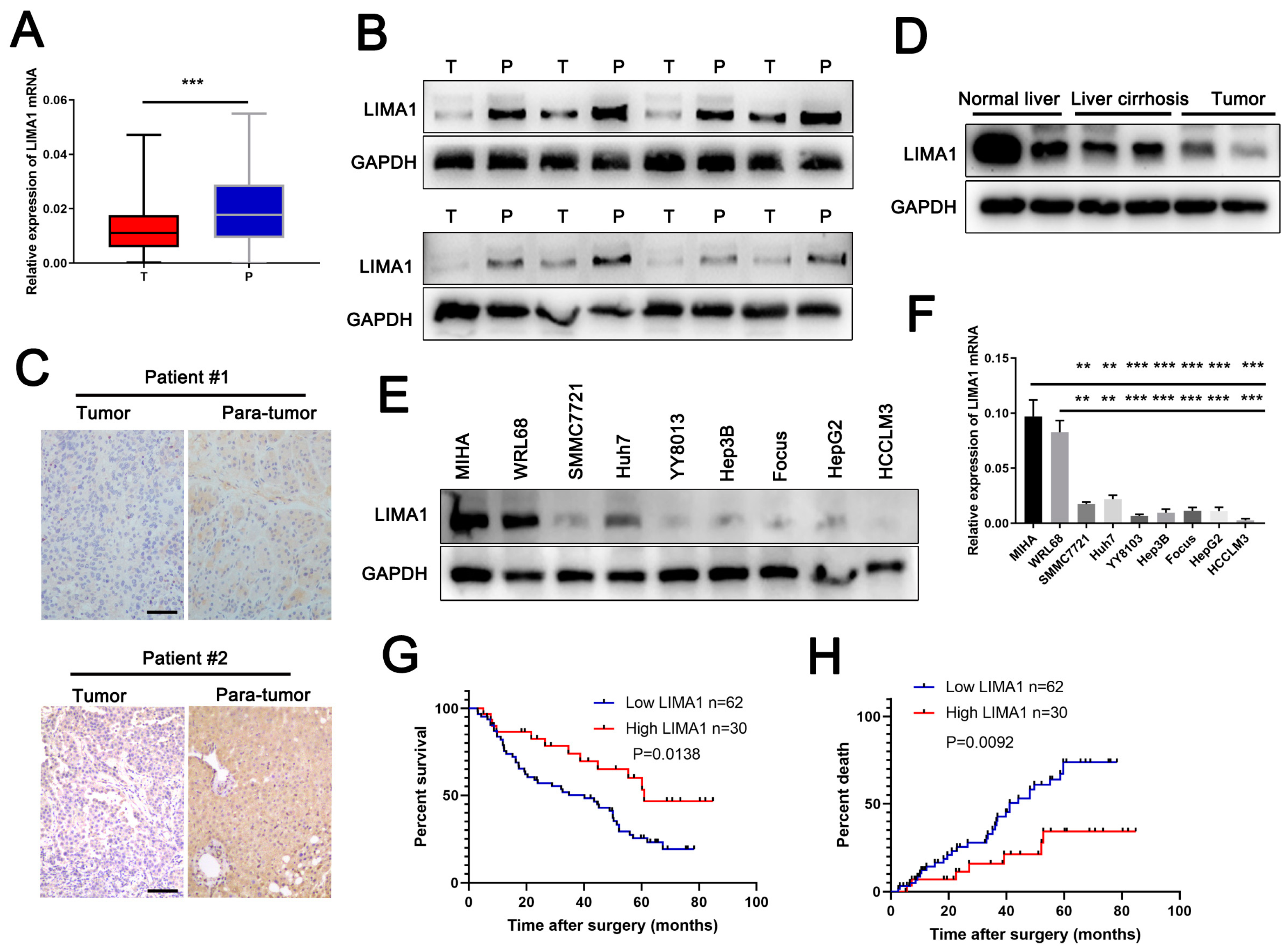
3.1. LIMA1 Was Downregulated in HCC and Positively Associated with Survival
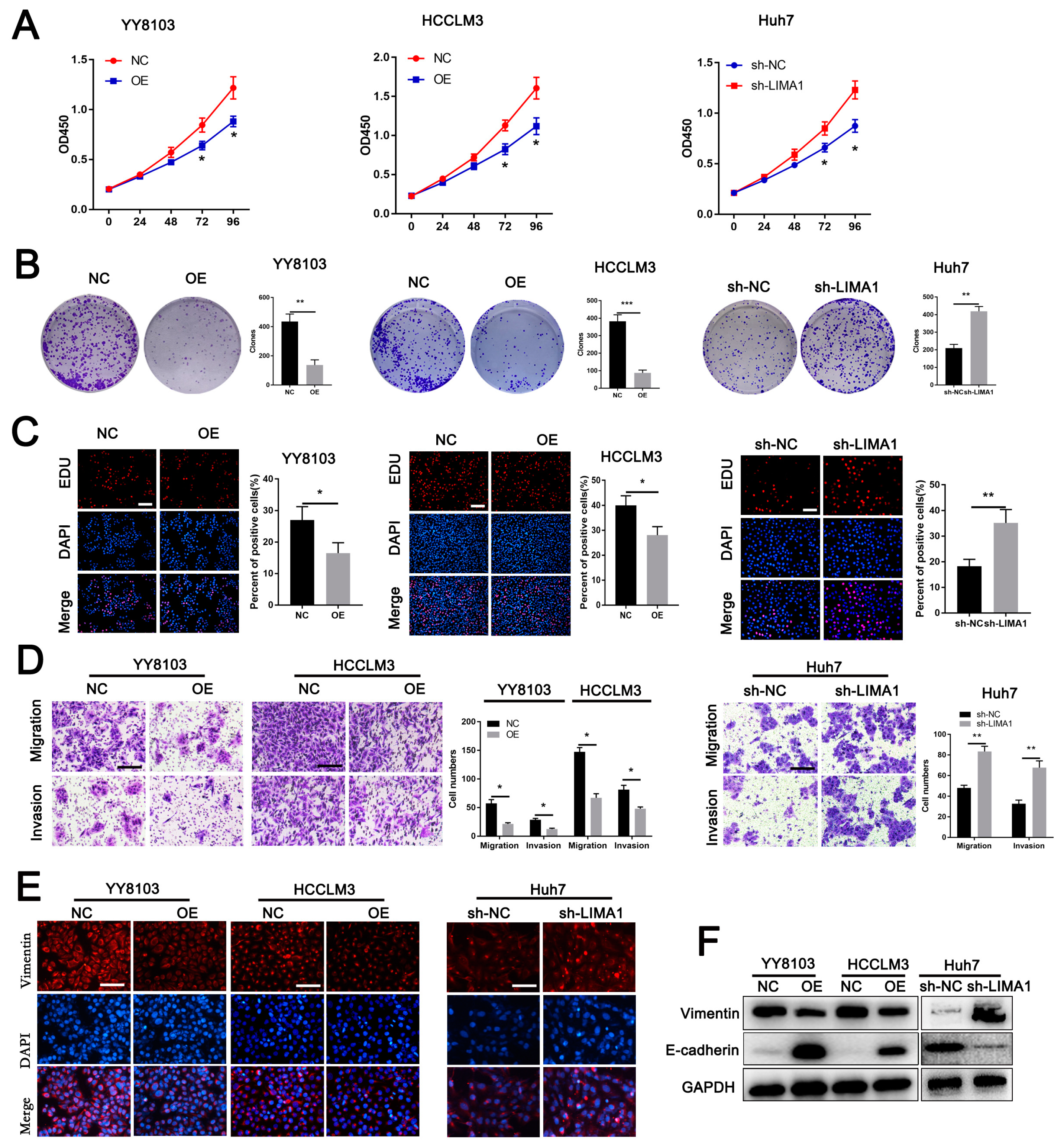
3.2. LIMA1 Suppressed HCC Cell Proliferation, Metastasis and EMT
3.3. LIMA1 Inhibited HCC Tumorigenesis In Vivo
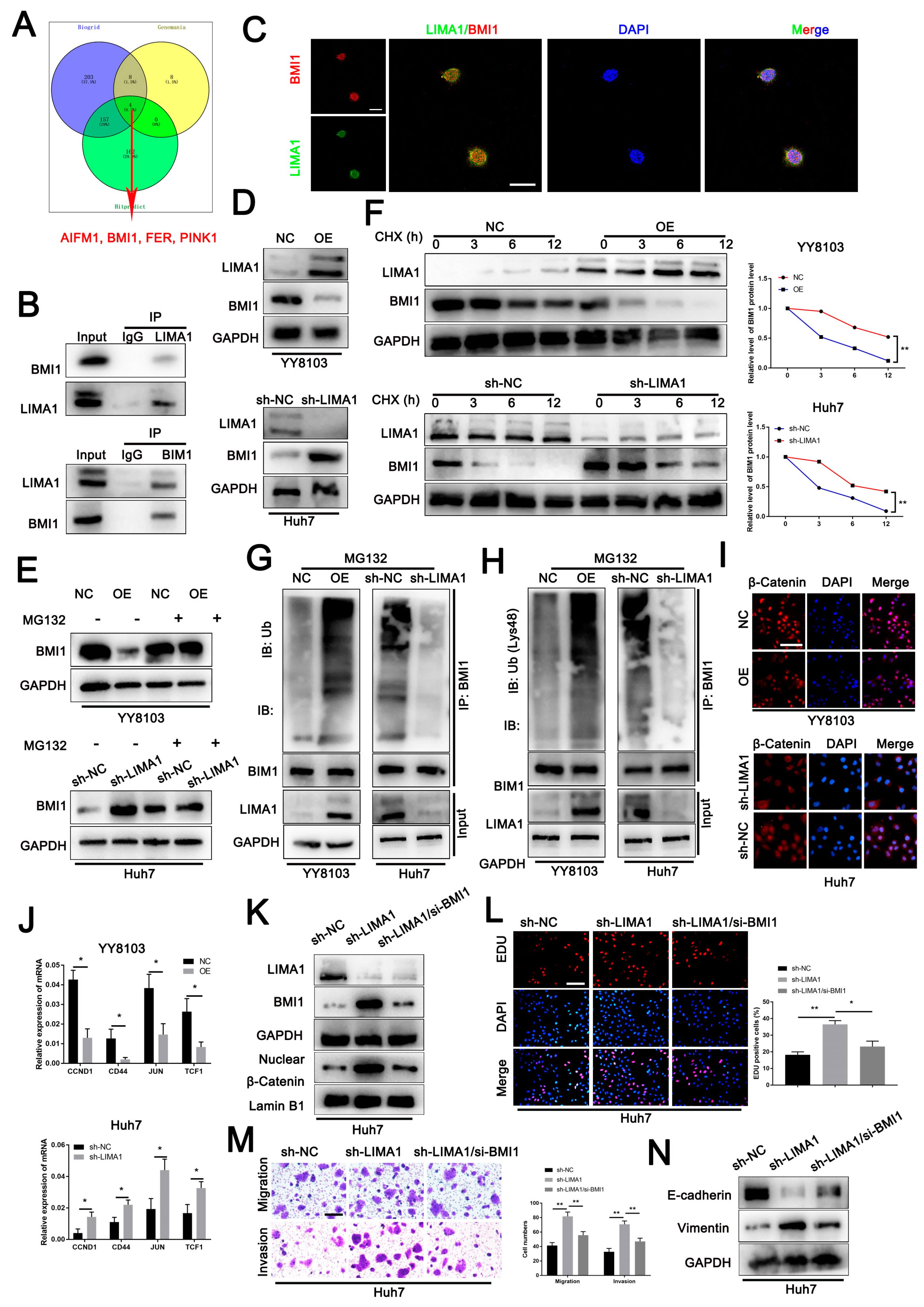
3.4. LIMA1 Impaired the Wnt/β-Catenin Signalling Pathway through BMI1
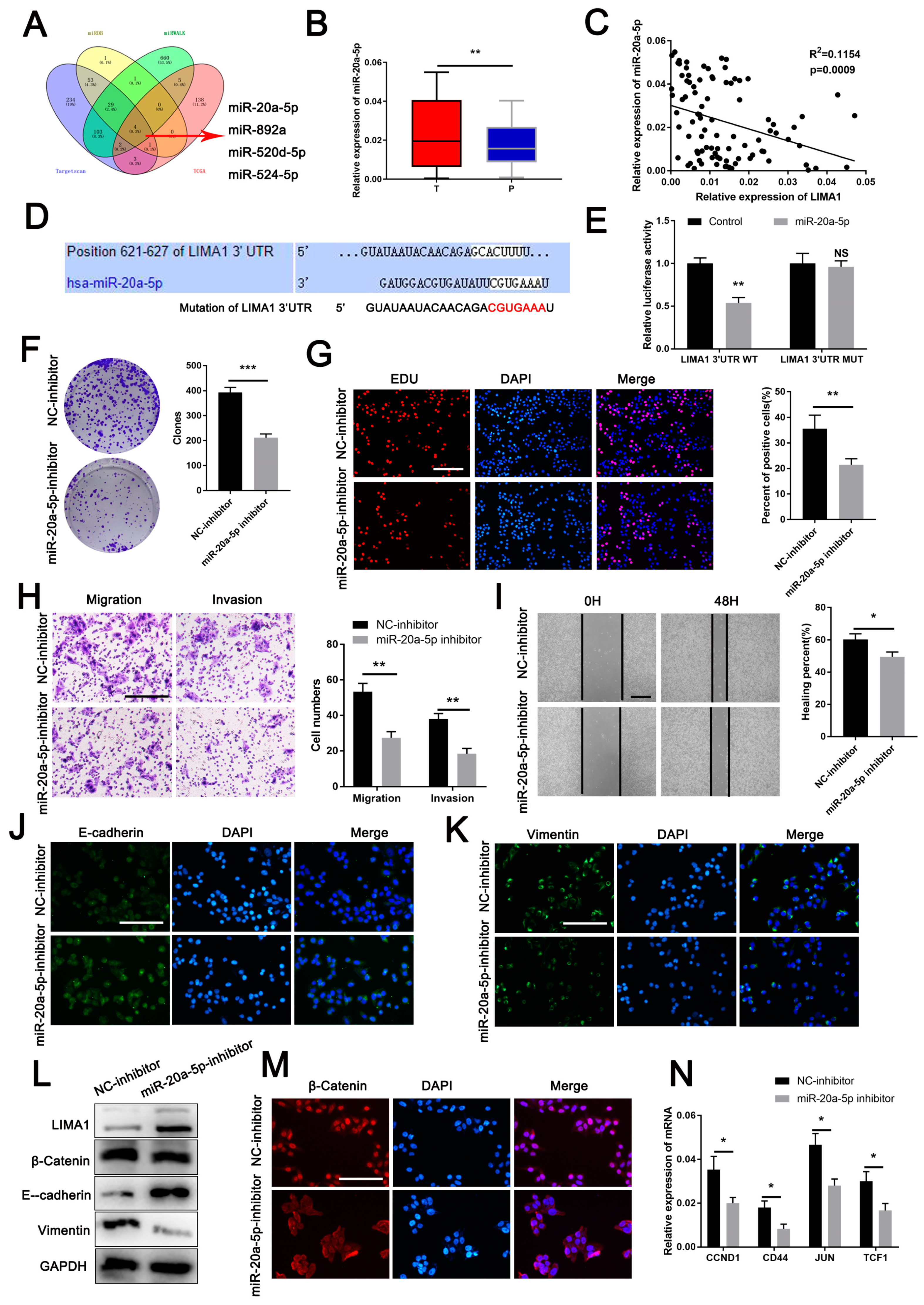
3.5. MiR-20a-5p Is an Oncogene Targeting LIMA1
3.6. LIMA1 Knockdown Restored the Oncogenic Role of miR-20a-5p
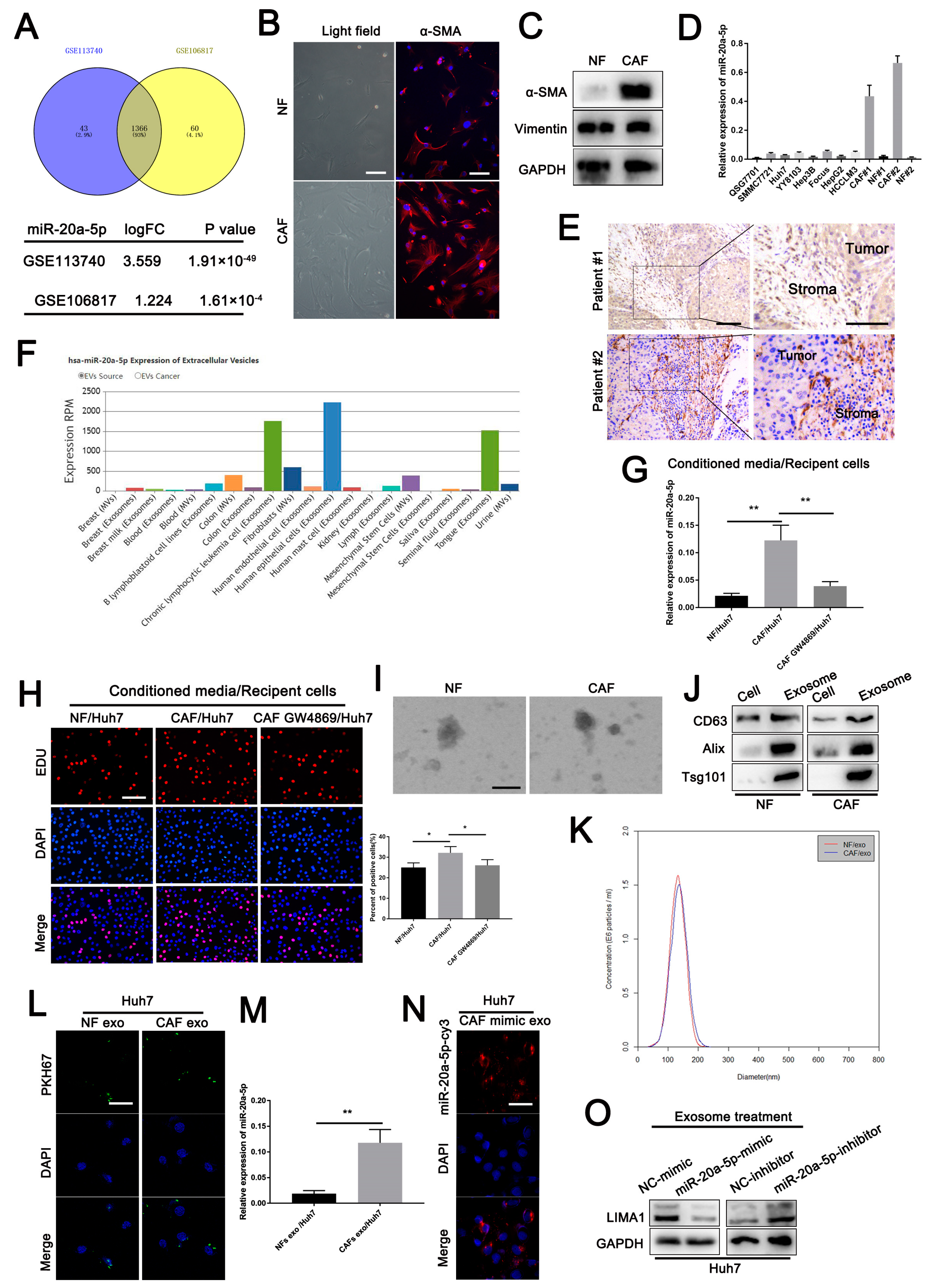
3.7. CAF-Derived Exosomes Carrying miR-20a-5p Facilitated HCC Cell Progression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kulik, L.; El-Serag, H.B. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019, 156, 477–491.e1. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, K.A.; Petrick, J.L.; El-Serag, H.B. Epidemiology of Hepatocellular Carcinoma. Hepatology 2021, 73 (Suppl. 1), 4–13. [Google Scholar] [CrossRef] [PubMed]
- Valery, P.C.; Laversanne, M.; Clark, P.J.; Petrick, J.L.; McGlynn, K.A.; Bray, F. Projections of primary liver cancer to 2030 in 30 countries worldwide. Hepatology 2018, 67, 600–611. [Google Scholar] [CrossRef] [PubMed]
- Zhan, H.; Zhao, X.; Lu, Z.; Yao, Y.; Zhang, X. Correlation and Survival Analysis of Distant Metastasis Site and Prognosis in Patients With Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 652768. [Google Scholar] [CrossRef] [PubMed]
- Maul, R.S.; Chang, D.D. EPLIN, epithelial protein lost in neoplasm. Oncogene 1999, 18, 7838–7841. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, T.; Idogawa, M.; Sasaki, Y.; Tokino, T. p53 mediates the suppression of cancer cell invasion by inducing LIMA1/EPLIN. Cancer Lett. 2017, 390, 58–66. [Google Scholar] [CrossRef]
- Gong, W.; Zeng, J.; Ji, J.; Jia, Y.; Jia, S.; Sanders, A.J.; Jiang, W.G. EPLIN Expression in Gastric Cancer and Impact on Prognosis and Chemoresistance. Biomolecules 2021, 11, 547. [Google Scholar] [CrossRef]
- Duethorn, B.; Groll, F.; Rieger, B.; Drexler, H.C.A.; Brinkmann, H.; Kremer, L.; Stehling, M.; Borowski, M.T.; Mildner, K.; Zeuschner, D.; et al. Lima1 mediates the pluripotency control of membrane dynamics and cellular metabolism. Nat. Commun. 2022, 13, 610. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Yin, Z.; Dong, C.; Jiang, K.; Xu, Z.; Li, R.; Guo, K.; Shao, S.; Wang, L. Heterogeneity of cancer-associated fibroblasts and roles in the progression, prognosis, and therapy of hepatocellular carcinoma. J. Hematol. Oncol. 2019, 12, 101. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, T.; Liu, R.; Ning, T.; Yang, H.; Liu, D.; Zhang, Q.; Lin, D.; Ge, S.; Bai, M.; et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol. Cancer 2020, 19, 43. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef]
- Yang, F.; Ning, Z.; Ma, L.; Liu, W.; Shao, C.; Shu, Y.; Shen, H. Exosomal miRNAs and miRNA dysregulation in cancer-associated fibroblasts. Mol. Cancer 2017, 16, 148. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef]
- Ludwig, A.K.; Giebel, B. Exosomes: Small vesicles participating in intercellular communication. Int. J. Biochem. Cell Biol. 2012, 44, 11–15. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Takahashi, A.; Okada, R.; Nagao, K.; Kawamata, Y.; Hanyu, A.; Yoshimoto, S.; Takasugi, M.; Watanabe, S.; Kanemaki, M.T.; Obuse, C.; et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2017, 8, 15287. [Google Scholar] [CrossRef]
- Gao, J.; Li, S.; Xu, Q.; Zhang, X.; Huang, M.; Dai, X.; Liu, L. Exosomes Promote Pre-Metastatic Niche Formation in Gastric Cancer. Front. Oncol. 2021, 11, 652378. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef]
- Guo, L.; Zhu, Y.; Li, L.; Zhou, S.; Yin, G.; Yu, G.; Cui, H. Breast cancer cell-derived exosomal miR-20a-5p promotes the proliferation and differentiation of osteoclasts by targeting SRCIN1. Cancer Med. 2019, 8, 5687–5701. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; He, J.; Disoma, C.; Hu, Y.; Li, J.; Chen, G.; Sheng, Y.; Cai, X.; Li, C.; Cheng, K.; et al. Hsa_circ_0107593 Suppresses the Progression of Cervical Cancer via Sponging hsa-miR-20a-5p/93-5p/106b-5p. Front. Oncol. 2020, 10, 590627. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Zhou, M.; Lv, H.; Mao, X.; Li, X.; Guo, H.; Li, L.; Xing, H. Long noncoding RNA LINC00657 inhibits cervical cancer development by sponging miR-20a-5p and targeting RUNX3. Cancer Lett. 2021, 498, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Zeng, H.; Chen, J.; Fu, S.; Huang, Q.; Xu, Y.; Xu, A.; Lan, H.Y.; Tang, Y. miR-20a-5p is enriched in hypoxia-derived tubular exosomes and protects against acute tubular injury. Clin. Sci. 2020, 134, 2223–2234. [Google Scholar] [CrossRef]
- Steder, M.; Alla, V.; Meier, C.; Spitschak, A.; Pahnke, J.; Furst, K.; Kowtharapu, B.S.; Engelmann, D.; Petigk, J.; Egberts, F.; et al. DNp73 exerts function in metastasis initiation by disconnecting the inhibitory role of EPLIN on IGF1R-AKT/STAT3 signaling. Cancer Cell 2013, 24, 512–527. [Google Scholar] [CrossRef]
- Wang, B.J.; Liu, D.C.; Guo, Q.Y.; Han, X.W.; Bi, X.M.; Wang, H.; Wu, Z.S.; Wu, W.Y. NUDT21 Suppresses Breast Cancer Tumorigenesis Through Regulating CPSF6 Expression. Cancer Manag. Res. 2020, 12, 3069–3078. [Google Scholar] [CrossRef]
- Song, M.; He, J.; Pan, Q.Z.; Yang, J.; Zhao, J.; Zhang, Y.J.; Huang, Y.; Tang, Y.; Wang, Q.; He, J.; et al. Cancer-Associated Fibroblast-Mediated Cellular Crosstalk Supports Hepatocellular Carcinoma Progression. Hepatology 2021, 73, 1717–1735. [Google Scholar] [CrossRef]
- Wu, D.; He, X.; Wang, W.; Hu, X.; Wang, K.; Wang, M. Long noncoding RNA SNHG12 induces proliferation, migration, epithelial-mesenchymal transition, and stemness of esophageal squamous cell carcinoma cells via post-transcriptional regulation of BMI1 and CTNNB1. Mol. Oncol. 2020, 14, 2332–2351. [Google Scholar] [CrossRef]
- Sahasrabuddhe, A.A.; Dimri, M.; Bommi, P.V.; Dimri, G.P. βTrCP regulates BMI1 protein turnover via ubiquitination and degradation. Cell Cycle 2011, 10, 1322–1330. [Google Scholar] [CrossRef]
- Li, X.G.; Wang, Z.; Chen, R.Q.; Fu, H.L.; Gao, C.Q.; Yan, H.C.; Xing, G.X.; Wang, X.Q. LGR5 and BMI1 Increase Pig Intestinal Epithelial Cell Proliferation by Stimulating WNT/β-Catenin Signaling. Int. J. Mol. Sci. 2018, 19, 1036. [Google Scholar] [CrossRef]
- Yu, H.; Gao, R.; Chen, S.; Liu, X.; Wang, Q.; Cai, W.; Vemula, S.; Fahey, A.C.; Henley, D.; Kobayashi, M.; et al. Bmi1 Regulates Wnt Signaling in Hematopoietic Stem and Progenitor Cells. Stem Cell Rev. Rep. 2021, 17, 2304–2313. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Fu, L.S.; Zhang, F.; Yang, Y.; Wu, X.Z. LncAY controls BMI1 expression and activates BMI1/Wnt/β-catenin signaling axis in hepatocellular carcinoma. Life Sci. 2021, 280, 119748. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, J.; Cairns, M.J. Identifying miRNAs, targets and functions. Brief. Bioinform. 2014, 15, 1–19. [Google Scholar] [CrossRef]
- Zuo, X.; Chen, Z.; Cai, J.; Gao, W.; Zhang, Y.; Han, G.; Pu, L.; Wu, Z.; You, W.; Qin, J.; et al. 5-Hydroxytryptamine Receptor 1D Aggravates Hepatocellular Carcinoma Progression Through FoxO6 in AKT-Dependent and Independent Manners. Hepatology 2019, 69, 2031–2047. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Dutta, A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009, 4, 199–227. [Google Scholar] [CrossRef] [PubMed]
- Herrera, M.; Llorens, C.; Rodriguez, M.; Herrera, A.; Ramos, R.; Gil, B.; Candia, A.; Larriba, M.J.; Garre, P.; Earl, J.; et al. Differential distribution and enrichment of non-coding RNAs in exosomes from normal and Cancer-associated fibroblasts in colorectal cancer. Mol. Cancer 2018, 17, 114. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, Y.; Zou, L.; Zhu, Z. Role of Exosomes in Crosstalk Between Cancer-Associated Fibroblasts and Cancer Cells. Front. Oncol. 2019, 9, 356. [Google Scholar] [CrossRef]
- Shin, K.; Lim, A.; Zhao, C.; Sahoo, D.; Pan, Y.; Spiekerkoetter, E.; Liao, J.C.; Beachy, P.A. Hedgehog signaling restrains bladder cancer progression by eliciting stromal production of urothelial differentiation factors. Cancer Cell 2014, 26, 521–533. [Google Scholar] [CrossRef]
- Gerling, M.; Buller, N.V.; Kirn, L.M.; Joost, S.; Frings, O.; Englert, B.; Bergstrom, A.; Kuiper, R.V.; Blaas, L.; Wielenga, M.C.; et al. Stromal Hedgehog signalling is downregulated in colon cancer and its restoration restrains tumour growth. Nat. Commun. 2016, 7, 12321. [Google Scholar] [CrossRef]


| Features | LIMA1 Expression | No. | p Value | |
|---|---|---|---|---|
| Low (62) | High (30) | |||
| Age (Years) | ||||
| ≤60 | 32 | 15 | 47 | 0.885 |
| >60 | 30 | 15 | 45 | |
| Gender | ||||
| Male | 37 | 18 | 55 | 0.976 |
| Female | 25 | 12 | 37 | |
| HbsAg | ||||
| Positive | 53 | 20 | 73 | 0.037 * |
| Negative | 9 | 10 | 19 | |
| Liver cirrhosis | ||||
| Yes | 49 | 20 | 69 | 0.199 |
| No | 13 | 10 | 23 | |
| AFP (ng/mL) | ||||
| >20 | 49 | 25 | 74 | 0.626 |
| ≤20 | 13 | 5 | 18 | |
| Tumour size (cm) | ||||
| >5 | 50 | 18 | 68 | 0.035 * |
| ≤5 | 12 | 12 | 24 | |
| Tumour multiplicity | ||||
| Single | 50 | 29 | 79 | 0.039 * |
| Multiple | 12 | 1 | 13 | |
| Stage | ||||
| I-II | 24 | 13 | 37 | 0.672 |
| III-IV | 38 | 17 | 55 | |
| Vascular invasion | ||||
| Yes | 13 | 11 | 24 | 0.108 |
| No | 49 | 19 | 68 | |
| Parameters | OS | RFS | ||
|---|---|---|---|---|
| p Value | HR (95% CI) | p Value | HR (95% CI) | |
| Univariate Analysis | ||||
| Age (>60 vs. ≤60) | 0.774 | 1.079 (0.641–1.817) | 0.284 | 1.424 (0.746–2.717) |
| Gender (male vs. female) | 0.488 | 1.210 (0.706–2.074) | 0.236 | 1.501 (0.766–2.940) |
| Tumor number (single vs. multiple) | 0.318 | 1.418 (0.714–2.815) | 0.101 | 1.924 (0.880–4.208) |
| Tumor size (>5 cm vs. ≤5 cm) | 0.001 * | 3.180 (1.588–6.366) | 0.002 * | 4.046 (1.675–9.775) |
| AFP (>20 vs. ≤20) | 0.012 * | 2.646 (1.237–5.662) | 0.067 | 2.273 (0.943–5.474) |
| HBsAg (positive vs. negative) | 0.017 * | 2.638 (1.185–5.869) | 0.008 * | 4.938 (1.506–16.187) |
| Liver cirrhosis | 0.055 | 1.913 (0.987–3.707) | 0.229 | 1.588 (0.747–3.378) |
| TNM stage | 0.002 * | 2.539 (1.420–4.540) | 0.034 * | 2.075 (1.056–4.079) |
| Vascular invasion (yes vs. no) | 0.004 * | 2.360 (1.310–4.254) | 0.133 | 1.868 (0.827–4.221) |
| LIMA1 | 0.017 * | 0.458 (0.242–0.868) | 0.013 * | 0.353 (0.155–0.803) |
| MiR-20a-5p | 0.043 * | 1.748 (1.019–3.000) | 0.004 * | 2.786 (1.379–5.629) |
| Multivariate analysis | ||||
| Tumor size (>5 cm vs. ≤5 cm) | 0.035 * | 2.151 (1.056–4.381) | 0.005 * | 3.620 (1.484–8.830) |
| TNM stage | 0.013 * | 2.126 (1.175–3.844) | - | - |
| Vascular invasion (yes vs. no) | 0.020 * | 2.042 (1.121–3.719) | - | - |
| LIMA1 | 0.032 * | 0.488 (0.253–0.940) | - | - |
| MiR-20a-5p | - | - | 0.014 * | 2.425 (1.193–4.928) |
| Features | miR-20a-5p Expression | No. | p Value | |
|---|---|---|---|---|
| Low (40) | High (52) | |||
| Age (Years) | ||||
| ≤60 | 22 | 25 | 47 | 0.510 |
| >60 | 18 | 27 | 45 | |
| Gender | ||||
| Male | 22 | 33 | 55 | 0.412 |
| Female | 18 | 19 | 37 | |
| HbsAg | ||||
| Positive | 28 | 45 | 73 | 0.052 |
| Negative | 12 | 7 | 19 | |
| Liver cirrhosis | ||||
| Yes | 27 | 42 | 69 | 0.145 |
| No | 13 | 10 | 23 | |
| AFP (ng/mL) | ||||
| >20 | 29 | 45 | 74 | 0.092 |
| ≤20 | 11 | 7 | 18 | |
| Tumour size (cm) | ||||
| >5 | 24 | 44 | 68 | 0.008* |
| ≤5 | 16 | 8 | 24 | |
| Tumour multiplicity | ||||
| Single | 37 | 42 | 79 | 0.109 |
| Multiple | 3 | 10 | 13 | |
| Stage | ||||
| I-II | 21 | 16 | 37 | 0.035 * |
| III-IV | 19 | 36 | 55 | |
| Vascular invasion | ||||
| Yes | 8 | 16 | 24 | 0.244 |
| No | 32 | 36 | 68 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, Y.; Wang, H.; Zhang, Q.; Liu, Z.; Wang, T.; Wu, Z.; Wu, W. CAF-Released Exosomal miR-20a-5p Facilitates HCC Progression via the LIMA1-Mediated β-Catenin Pathway. Cells 2022, 11, 3857. https://doi.org/10.3390/cells11233857
Qi Y, Wang H, Zhang Q, Liu Z, Wang T, Wu Z, Wu W. CAF-Released Exosomal miR-20a-5p Facilitates HCC Progression via the LIMA1-Mediated β-Catenin Pathway. Cells. 2022; 11(23):3857. https://doi.org/10.3390/cells11233857
Chicago/Turabian StyleQi, Yong, Haibo Wang, Qikun Zhang, Zhiqiang Liu, Tianbing Wang, Zhengsheng Wu, and Wenyong Wu. 2022. "CAF-Released Exosomal miR-20a-5p Facilitates HCC Progression via the LIMA1-Mediated β-Catenin Pathway" Cells 11, no. 23: 3857. https://doi.org/10.3390/cells11233857
APA StyleQi, Y., Wang, H., Zhang, Q., Liu, Z., Wang, T., Wu, Z., & Wu, W. (2022). CAF-Released Exosomal miR-20a-5p Facilitates HCC Progression via the LIMA1-Mediated β-Catenin Pathway. Cells, 11(23), 3857. https://doi.org/10.3390/cells11233857