Circulating Small Extracellular Vesicle-Derived miR-342-5p Ameliorates Beta-Amyloid Formation via Targeting Beta-site APP Cleaving Enzyme 1 in Alzheimer’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects and Sample Preparation
2.2. Isolation and Characterization of Exosome from the Peripheral Blood
2.3. Cell Culture and RNA Transfection
2.4. RNA Extraction and Quantitative RT-PCR
2.5. Western Blotting
2.6. Dual-Luciferase Reporter Assay
2.7. Mouse Hippocampus Exosome Isolation and MiR Mimics Transfection
2.8. Incubation of HT22 Cells with Dil-Labeled sEVs
2.9. Statistical Analysis
3. Results
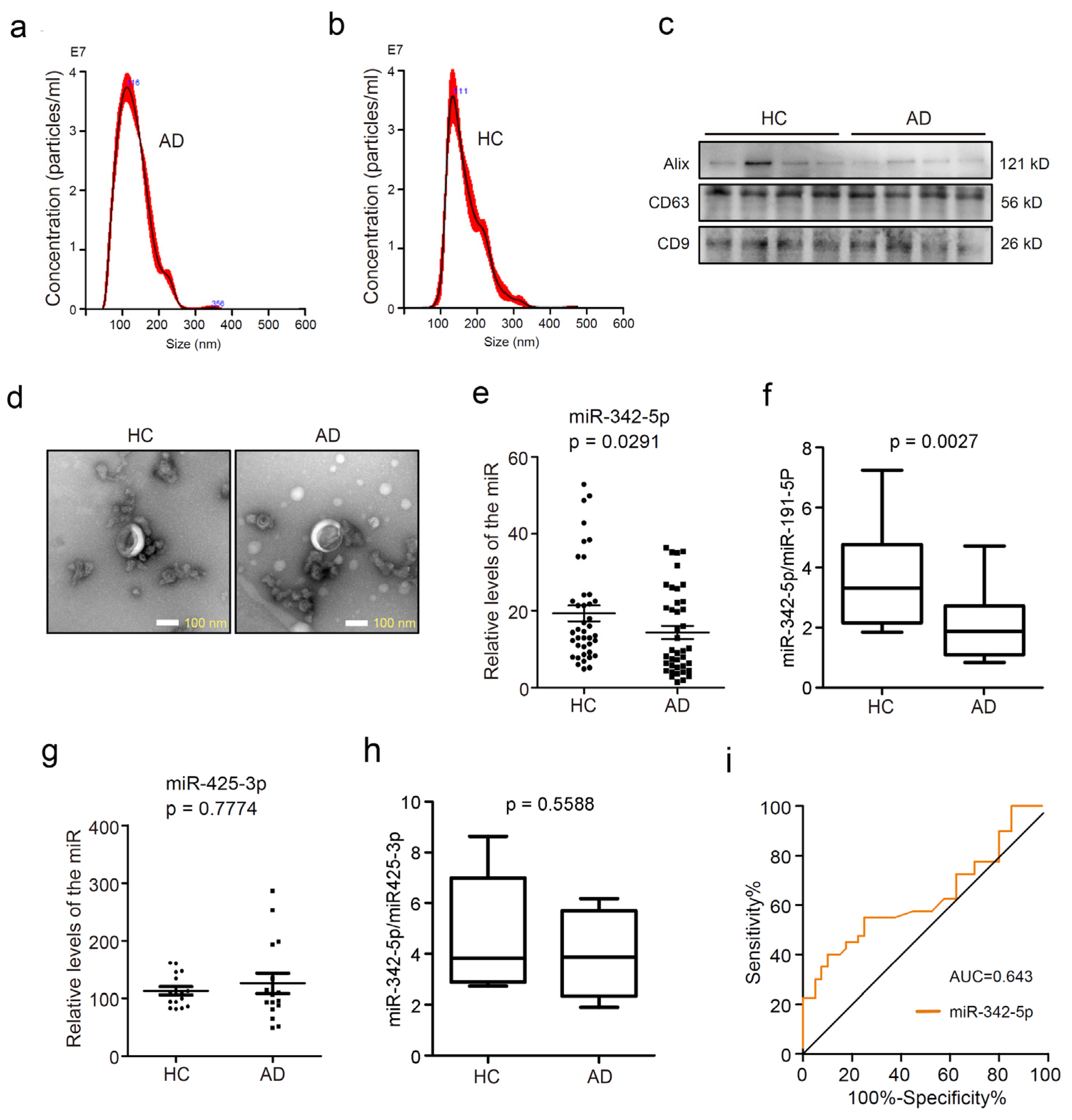
3.1. MiR-342-5p was Dysregulated in Serum sEVs from AD Patients
3.2. MiR-342-5p Targets Bace1 in Mouse Hippocampal HT22 Neurons
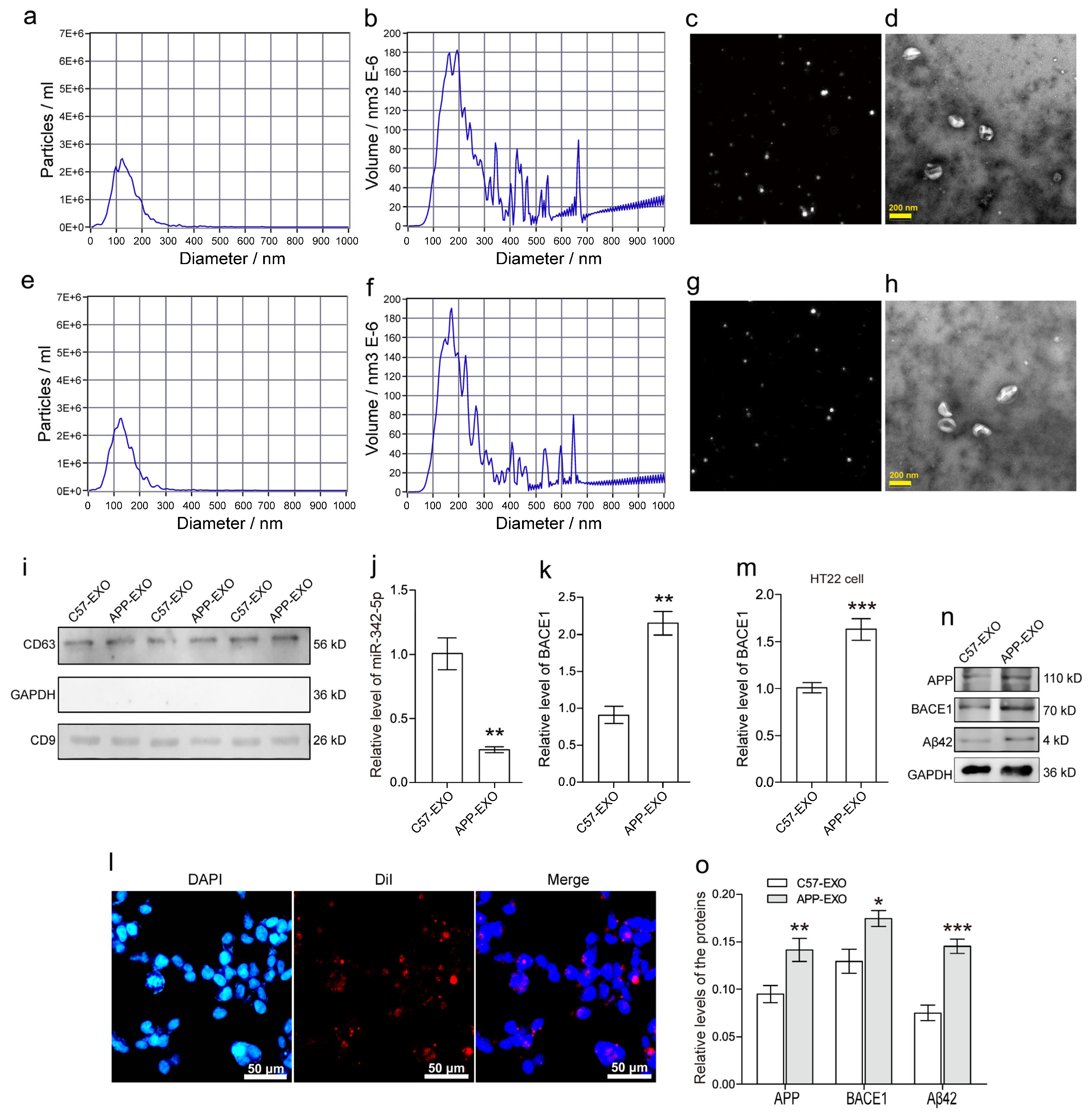
3.3. MiR-342-5p-Dysregulated sEVs from Hippocampus of APP Mouse Exacerbate Aβ42 Formation in HT22 Neurons
3.4. MiR-342-5p Enrichment in Hippocampal sEVs Rescued Aβ42 Formation in Recipient HT22 Neurons
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Song, C.; Shi, J.; Zhang, P.; Zhang, Y.; Xu, J.; Zhao, L.; Zhang, R.; Wang, H.; Chen, H. Immunotherapy for Alzheimer’s disease: Targeting beta-amyloid and beyond. Transl. Neurodegener. 2022, 11, 18. [Google Scholar] [CrossRef]
- Lazarov, O.; Lee, M.; Peterson, D.A.; Sisodia, S.S. Evidence that synaptically released beta-amyloid accumulates as extracellular deposits in the hippocampus of transgenic mice. J. Neurosci. 2002, 22, 9785–9793. [Google Scholar] [CrossRef]
- Vassar, R.; Bennett, B.D.; Babu-Khan, S.; Kahn, S.; Mendiaz, E.A.; Denis, P.; Teplow, D.B.; Ross, S.; Amarante, P.; Loeloff, R.; et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 1999, 286, 735–741. [Google Scholar] [CrossRef]
- Liu, L.; Ding, L.; Rovere, M.; Wolfe, M.S.; Selkoe, D.J. A cellular complex of BACE1 and gamma-secretase sequentially generates Abeta from its full-length precursor. J. Cell Biol. 2019, 218, 644–663. [Google Scholar] [CrossRef]
- Lopez-Toledano, M.A.; Shelanski, M.L. Neurogenic effect of beta-amyloid peptide in the development of neural stem cells. J. Neurosci. 2004, 24, 5439–5444. [Google Scholar] [CrossRef]
- Plant, L.D.; Boyle, J.P.; Smith, I.F.; Peers, C.; Pearson, H.A. The production of amyloid beta peptide is a critical requirement for the viability of central neurons. J. Neurosci. 2003, 23, 5531–5535. [Google Scholar] [CrossRef] [PubMed]
- Kamenetz, F.; Tomita, T.; Hsieh, H.; Seabrook, G.; Borchelt, D.; Iwatsubo, T.; Sisodia, S.; Malinow, R. APP processing and synaptic function. Neuron 2003, 37, 925–937. [Google Scholar] [CrossRef]
- Abramov, E.; Dolev, I.; Fogel, H.; Ciccotosto, G.D.; Ruff, E.; Slutsky, I. Amyloid-beta as a positive endogenous regulator of release probability at hippocampal synapses. Nat. Neurosci. 2009, 12, 1567–1576. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, J.L.; Tong, M.; Alfulaij, N.; Sherrin, T.; Contarino, M.; White, M.M.; Bellinger, F.P.; Todorovic, C.; Nichols, R.A. Regulation of presynaptic Ca2+, synaptic plasticity and contextual fear conditioning by a N-terminal beta-amyloid fragment. J. Neurosci. 2014, 34, 14210–14218. [Google Scholar] [CrossRef]
- Morley, J.E.; Farr, S.A.; Banks, W.A.; Johnson, S.N.; Yamada, K.A.; Xu, L. A physiological role for amyloid-beta protein:enhancement of learning and memory. J. Alzheimer’s Dis. 2010, 19, 441–449. [Google Scholar] [CrossRef]
- Puzzo, D.; Privitera, L.; Fa, M.; Staniszewski, A.; Hashimoto, G.; Aziz, F.; Sakurai, M.; Ribe, E.M.; Troy, C.M.; Mercken, M.; et al. Endogenous amyloid-beta is necessary for hippocampal synaptic plasticity and memory. Ann. Neurol. 2011, 69, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Palmeri, A.; Ricciarelli, R.; Gulisano, W.; Rivera, D.; Rebosio, C.; Calcagno, E.; Tropea, M.R.; Conti, S.; Das, U.; Roy, S.; et al. Amyloid-beta Peptide Is Needed for cGMP-Induced Long-Term Potentiation and Memory. J. Neurosci. 2017, 37, 6926–6937. [Google Scholar] [CrossRef] [PubMed]
- Meier-Stephenson, F.S.; Meier-Stephenson, V.C.; Carter, M.D.; Meek, A.R.; Wang, Y.; Pan, L.; Chen, Q.; Jacobo, S.; Wu, F.; Lu, E.; et al. Alzheimer’s disease as an autoimmune disorder of innate immunity endogenously modulated by tryptophan metabolites. Alzheimer’s Dement. 2022, 8, e12283. [Google Scholar] [CrossRef] [PubMed]
- Gosztyla, M.L.; Brothers, H.M.; Robinson, S.R. Alzheimer’s Amyloid-beta is an Antimicrobial Peptide: A Review of the Evidence. J. Alzheimer’s Dis. 2018, 62, 1495–1506. [Google Scholar] [CrossRef]
- Smith, M.A.; Casadesus, G.; Joseph, J.A.; Perry, G. Amyloid-beta and tau serve antioxidant functions in the aging and Alzheimer brain. Free. Radic. Biol. Med. 2002, 33, 1194–1199. [Google Scholar] [CrossRef]
- Rockenstein, E.; Mante, M.; Alford, M.; Adame, A.; Crews, L.; Hashimoto, M.; Esposito, L.; Mucke, L.; Masliah, E. High beta-secretase activity elicits neurodegeneration in transgenic mice despite reductions in amyloid-beta levels: Implications for the treatment of Alzheimer disease. J. Biol. Chem. 2005, 280, 32957–32967. [Google Scholar] [CrossRef]
- McConlogue, L.; Buttini, M.; Anderson, J.P.; Brigham, E.F.; Chen, K.S.; Freedman, S.B.; Games, D.; Johnson-Wood, K.; Lee, M.; Zeller, M.; et al. Partial reduction of BACE1 has dramatic effects on Alzheimer plaque and synaptic pathology in APP Transgenic Mice. J. Biol. Chem. 2007, 282, 26326–26334. [Google Scholar] [CrossRef]
- Jiang, Y.; Rigoglioso, A.; Peterhoff, C.M.; Pawlik, M.; Sato, Y.; Bleiwas, C.; Stavrides, P.; Smiley, J.F.; Ginsberg, S.D.; Mathews, P.M.; et al. Partial BACE1 reduction in a Down syndrome mouse model blocks Alzheimer-related endosomal anomalies and cholinergic neurodegeneration: Role of APP-CTF. Neurobiol. Aging 2016, 39, 90–98. [Google Scholar] [CrossRef]
- Putteeraj, M.; Fairuz, Y.M.; Teoh, S.L. MicroRNA Dysregulation in Alzheimer’s Disease. CNS Neurol. Disord. Drug Targets 2017, 16, 1000–1009. [Google Scholar] [CrossRef]
- Wang, L.L.; Min, L.; Guo, Q.D.; Zhang, J.X.; Jiang, H.L.; Shao, S.; Xing, J.G.; Yin, L.L.; Liu, J.H.; Liu, R.; et al. Profiling microRNA from Brain by Microarray in a Transgenic Mouse Model of Alzheimer’s Disease. BioMed Res. Int. 2017, 2017, 8030369. [Google Scholar] [CrossRef]
- Sun, X.; Wu, Y.; Gu, M.; Zhang, Y. miR-342-5p decreases ankyrin G levels in Alzheimer’s disease transgenic mouse models. Cell Rep. 2014, 6, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wu, Y.; Gu, M.; Liu, Z.; Ma, Y.; Li, J.; Zhang, Y. Selective filtering defect at the axon initial segment in Alzheimer’s disease mouse models. Proc. Natl. Acad. Sci. USA 2014, 111, 14271–14276. [Google Scholar] [CrossRef] [PubMed]
- Lugli, G.; Cohen, A.M.; Bennett, D.A.; Shah, R.C.; Fields, C.J.; Hernandez, A.G.; Smalheiser, N.R. Plasma Exosomal miRNAs in Persons with and without Alzheimer Disease: Altered Expression and Prospects for Biomarkers. PLoS ONE 2015, 10, e0139233. [Google Scholar] [CrossRef] [PubMed]
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R., Jr.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.J.; Mayeux, R.; et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011, 7, 263–269. [Google Scholar] [CrossRef]
- Dong, Z.; Gu, H.; Guo, Q.; Liang, S.; Xue, J.; Yao, F.; Liu, X.; Li, F.; Liu, H.; Sun, L.; et al. Profiling of Serum Exosome MiRNA Reveals the Potential of a MiRNA Panel as Diagnostic Biomarker for Alzheimer’s Disease. Mol. Neurobiol. 2021, 58, 3084–3094. [Google Scholar] [CrossRef] [PubMed]
- Polanco, J.C.; Scicluna, B.J.; Hill, A.F.; Gotz, J. Extracellular Vesicles Isolated from the Brains of rTg4510 Mice Seed Tau Protein Aggregation in a Threshold-dependent Manner. J. Biol. Chem. 2016, 291, 12445–12466. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Gu, J.; Jiang, P.; Qian, H.; Xu, W.; Zhang, X. Exosomes in gastric cancer: Roles, mechanisms, and applications. Mol. Cancer 2019, 18, 41. [Google Scholar] [CrossRef]
- Dakterzada, F.; Targa, A.; Benitez, I.D.; Romero-ElKhayat, L.; de Gonzalo-Calvo, D.; Torres, G.; Moncusi-Moix, A.; Huerto, R.; Sanchez-de-la-Torre, M.; Barbe, F.; et al. Identification and validation of endogenous control miRNAs in plasma samples for normalization of qPCR data for Alzheimer’s disease. Alzheimer’s Res. Ther. 2020, 12, 163. [Google Scholar] [CrossRef]
- Dakterzada, F.; David Benitez, I.; Targa, A.; Llado, A.; Torres, G.; Romero, L.; de Gonzalo-Calvo, D.; Moncusi-Moix, A.; Tort-Merino, A.; Huerto, R.; et al. Reduced Levels of miR-342-5p in Plasma Are Associated with Worse Cognitive Evolution in Patients With Mild Alzheimer’s Disease. Front. Aging Neurosci. 2021, 13, 705989. [Google Scholar] [CrossRef]
- Fevrier, B.; Raposo, G. Exosomes: Endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 2004, 16, 415–421. [Google Scholar] [CrossRef]
- Denzer, K.; Kleijmeer, M.J.; Heijnen, H.F.; Stoorvogel, W.; Geuze, H.J. Exosome: From internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 2000, 113 Pt 19, 3365–3374. [Google Scholar] [CrossRef]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009, 11, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Mukherjee, K.; Chakrabarty, Y.; Chatterjee, S.; Ghoshal, B.; Bhattacharyya, S.N. GW182 Proteins Restrict Extracellular Vesicle-Mediated Export of MicroRNAs in Mammalian Cancer Cells. Mol. Cell. Biol. 2021, 41, e00483-20. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Kim, G.; Kim, M.; Lee, Y.; Byun, J.W.; Hwang, D.W.; Lee, M. Systemic delivery of microRNA-21 antisense oligonucleotides to the brain using T7-peptide decorated exosomes. J. Control. Release 2020, 317, 273–281. [Google Scholar] [CrossRef]
- Marcus, M.E.; Leonard, J.N. FedExosomes: Engineering Therapeutic Biological Nanoparticles that Truly Deliver. Pharmaceuticals 2013, 6, 659–680. [Google Scholar] [CrossRef] [PubMed]
- Cataldo, A.M.; Petanceska, S.; Terio, N.B.; Peterhoff, C.M.; Durham, R.; Mercken, M.; Mehta, P.D.; Buxbaum, J.; Haroutunian, V.; Nixon, R.A. Abeta localization in abnormal endosomes: Association with earliest Abeta elevations in AD and Down syndrome. Neurobiol. Aging 2004, 25, 1263–1272. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Feng, T.; Tammineni, P.; Chang, Q.; Jeong, Y.Y.; Margolis, D.J.; Cai, H.; Kusnecov, A.; Cai, Q. Regulation of Synaptic Amyloid-beta Generation through BACE1 Retrograde Transport in a Mouse Model of Alzheimer’s Disease. J. Neurosci. 2017, 37, 2639–2655. [Google Scholar] [CrossRef] [PubMed]
- Sharples, R.A.; Vella, L.J.; Nisbet, R.M.; Naylor, R.; Perez, K.; Barnham, K.J.; Masters, C.L.; Hill, A.F. Inhibition of gamma-secretase causes increased secretion of amyloid precursor protein C-terminal fragments in association with exosomes. FASEB J. 2008, 22, 1469–1478. [Google Scholar] [CrossRef]
- Campos-Pena, V.; Pichardo-Rojas, P.; Sanchez-Barbosa, T.; Ortiz-Islas, E.; Rodriguez-Perez, C.E.; Montes, P.; Ramos-Palacios, G.; Silva-Adaya, D.; Valencia-Quintana, R.; Cerna-Cortes, J.F.; et al. Amyloid beta, Lipid Metabolism, Basal Cholinergic System, and Therapeutics in Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 12092. [Google Scholar] [CrossRef]
- Modler, A.J.; Gast, K.; Lutsch, G.; Damaschun, G. Assembly of amyloid protofibrils via critical oligomers—A novel pathway of amyloid formation. J. Mol. Biol. 2003, 325, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Sarroukh, R.; Cerf, E.; Derclaye, S.; Dufrene, Y.F.; Goormaghtigh, E.; Ruysschaert, J.M.; Raussens, V. Transformation of amyloid beta (1-40) oligomers into fibrils is characterized by a major change in secondary structure. Cell. Mol. Life Sci. 2011, 68, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- More, S.S.; Vince, R. Hyperspectral imaging signatures detect amyloidopathy in Alzheimer’s mouse retina well before onset of cognitive decline. ACS Chem.Neurosci. 2015, 6, 306–315. [Google Scholar] [CrossRef]
- Panza, F.; Lozupone, M.; Logroscino, G.; Imbimbo, B.P. A critical appraisal of amyloid-beta-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 2019, 15, 73–88. [Google Scholar] [CrossRef]
- Ben Halima, S.; Mishra, S.; Raja, K.M.P.; Willem, M.; Baici, A.; Simons, K.; Brustle, O.; Koch, P.; Haass, C.; Caflisch, A.; et al. Specific Inhibition of beta-Secretase Processing of the Alzheimer Disease Amyloid Precursor Protein. Cell Rep. 2016, 14, 2127–2141. [Google Scholar] [CrossRef] [PubMed]
- Sannerud, R.; Declerck, I.; Peric, A.; Raemaekers, T.; Menendez, G.; Zhou, L.; Veerle, B.; Coen, K.; Munck, S.; De Strooper, B.; et al. ADP ribosylation factor 6 (ARF6) controls amyloid precursor protein (APP) processing by mediating the endosomal sorting of BACE1. Proc. Natl. Acad. Sci. USA 2011, 108, E559–E568. [Google Scholar] [CrossRef] [PubMed]
- Toh, W.H.; Chia, P.Z.C.; Hossain, M.I.; Gleeson, P.A. GGA1 regulates signal-dependent sorting of BACE1 to recycling endosomes, which moderates Abeta production. Mol. Biol. Cell 2018, 29, 191–208. [Google Scholar] [CrossRef]


| Characteristics | n (m/f) | Mean Age | MMSE Score |
|---|---|---|---|
| HC | 40 (20/20) | 77.1 ± 6.2 | n.d. |
| AD | 40 (20/20) | 75.1 ± 7.5 | 6.55 ± 2.86 |
| Genes | Primer Sequences (5′ to 3′) |
|---|---|
| hsa-miR-342-5p | AGGGGTGCTATCTGTGAAAAA |
| hsa-U6 | TTCGTGAAGCGTTCCATATTTT |
| mmu-GAPDH -F | CAAAATGGTGAAGGTCGGTGT |
| mmu-GAPDH -R | GAGGTCAATGAAGGGGTCGTT |
| mmu-miR-342-5p-F | CGCAGAGGGGTGCTATCTGT |
| mmu-miR-342-5p-R | AGTGCGTGTCGTGGAGTCG |
| mmu-U6-F | CGATACAGAGAAGATTAGCATGGC |
| mmu-U6-R | AACGCTTCACGAATTTGCGT |
| mmu-BACE1-F | GACCACTCGCTATACACGGG |
| mmu-BACE1-R | CTTCTCCGTCTCCTTGCAGT |
| Ortholog of Target Gene | Representative Transcript | Gene Name | Cumulative Weighted Context++ Score | Total Context++ Score |
|---|---|---|---|---|
| Nsmf | ENSMUST00000100334.5 | NMDA receptor synaptonuclear signaling and neuronal migration factor | −0.43 | −0.43 |
| Smpd3 | ENSMUST00000067512.7 | sphingomyelin phosphodiesterase 3, neutral | −0.02 | −0.04 |
| Bace1 | ENSMUST00000034591.5 | beta-site APP cleaving enzyme 1 | −0.2 | −0.38 |
| Nptxr | ENSMUST00000175858.3 | neuronal pentraxin receptor | −0.25 | −0.65 |
| Syp | ENSMUST00000069520.5 | synaptophysin | −0.11 | −0.11 |
| Syngap1 | ENSMUST00000081285.4 | synaptic Ras GTPase activating protein 1 homolog (rat) | −0.62 | −0.62 |
| Stxbp1 | ENSMUST00000077458.4 | syntaxin binding protein 1 | −0.37 | −0.38 |
| Stx1b | ENSMUST00000106267.3 | syntaxin 1B | −0.24 | −0.24 |
| Nf2 | ENSMUST00000056290.7 | neurofibromatosis 2 | −0.1 | −0.1 |
| Nptx1 | ENSMUST00000026670.4 | neuronal pentraxin 1 | −0.14 | −0.14 |
| Nrxn2 | ENSMUST00000113462.2 | neurexin II | −0.24 | −0.24 |
| Ngb | ENSMUST00000110176.1 | neuroglobin | −0.21 | −0.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Z.; Gu, H.; Guo, Q.; Liu, X.; Li, F.; Liu, H.; Sun, L.; Ma, H.; Zhao, K. Circulating Small Extracellular Vesicle-Derived miR-342-5p Ameliorates Beta-Amyloid Formation via Targeting Beta-site APP Cleaving Enzyme 1 in Alzheimer’s Disease. Cells 2022, 11, 3830. https://doi.org/10.3390/cells11233830
Dong Z, Gu H, Guo Q, Liu X, Li F, Liu H, Sun L, Ma H, Zhao K. Circulating Small Extracellular Vesicle-Derived miR-342-5p Ameliorates Beta-Amyloid Formation via Targeting Beta-site APP Cleaving Enzyme 1 in Alzheimer’s Disease. Cells. 2022; 11(23):3830. https://doi.org/10.3390/cells11233830
Chicago/Turabian StyleDong, Zhiwu, Hongjun Gu, Qiang Guo, Xianglu Liu, Feifei Li, Huiling Liu, Li Sun, Huimin Ma, and Kewen Zhao. 2022. "Circulating Small Extracellular Vesicle-Derived miR-342-5p Ameliorates Beta-Amyloid Formation via Targeting Beta-site APP Cleaving Enzyme 1 in Alzheimer’s Disease" Cells 11, no. 23: 3830. https://doi.org/10.3390/cells11233830
APA StyleDong, Z., Gu, H., Guo, Q., Liu, X., Li, F., Liu, H., Sun, L., Ma, H., & Zhao, K. (2022). Circulating Small Extracellular Vesicle-Derived miR-342-5p Ameliorates Beta-Amyloid Formation via Targeting Beta-site APP Cleaving Enzyme 1 in Alzheimer’s Disease. Cells, 11(23), 3830. https://doi.org/10.3390/cells11233830





