SAPAP Scaffold Proteins: From Synaptic Function to Neuropsychiatric Disorders
Abstract
1. Introduction
2. SAPAP Family
2.1. SAPAP-Interacting Proteins
2.2. Expression of SAPAPs in the CNS
3. Roles of SAPAPs in Synaptic Structure and Function
3.1. Synaptic Formation and Maturation
3.2. Synaptic Transmission and Plasticity
4. SAPAPs’ Expression, FMRP, and Neuropsychiatric Disorders
5. Human Molecular Genetic Analysis of Neuropsychiatric Disorders Linked to SAPAP
5.1. SAPAP1 and Neuropsychiatric Disorders
5.2. SAPAP2 and Neuropsychiatric Disorders
| Gene | Schizophrenia | ASD | OCD and Related Disorders | AD and Other Cognitive Disorders | ADHD and MDD |
|---|---|---|---|---|---|
| SAPAP1 | Rare missense mutation c.1922A > G (a benign variant) [132] | Associated with the IGAP (International Genomics of Alzheimer’s Project) SNP rs8093731 in Desmoglein-2/DSG2; underexpressed in the entorhinal cortex, hippocampus, and frontal and temporal cortex of AD cases [28] | SNPs rs1116345 and rs34248 [144] | c.1397A > G, p. Asp466Gly, exon 8 in subcortical heterotopias [34] | SNPs rs2049161and rs16946051 associated with cognitive flexibility in ADHD [27] SNP rs12455524 in recurrent MDD [134] |
| De novo CNV deletion [24] | Identified as an ASD-associated gene in a genome-wide network analysis [133] | Two SNPs located within an intron of SAPAP1 [25] | N/A | N/A | |
| N/A | N/A | Rare CNVs (62 kb duplication) [26] | N/A | N/A | |
| SAPAP2 | Damaging missense [147] | CNV 8:1626547:G:C [30] | SNPs rs6558484 and rs7014992 associated with OFC white matter volume [144] | 8p23.3 deletion in DD/mental retardation [145] | N/A |
| CNV duplication [146] | Rare de novo CNV duplications(704383-1521910) [29] | Rare CNVs (16 kb deletion) [26] | SNP rs34130287C within the first intron [149] | N/A | |
| SNV c.-69+9C.T, c.-69+13C.T, c.-69+47C.T, c.-69+55C.T [20] | SNP rs2906569 at intron 1, rs2301963 (P384Q) at exon 3 and several nonsynonymous variants [137] | N/A | SNP rs6992443 [150] | N/A | |
| N/A | Rare de novo CMV duplication 8:704383-1521910 [138] | N/A | SNP rs2957061 SNP chr8:1316870; minor allele frequency (a locus within SAPAP2) [22] | N/A | |
| Gene | Schizophrenia | ASD | OCD and Related Disorders | AD and Other Cognitive Disorders | |
| SAPAP2 | CNV deletion, 8p 23.2 and 8p 23.1 [139] | CNV deletion 8p 23.3 [151] | N/A | N/A | |
| N/A | CNV deletion 8p 23.2 and 8p 23.1 [139] | N/A | N/A | ||
| CNV deletion, 8p23.3-p23.1 [148] | CNV duplication 8p23.3 (patients with de novo rearrangements) [140] | N/A | N/A | ||
| N/A | CNV duplication,8p23.3 (1, 499, 963–1, 854, 917) [141] | N/A | N/A | ||
| SAPAP3 | SNVs c.1141G > A; c.1759G > C; c.2309G > T; c.2578-11C > T [20] | CNV 1:35365700:G:A [30] | Increased frequency of rare nonsynonymous coding variants (in OCD and TTM) [152] | N/A | |
| N/A | N/A | SNPs rs6662980–rs4652867 in grooming disorder [31] | N/A | ||
| N/A | N/A | SNP rs11264126 and two haplotypes containing rs11264126 and rs12141243 in TS [33] | N/A | ||
| N/A | N/A | SNPs rs11583978 and rs6682829 in early-onset OCD [32] | N/A | ||
| SAPAP4 | N/A | SNP located at the 20q11.21–q13.12 locus comprising the Sapap4 gene [153] | N/A | De novo frameshift SNVs (c.2714_2715insCAGCTGG) insertion, N905Qfs, exon 12; c.2893T > G, p. Ser965Ala, exon 13 in subcortical heterotopias [34] | |
5.3. SAPAP3 and Neuropsychiatric Disorders
5.4. SAPAP4 and Neuropsychiatric Disorders
6. Mutational Studies in Murine SAPAP Models
| Target Protein | Murine Model Details and Background | Phenotype | |||||
|---|---|---|---|---|---|---|---|
| Social Behaviors and PPI | Locomotion and Motor Ability | Compulsive and Repetitive Behaviors | Cognitive Function/ Learning and Memory | Anxiety, Depression, and Reward-Related Behaviors | Other Phenotypes | ||
| SAPAP1 | Dlgap1 KO (129/C57BL/6J) |
|
|
| N/A |
|
|
| Dlgap1 KO (129 S5/C57BL/6J) | N/A |
| N/A |
| N/A | N/A | |
| SAPAP2 | Dlgap2 KO exon 6 mutated (C57BL/6) |
|
| NA |
| N/A | N/A |
| Dlgap2 KO (129 S5/C57BL/6J) | N/A |
| N/A | N/A | N/A | ||
| Dlgap2 KO exon 6 mutated (C57BL/6J) | N/A | N/A | N/A | N/A | N/A | ||
| Target Protein | Murine Model Details and Background | Phenotype | |||||
| Social Behaviors and PPI | Locomotion and Motor Ability | Compulsive and Repetitive Behaviors | Cognitive Function/Learning and Memory | Anxiety, Depression, and Reward-Related Behaviors | Other Phenotypes | ||
| SAPAP3 | Sapap3 KO exon 3 mutated (C57BL/6J) |
| |||||
| Sapap3 KO, details not mentioned (C57BL/6J) | N/A | N/A |
|
| N/A | N/A | |
| Target Protein | Murine Model Details and Background | Phenotype | |||||
| Social Behaviors and PPI | Locomotion and Motor Ability | Compulsive and Repetitive Behaviors | Cognitive Function/Learning and Memory | Anxiety, Depression, and Reward-Related Behaviors | Other Phenotypes | ||
| SAPAP4 | Sapap4 KO replaced exons 3–6 with an ATG-YFP-STOP cassette (C57BL/6J) |
|
| N/A |
|
| |
| Dlgap4geo/geo exon trap vector integrated in intron 7 of the Dlgap4 gene (C57BL/6J) |
|
|
|
|
| ||
| Dlgap4 KO exon 8 mutated (C57BL/6N) | N/A | N/A | N/A | N/A | N/A |
| |
| Target protein | Murine Model Details and Background | Phenotype | |||||
| Synaptic Morphology | Synaptic and Circuity Functions | mRNA or Protein Level | |||||
| SAPAP1 | Dlgap1 KO (129/C57BL/6J) | N/A | N/A | Cortex (total proteins)
| |||
| SAPAP1 | Dlgap1 KO (129 S5/C57BL/6J) | N/A | N/A | N/A [156] | |||
| SAPAP2 | Dlgap2 KO exon 6 mutated (C57BL/6) | OFC
| OFC
| Cortex (synaptosome proteins) | |||
| Dlgap2 KO (129 S5/C57BL/6J) | N/A | N/A | N/A [156] | ||||
| Dlgap2 KO exon 6 mutated (C57BL/6J) | N/A | N/A | N/A [21] | ||||
| Target Protein | Murine Model Details and Background | Phenotype | |||||
| Synaptic Morphology | Synaptic and Circuity Functions | mRNA or Protein Level | |||||
| SAPAP3 | Sapap3 KO exon 3 mutated (C57BL/6J) | Striatum
| Striatum
| Striatum
↓ rectification index (D2-MSNs), ↓ cumulative probability (M1/M2-to-DLS AMPAR currents) [104] LOFC | Striatum (PSD proteins)
| ||
| Target Protein | Murine model Details and Background | Phenotype | |||||
| Synaptic Morphology | Synaptic and Circuity Functions | mRNA or Protein Level | |||||
| SAPAP3 | Sapap3 KO details not mentioned (C57BL/6J) | N/A |
| Distinct patterns of abnormal LOFC activity during grooming and reverse learning [163] | N/A | ||
| SAPAP4 | Sapap4 KO replace exons 3–6 with an ATG-YFP-STOP cassette (C57BL/6J) | mPFC
| NAc (shell)
| N/A | PFC
| ||
| Dlgap4geo/geo exon trap vector integrated in intron 7 of the Dlgap4 gene (C57BL/6J) | Hippocampus (CA1)
| Hippocampus (CA1)
| N/A | Hippocampus (PSD proteins)
| |||
| Dlgap4 KO exon 8 mutated(C57BL/6N) | N/A | N/A | N/A | Sapap4 mRNA expression throughout the murine cortical wall at E14.5 [34] | |||
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ting, J.T.; Peca, J.; Feng, G. Functional consequences of mutations in postsynaptic scaffolding proteins and relevance to psychiatric disorders. Annu. Rev. Neurosci. 2012, 35, 49–71. [Google Scholar] [CrossRef]
- Soler, J.; Fananas, L.; Parellada, M.; Krebs, M.O.; Rouleau, G.A.; Fatjo-Vilas, M. Genetic variability in scaffolding proteins and risk for schizophrenia and autism-spectrum disorders: A systematic review. J. Psychiatry Neurosci. 2018, 43, 223–244. [Google Scholar] [CrossRef]
- Lee, Y.; Zhang, Y.; Kim, S.; Han, K. Excitatory and inhibitory synaptic dysfunction in mania: An emerging hypothesis from animal model studies. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, A.H.; Rasmussen, H.B.; Silahtaroglu, A. The DLGAP family: Neuronal expression, function and role in brain disorders. Mol. Brain 2017, 10, 43. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, P.; Feng, G. Learning from animal models of obsessive-compulsive disorder. Biol. Psychiatry 2016, 79, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, P.; Feng, G. SHANK proteins: Roles at the synapse and in autism spectrum disorder. Nat. Rev. Neurosci. 2017, 18, 147–157. [Google Scholar] [CrossRef]
- de Bartolomeis, A.; Barone, A.; Buonaguro, E.F.; Tomasetti, C.; Vellucci, L.; Iasevoli, F. The Homer1 family of proteins at the crossroad of dopamine-glutamate signaling: An emerging molecular “Lego” in the pathophysiology of psychiatric disorders. A systematic review and translational insight. Neurosci. Biobehav. Rev. 2022, 136, 104596. [Google Scholar] [CrossRef] [PubMed]
- Sheng, M.; Hoogenraad, C.C. The postsynaptic architecture of excitatory synapses: A more quantitative view. Annu. Rev. Biochem. 2007, 76, 823–847. [Google Scholar] [CrossRef] [PubMed]
- Sheng, M.; Kim, E. The postsynaptic organization of synapses. Cold Spring Harb. Perspect. Biol. 2011, 3, a005678. [Google Scholar] [CrossRef]
- Sinnen, B.L.; Bowen, A.B.; Forte, J.S.; Hiester, B.G.; Crosby, K.C.; Gibson, E.S.; Dell’Acqua, M.L.; Kennedy, M.J. Optogenetic Control of Synaptic Composition and Function. Neuron 2017, 93, 646–660.e645. [Google Scholar] [CrossRef] [PubMed]
- Scannevin, R.H.; Huganir, R.L. Postsynaptic organization and regulation of excitatory synapses. Nat. Rev. Neurosci. 2000, 1, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Kaizuka, T.; Takumi, T. Postsynaptic density proteins and their involvement in neurodevelopmental disorders. J. Biochem. 2018, 163, 447–455. [Google Scholar] [CrossRef]
- Welch, J.M.; Lu, J.; Rodriguiz, R.M.; Trotta, N.C.; Peca, J.; Ding, J.D.; Feliciano, C.; Chen, M.; Adams, J.P.; Luo, J.; et al. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature 2007, 448, 894–900. [Google Scholar] [CrossRef] [PubMed]
- Jiang-Xie, L.F.; Liao, H.M.; Chen, C.H.; Chen, Y.T.; Ho, S.Y.; Lu, D.H.; Lee, L.J.; Liou, H.H.; Fu, W.M.; Gau, S.S. Autism-associated gene Dlgap2 mutant mice demonstrate exacerbated aggressive behaviors and orbitofrontal cortex deficits. Mol. Autism 2014, 5, 32. [Google Scholar] [CrossRef] [PubMed]
- Coba, M.P.; Ramaker, M.J.; Ho, E.V.; Thompson, S.L.; Komiyama, N.H.; Grant, S.G.N.; Knowles, J.A.; Dulawa, S.C. Dlgap1 knockout mice exhibit alterations of the postsynaptic density and selective reductions in sociability. Sci. Rep. 2018, 8, 2281. [Google Scholar] [CrossRef] [PubMed]
- Schob, C.; Morellini, F.; Ohana, O.; Bakota, L.; Hrynchak, M.V.; Brandt, R.; Brockmann, M.D.; Cichon, N.; Hartung, H.; Hanganu-Opatz, I.L.; et al. Cognitive impairment and autistic-like behaviour in SAPAP4-deficient mice. Transl. Psychiatry 2019, 9, 7. [Google Scholar] [CrossRef]
- Wang, T.; Bai, Y.; Zheng, X.; Liu, X.; Xing, S.; Wang, L.; Wang, H.; Feng, G.; Li, C. Sapap4 deficiency leads to postsynaptic defects and abnormal behaviors relevant to hyperkinetic neuropsychiatric disorder in mice. Cereb Cortex 2022. [Google Scholar] [CrossRef]
- Welch, J.M.; Wang, D.; Feng, G. Differential mRNA expression and protein localization of the SAP90/PSD-95-associated proteins (SAPAPs) in the nervous system of the mouse. J. Comp. Neurol. 2004, 472, 24–39. [Google Scholar] [CrossRef]
- Kindler, S.; Rehbein, M.; Classen, B.; Richter, D.; Bockers, T.M. Distinct spatiotemporal expression of SAPAP transcripts in the developing rat brain: A novel dendritically localized mRNA. Brain Res. Mol. Brain Res. 2004, 126, 14–21. [Google Scholar] [CrossRef]
- Li, J.M.; Lu, C.L.; Cheng, M.C.; Luu, S.U.; Hsu, S.H.; Hu, T.M.; Tsai, H.Y.; Chen, C.H. Role of the DLGAP2 gene encoding the SAP90/PSD-95-associated protein 2 in schizophrenia. PLoS ONE 2014, 9, e85373. [Google Scholar] [CrossRef]
- Meng, W.; Sjoholm, L.K.; Kononenko, O.; Tay, N.; Zhang, D.; Sarkisyan, D.; Geske, J.R.; Ing, A.; Qiu, W.; Watanabe, H.; et al. Genotype-dependent epigenetic regulation of DLGAP2 in alcohol use and dependence. Mol. Psychiatry 2021, 26, 4367–4382. [Google Scholar] [CrossRef] [PubMed]
- Ouellette, A.R.; Neuner, S.M.; Dumitrescu, L.; Anderson, L.C.; Gatti, D.M.; Mahoney, E.R.; Bubier, J.A.; Churchill, G.; Peters, L.; Huentelman, M.J.; et al. Cross-species analyses identify dlgap2 as a regulator of age-related cognitive decline and alzheimer’s dementia. Cell Rep. 2020, 32, 108091. [Google Scholar] [CrossRef] [PubMed]
- Minocherhomji, S.; Hansen, C.; Kim, H.G.; Mang, Y.; Bak, M.; Guldberg, P.; Papadopoulos, N.; Eiberg, H.; Doh, G.D.; Mollgard, K.; et al. Epigenetic remodelling and dysregulation of DLGAP4 is linked with early-onset cerebellar ataxia. Hum. Mol. Genet. 2014, 23, 6163–6176. [Google Scholar] [CrossRef] [PubMed]
- Kirov, G.; Pocklington, A.J.; Holmans, P.; Ivanov, D.; Ikeda, M.; Ruderfer, D.; Moran, J.; Chambert, K.; Toncheva, D.; Georgieva, L.; et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol. Psychiatry 2012, 17, 142–153. [Google Scholar] [CrossRef]
- Stewart, S.E.; Yu, D.; Scharf, J.M.; Neale, B.M.; Fagerness, J.A.; Mathews, C.A.; Arnold, P.D.; Evans, P.D.; Gamazon, E.R.; Davis, L.K.; et al. Genome-wide association study of obsessive-compulsive disorder. Mol. Psychiatry 2013, 18, 788–798. [Google Scholar] [CrossRef]
- Gazzellone, M.J.; Zarrei, M.; Burton, C.L.; Walker, S.; Uddin, M.; Shaheen, S.M.; Coste, J.; Rajendram, R.; Schachter, R.J.; Colasanto, M.; et al. Uncovering obsessive-compulsive disorder risk genes in a pediatric cohort by high-resolution analysis of copy number variation. J. Neurodev. Disord. 2016, 8, 36. [Google Scholar] [CrossRef]
- Fan, Z.; Qian, Y.; Lu, Q.; Wang, Y.; Chang, S.; Yang, L. DLGAP1 and NMDA receptor-associated postsynaptic density protein genes influence executive function in attention deficit hyperactivity disorder. Brain Behav. 2018, 8, e00914. [Google Scholar] [CrossRef]
- Katsumata, Y.; Nelson, P.T.; Estus, S.; Alzheimer’s Disease Neuroimaging, I.; Fardo, D.W. Translating Alzheimer’s disease-associated polymorphisms into functional candidates: A survey of IGAP genes and SNPs. Neurobiol. Aging 2019, 74, 135–146. [Google Scholar] [CrossRef]
- Pinto, D.; Pagnamenta, A.T.; Klei, L.; Anney, R.; Merico, D.; Regan, R.; Conroy, J.; Magalhaes, T.R.; Correia, C.; Abrahams, B.S.; et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 2010, 466, 368–372. [Google Scholar] [CrossRef]
- Iossifov, I.; O’Roak, B.J.; Sanders, S.J.; Ronemus, M.; Krumm, N.; Levy, D.; Stessman, H.A.; Witherspoon, K.T.; Vives, L.; Patterson, K.E.; et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014, 515, 216–221. [Google Scholar] [CrossRef]
- Bienvenu, O.J.; Wang, Y.; Shugart, Y.Y.; Welch, J.M.; Grados, M.A.; Fyer, A.J.; Rauch, S.L.; McCracken, J.T.; Rasmussen, S.A.; Murphy, D.L.; et al. Sapap3 and pathological grooming in humans: Results from the OCD collaborative genetics study. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009, 150B, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Boardman, L.; van der Merwe, L.; Lochner, C.; Kinnear, C.J.; Seedat, S.; Stein, D.J.; Moolman-Smook, J.C.; Hemmings, S.M. Investigating SAPAP3 variants in the etiology of obsessive-compulsive disorder and trichotillomania in the South African white population. Compr. Psychiatry 2011, 52, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Crane, J.; Fagerness, J.; Osiecki, L.; Gunnell, B.; Stewart, S.E.; Pauls, D.L.; Scharf, J.M.; Tourette Syndrome International Consortium for, G. Family-based genetic association study of DLGAP3 in Tourette Syndrome. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2011, 156B, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.M.; Poirier, K.; Belvindrah, R.; Moutkine, I.; Houllier, A.; LeMoing, A.G.; Petit, F.; Boland, A.; Collins, S.C.; Soiza-Reilly, M.; et al. Novel role of the synaptic scaffold protein Dlgap4 in ventricular surface integrity and neuronal migration during cortical development. Nat. Commun. 2022, 13, 2746. [Google Scholar] [CrossRef]
- Wu, B.; Li, C.; Lei, H. SAPAP4 deletion causes synaptic dysfunction in the nucleus accumbens. Biochem. Biophys. Res. Commun. 2018, 505, 1223–1227. [Google Scholar] [CrossRef]
- Kim, E.; Naisbitt, S.; Hsueh, Y.P.; Rao, A.; Rothschild, A.; Craig, A.M.; Sheng, M. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J. Cell Biol. 1997, 136, 669–678. [Google Scholar] [CrossRef]
- Takeuchi, M.; Hata, Y.; Hirao, K.; Toyoda, A.; Irie, M.; Takai, Y. SAPAPs. A family of PSD-95/SAP90-associated proteins localized at postsynaptic density. J. Biol. Chem. 1997, 272, 11943–11951. [Google Scholar] [CrossRef]
- Naisbitt, S.; Valtschanoff, J.; Allison, D.W.; Sala, C.; Kim, E.; Craig, A.M.; Weinberg, R.J.; Sheng, M. Interaction of the postsynaptic density-95/guanylate kinase domain-associated protein complex with a light chain of myosin-V and dynein. J. Neurosci. 2000, 20, 4524–4534. [Google Scholar] [CrossRef]
- Irie, M.; Hata, Y.; Takeuchi, M.; Ichtchenko, K.; Toyoda, A.; Hirao, K.; Takai, Y.; Rosahl, T.W.; Sudhof, T.C. Binding of neuroligins to PSD-95. Science 1997, 277, 1511–1515. [Google Scholar] [CrossRef]
- Mondin, M.; Labrousse, V.; Hosy, E.; Heine, M.; Tessier, B.; Levet, F.; Poujol, C.; Blanchet, C.; Choquet, D.; Thoumine, O. Neurexin-neuroligin adhesions capture surface-diffusing AMPA receptors through PSD-95 scaffolds. J. Neurosci. 2011, 31, 13500–13515. [Google Scholar] [CrossRef]
- Chen, L.; Chetkovich, D.M.; Petralia, R.S.; Sweeney, N.T.; Kawasaki, Y.; Wenthold, R.J.; Bredt, D.S.; Nicoll, R.A. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 2000, 408, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Bats, C.; Groc, L.; Choquet, D. The interaction between Stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron 2007, 53, 719–734. [Google Scholar] [CrossRef] [PubMed]
- Dakoji, S.; Tomita, S.; Karimzadegan, S.; Nicoll, R.A.; Bredt, D.S. Interaction of transmembrane AMPA receptor regulatory proteins with multiple membrane associated guanylate kinases. Neuropharmacology 2003, 45, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Schwenk, J.; Harmel, N.; Brechet, A.; Zolles, G.; Berkefeld, H.; Muller, C.S.; Bildl, W.; Baehrens, D.; Huber, B.; Kulik, A.; et al. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron 2012, 74, 621–633. [Google Scholar] [CrossRef]
- Elias, G.M.; Elias, L.A.; Apostolides, P.F.; Kriegstein, A.R.; Nicoll, R.A. Differential trafficking of AMPA and NMDA receptors by SAP102 and PSD-95 underlies synapse development. Proc. Natl. Acad. Sci. USA 2008, 105, 20953–20958. [Google Scholar] [CrossRef]
- Bissen, D.; Foss, F.; Acker-Palmer, A. AMPA receptors and their minions: Auxiliary proteins in AMPA receptor trafficking. Cell Mol. Life Sci. 2019, 76, 2133–2169. [Google Scholar] [CrossRef]
- Kim, J.H.; Liao, D.; Lau, L.F.; Huganir, R.L. SynGAP: A synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron 1998, 20, 683–691. [Google Scholar] [CrossRef]
- Brenman, J.E.; Chao, D.S.; Gee, S.H.; McGee, A.W.; Craven, S.E.; Santillano, D.R.; Wu, Z.; Huang, F.; Xia, H.; Peters, M.F.; et al. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell 1996, 84, 757–767. [Google Scholar] [CrossRef]
- Garcia, E.P.; Mehta, S.; Blair, L.A.; Wells, D.G.; Shang, J.; Fukushima, T.; Fallon, J.R.; Garner, C.C.; Marshall, J. SAP90 binds and clusters kainate receptors causing incomplete desensitization. Neuron 1998, 21, 727–739. [Google Scholar] [CrossRef]
- Hu, T.M.; Wu, C.L.; Hsu, S.H.; Tsai, H.Y.; Cheng, F.Y.; Cheng, M.C. Ultrarare loss-of-function mutations in the genes encoding the ionotropic glutamate receptors of kainate subtypes associated with schizophrenia disrupt the interaction with PSD95. J. Pers. Med. 2022, 12, 783. [Google Scholar] [CrossRef]
- Hirao, K.; Hata, Y.; Ide, N.; Takeuchi, M.; Irie, M.; Yao, I.; Deguchi, M.; Toyoda, A.; Sudhof, T.C.; Takai, Y. A novel multiple PDZ domain-containing molecule interacting with N-methyl-D-aspartate receptors and neuronal cell adhesion proteins. J. Biol. Chem. 1998, 273, 21105–21110. [Google Scholar] [CrossRef] [PubMed]
- Sumita, K.; Sato, Y.; Iida, J.; Kawata, A.; Hamano, M.; Hirabayashi, S.; Ohno, K.; Peles, E.; Hata, Y. Synaptic scaffolding molecule (S-SCAM) membrane-associated guanylate kinase with inverted organization (MAGI)-2 is associated with cell adhesion molecules at inhibitory synapses in rat hippocampal neurons. J. Neurochem. 2007, 100, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Kitano, J.; Yamazaki, Y.; Kimura, K.; Masukado, T.; Nakajima, Y.; Nakanishi, S. Tamalin is a scaffold protein that interacts with multiple neuronal proteins in distinct modes of protein-protein association. J. Biol. Chem. 2003, 278, 14762–14768. [Google Scholar] [CrossRef] [PubMed]
- Hirao, K.; Hata, Y.; Deguchi, M.; Yao, I.; Ogura, M.; Rokukawa, C.; Kawabe, H.; Mizoguchi, A.; Takai, Y. Association of synapse-associated protein 90/ postsynaptic density-95-associated protein (SAPAP) with neurofilaments. Genes Cells 2000, 5, 203–210. [Google Scholar] [CrossRef]
- Naisbitt, S.; Kim, E.; Tu, J.C.; Xiao, B.; Sala, C.; Valtschanoff, J.; Weinberg, R.J.; Worley, P.F.; Sheng, M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron 1999, 23, 569–582. [Google Scholar] [CrossRef]
- Yao, I.; Hata, Y.; Hirao, K.; Deguchi, M.; Ide, N.; Takeuchi, M.; Takai, Y. Synamon, a novel neuronal protein interacting with synapse-associated protein 90/postsynaptic density-95-associated protein. J. Biol. Chem. 1999, 274, 27463–27466. [Google Scholar] [CrossRef]
- Jung, S.; Park, M. Shank postsynaptic scaffolding proteins in autism spectrum disorder: Mouse models and their dysfunctions in behaviors, synapses, and molecules. Pharm. Res. 2022, 182, 106340. [Google Scholar] [CrossRef]
- Du, Y.; Weed, S.A.; Xiong, W.C.; Marshall, T.D.; Parsons, J.T. Identification of a novel cortactin SH3 domain-binding protein and its localization to growth cones of cultured neurons. Mol. Cell. Biol. 1998, 18, 5838–5851. [Google Scholar] [CrossRef]
- Kim, E.; Sheng, M. PDZ domain proteins of synapses. Nat. Rev. Neurosci. 2004, 5, 771–781. [Google Scholar] [CrossRef]
- Zeng, M.; Shang, Y.; Guo, T.; He, Q.; Yung, W.H.; Liu, K.; Zhang, M. A binding site outside the canonical PDZ domain determines the specific interaction between Shank and SAPAP and their function. Proc. Natl. Acad. Sci. USA 2016, 113, E3081–E3090. [Google Scholar] [CrossRef]
- Li, J.; Zhang, W.; Yang, H.; Howrigan, D.P.; Wilkinson, B.; Souaiaia, T.; Evgrafov, O.V.; Genovese, G.; Clementel, V.A.; Tudor, J.C.; et al. Spatiotemporal profile of postsynaptic interactomes integrates components of complex brain disorders. Nat. Neurosci. 2017, 20, 1150–1161. [Google Scholar] [CrossRef] [PubMed]
- Kawabe, H.; Hata, Y.; Takeuchi, M.; Ide, N.; Mizoguchi, A.; Takai, Y. nArgBP2, a novel neural member of ponsin/ArgBP2/vinexin family that interacts with synapse-associated protein 90/postsynaptic density-95-associated protein (SAPAP). J. Biol. Chem. 1999, 274, 30914–30918. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, S.E.; Kim, Y.; Han, J.K.; Park, H.; Lee, U.; Na, M.; Jeong, S.; Chung, C.; Cestra, G.; Chang, S. nArgBP2 regulates excitatory synapse formation by controlling dendritic spine morphology. Proc. Natl. Acad. Sci. USA 2016, 113, 6749–6754. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Gao, X.; Li, C.; Feliciano, C.; Wang, D.; Zhou, D.; Mei, Y.; Monteiro, P.; Anand, M.; Itohara, S.; et al. Impaired dendritic development and memory in sorbs2 knock-out mice. J. Neurosci. 2016, 36, 2247–2260. [Google Scholar] [CrossRef]
- Cestra, G.; Toomre, D.; Chang, S.; De Camilli, P. The Abl/Arg substrate ArgBP2/nArgBP2 coordinates the function of multiple regulatory mechanisms converging on the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 2005, 102, 1731–1736. [Google Scholar] [CrossRef]
- Rodriguez-Crespo, I.; Yelamos, B.; Roncal, F.; Albar, J.P.; Ortiz de Montellano, P.R.; Gavilanes, F. Identification of novel cellular proteins that bind to the LC8 dynein light chain using a pepscan technique. FEBS Lett. 2001, 503, 135–141. [Google Scholar] [CrossRef]
- Haraguchi, K.; Satoh, K.; Yanai, H.; Hamada, F.; Kawabuchi, M.; Akiyama, T. The hDLG-associated protein DAP interacts with dynein light chain and neuronal nitric oxide synthase. Genes Cells 2000, 5, 905–911. [Google Scholar] [CrossRef]
- Tong, J.; Yang, H.; Eom, S.H.; Chun, C.; Im, Y.J. Structure of the GH1 domain of guanylate kinase-associated protein from Rattus norvegicus. Biochem. Biophys. Res. Commun. 2014, 452, 130–135. [Google Scholar] [CrossRef]
- Morris, C.W.; Watkins, D.S.; Salek, A.B.; Edler, M.C.; Baucum, A.J., 2nd. The association of spinophilin with disks large-associated protein 3 (SAPAP3) is regulated by metabotropic glutamate receptor (mGluR) 5. Mol. Cell. Neurosci. 2018, 90, 60–69. [Google Scholar] [CrossRef]
- Roselli, F.; Livrea, P.; Almeida, O.F. CDK5 is essential for soluble amyloid beta-induced degradation of GKAP and remodeling of the synaptic actin cytoskeleton. PLoS ONE 2011, 6, e23097. [Google Scholar] [CrossRef]
- Chua, J.J.; Schob, C.; Rehbein, M.; Gkogkas, C.G.; Richter, D.; Kindler, S. Synthesis of two SAPAP3 isoforms from a single mRNA is mediated via alternative translational initiation. Sci. Rep. 2012, 2, 484. [Google Scholar] [CrossRef]
- Polioudakis, D.; de la Torre-Ubieta, L.; Langerman, J.; Elkins, A.G.; Shi, X.; Stein, J.L.; Vuong, C.K.; Nichterwitz, S.; Gevorgian, M.; Opland, C.K.; et al. A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron 2019, 103, 785–801.e788. [Google Scholar] [CrossRef]
- Hanus, C.; Schuman, E.M. Proteostasis in complex dendrites. Nat. Rev. Neurosci. 2013, 14, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Cajigas, I.J.; Tushev, G.; Will, T.J.; tom Dieck, S.; Fuerst, N.; Schuman, E.M. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 2012, 74, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Schuman, E.M. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science 1996, 273, 1402–1406. [Google Scholar] [CrossRef] [PubMed]
- Aakalu, G.; Smith, W.B.; Nguyen, N.; Jiang, C.; Schuman, E.M. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 2001, 30, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Hegde, A.N.; Goldberg, A.L.; Schwartz, J.H. Regulatory subunits of cAMP-dependent protein kinases are degraded after conjugation to ubiquitin: A molecular mechanism underlying long-term synaptic plasticity. Proc. Natl. Acad. Sci. USA 1993, 90, 7436–7440. [Google Scholar] [CrossRef]
- Steward, O.; Schuman, E.M. Compartmentalized synthesis and degradation of proteins in neurons. Neuron 2003, 40, 347–359. [Google Scholar] [CrossRef]
- Cajigas, I.J.; Will, T.; Schuman, E.M. Protein homeostasis and synaptic plasticity. EMBO J. 2010, 29, 2746–2752. [Google Scholar] [CrossRef]
- Mabb, A.M.; Ehlers, M.D. Ubiquitination in postsynaptic function and plasticity. Annu. Rev. Cell Dev. Biol. 2010, 26, 179–210. [Google Scholar] [CrossRef]
- Ehlers, M.D. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat. Neurosci. 2003, 6, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Ade, K.K.; Caffall, Z.; Ilcim Ozlu, M.; Eroglu, C.; Feng, G.; Calakos, N. Circuit-selective striatal synaptic dysfunction in the Sapap3 knockout mouse model of obsessive-compulsive disorder. Biol. Psychiatry 2014, 75, 623–630. [Google Scholar] [CrossRef]
- Kim, E.; Niethammer, M.; Rothschild, A.; Jan, Y.N.; Sheng, M. Clustering of shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature 1995, 378, 85. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Shang, Y.; Zhang, M. Mechanistic basis of MAGUK-organized complexes in synaptic development and signalling. Nat. Rev. Neurosci. 2016, 17, 209–223. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.O.; Yu, L.; Coba, M.P.; Husi, H.; Campuzano, I.; Blackstock, W.P.; Choudhary, J.S.; Grant, S.G. Proteomic analysis of in vivo phosphorylated synaptic proteins. J. Biol. Chem. 2005, 280, 5972–5982. [Google Scholar] [CrossRef]
- Shin, S.M.; Zhang, N.; Hansen, J.; Gerges, N.Z.; Pak, D.T.; Sheng, M.; Lee, S.H. GKAP orchestrates activity-dependent postsynaptic protein remodeling and homeostatic scaling. Nat. Neurosci. 2012, 15, 1655–1666. [Google Scholar] [CrossRef]
- Li, J.; Wilkinson, B.; Clementel, V.A.; Hou, J.; O’Dell, T.J.; Coba, M.P. Long-term potentiation modulates synaptic phosphorylation networks and reshapes the structure of the postsynaptic interactome. Sci. Signal 2016, 9, rs8. [Google Scholar] [CrossRef]
- Zhu, J.; Zhou, Q.; Shang, Y.; Li, H.; Peng, M.; Ke, X.; Weng, Z.; Zhang, R.; Huang, X.; Li, S.S.C.; et al. Synaptic targeting and function of SAPAPs mediated by phosphorylation-dependent binding to PSD-95 MAGUKs. Cell Rep. 2017, 21, 3781–3793. [Google Scholar] [CrossRef]
- Hung, A.Y.; Sung, C.C.; Brito, I.L.; Sheng, M. Degradation of postsynaptic scaffold GKAP and regulation of dendritic spine morphology by the TRIM3 ubiquitin ligase in rat hippocampal neurons. PLoS ONE 2010, 5, e9842. [Google Scholar] [CrossRef]
- Rao, A.; Kim, E.; Sheng, M.; Craig, A.M. Heterogeneity in the molecular composition of excitatory postsynaptic sites during development of hippocampal neurons in culture. J. Neurosci. 1998, 18, 1217–1229. [Google Scholar] [CrossRef]
- Yao, I.; Iida, J.; Nishimura, W.; Hata, Y. Synaptic localization of SAPAP1, a synaptic membrane-associated protein. Genes Cells 2003, 8, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Romorini, S.; Piccoli, G.; Jiang, M.; Grossano, P.; Tonna, N.; Passafaro, M.; Zhang, M.; Sala, C. A functional role of postsynaptic density-95-guanylate kinase-associated protein complex in regulating Shank assembly and stability to synapses. J. Neurosci. 2004, 24, 9391–9404. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moutin, E.; Raynaud, F.; Fagni, L.; Perroy, J. GKAP-DLC2 interaction organizes the postsynaptic scaffold complex to enhance synaptic NMDA receptor activity. J. Cell Sci. 2012, 125, 2030–2040. [Google Scholar] [CrossRef] [PubMed]
- Moutin, E.; Compan, V.; Raynaud, F.; Clerte, C.; Bouquier, N.; Labesse, G.; Ferguson, M.L.; Fagni, L.; Royer, C.A.; Perroy, J. The stoichiometry of scaffold complexes in living neurons-DLC2 functions as a dimerization engine for GKAP. J. Cell Sci. 2014, 127, 3451–3462. [Google Scholar] [CrossRef]
- Wan, Y.; Feng, G.; Calakos, N. Sapap3 deletion causes mGluR5-dependent silencing of AMPAR synapses. J. Neurosci. 2011, 31, 16685–16691. [Google Scholar] [CrossRef]
- Lousada, E.; Boudreau, M.; Cohen-Adad, J.; Nait Oumesmar, B.; Burguiere, E.; Schreiweis, C. Reduced axon calibre in the associative striatum of the sapap3 knockout mouse. Brain Sci. 2021, 11, 1353. [Google Scholar] [CrossRef]
- Joglekar, A.; Prjibelski, A.; Mahfouz, A.; Collier, P.; Lin, S.; Schlusche, A.K.; Marrocco, J.; Williams, S.R.; Haase, B.; Hayes, A.; et al. A spatially resolved brain region- and cell type-specific isoform atlas of the postnatal mouse brain. Nat. Commun. 2021, 12, 463. [Google Scholar] [CrossRef]
- Turrigiano, G. Homeostatic synaptic plasticity: Local and global mechanisms for stabilizing neuronal function. Cold Spring Harb. Perspect. Biol. 2012, 4, a005736. [Google Scholar] [CrossRef]
- Chen, M.; Wan, Y.; Ade, K.; Ting, J.; Feng, G.; Calakos, N. Sapap3 deletion anomalously activates short-term endocannabinoid-mediated synaptic plasticity. J. Neurosci. 2011, 31, 9563–9573. [Google Scholar] [CrossRef]
- Hung, A.Y.; Futai, K.; Sala, C.; Valtschanoff, J.G.; Ryu, J.; Woodworth, M.A.; Kidd, F.L.; Sung, C.C.; Miyakawa, T.; Bear, M.F.; et al. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J. Neurosci. 2008, 28, 1697–1708. [Google Scholar] [CrossRef]
- Ade, K.K.; Wan, Y.; Hamann, H.C.; O’Hare, J.K.; Guo, W.; Quian, A.; Kumar, S.; Bhagat, S.; Rodriguiz, R.M.; Wetsel, W.C.; et al. Increased metabotropic glutamate receptor 5 signaling underlies obsessive-compulsive disorder-like behavioral and striatal circuit abnormalities in mice. Biol. Psychiatry 2016, 80, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Burguiere, E.; Monteiro, P.; Feng, G.; Graybiel, A.M. Optogenetic stimulation of lateral orbitofronto-striatal pathway suppresses compulsive behaviors. Science 2013, 340, 1243–1246. [Google Scholar] [CrossRef] [PubMed]
- Corbit, V.L.; Manning, E.E.; Gittis, A.H.; Ahmari, S.E. Strengthened inputs from secondary motor cortex to striatum in a mouse model of compulsive behavior. J. Neurosci. 2019, 39, 2965–2975. [Google Scholar] [CrossRef] [PubMed]
- Hadjas, L.C.; Schartner, M.M.; Cand, J.; Creed, M.C.; Pascoli, V.; Luscher, C.; Simmler, L.D. Projection-specific deficits in synaptic transmission in adult Sapap3-knockout mice. Neuropsychopharmacology 2020, 45, 2020–2029. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Armenta, K.I.; Alatriste-Leon, H.; Verma-Rodriguez, A.K.; Llanos-Moreno, A.; Ramirez-Jarquin, J.O.; Tecuapetla, F. Optogenetic inhibition of indirect pathway neurons in the dorsomedial striatum reduces excessive grooming in Sapap3-knockout mice. Neuropsychopharmacology 2022, 47, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wu, G.; Liu, M.; Sun, X.; Xu, Q.; Zhang, C.; Lei, H. Dysfunction of orbitofrontal GABAergic interneurons leads to impaired reversal learning in a mouse model of obsessive-compulsive disorder. Curr. Biol. 2021, 31, 381–393.e384. [Google Scholar] [CrossRef]
- Davis, G.L.; Minerva, A.R.; Lario, A.; Simmler, L.D.; Rodriguez, C.I.; Gunaydin, L.A. Ketamine increases activity of a fronto-striatal projection that regulates compulsive behavior in SAPAP3 knockout mice. Nat. Commun. 2021, 12, 6040. [Google Scholar] [CrossRef]
- Wood, J.; LaPalombara, Z.; Ahmari, S.E. Monoamine abnormalities in the SAPAP3 knockout model of obsessive-compulsive disorder-related behaviour. Philos. Trans. R Soc. Lond. B Biol. Sci. 2018, 373, 20170023. [Google Scholar] [CrossRef]
- Manning, E.E.; Wang, A.Y.; Saikali, L.M.; Winner, A.S.; Ahmari, S.E. Disruption of prepulse inhibition is associated with compulsive behavior severity and nucleus accumbens dopamine receptor changes in Sapap3 knockout mice. Sci. Rep. 2021, 11, 9442. [Google Scholar] [CrossRef]
- Tomasetti, C.; Iasevoli, F.; Buonaguro, E.F.; De Berardis, D.; Fornaro, M.; Fiengo, A.L.; Martinotti, G.; Orsolini, L.; Valchera, A.; Di Giannantonio, M.; et al. Treating the synapse in major psychiatric disorders: The role of postsynaptic density network in dopamine-glutamate interplay and psychopharmacologic drugs molecular actions. Int. J. Mol. Sci. 2017, 18, 135. [Google Scholar] [CrossRef]
- Zai, G.; Barta, C.; Cath, D.; Eapen, V.; Geller, D.; Grunblatt, E. New insights and perspectives on the genetics of obsessive-compulsive disorder. Psychiatr. Genet. 2019, 29, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Chertkow-Deutsher, Y.; Cohen, H.; Klein, E.; Ben-Shachar, D. DNA methylation in vulnerability to post-traumatic stress in rats: Evidence for the role of the post-synaptic density protein Dlgap2. Int. J. Neuropsychopharmacol. 2010, 13, 347–359. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, Y.; Wang, K.; Zhang, P.; Huang, J.; An, H.; Wang, N.; De Yang, F.; Wang, Z.; Tan, S.; Chen, S.; et al. Quantitative DNA methylation analysis of DLGAP2 gene using pyrosequencing in schizophrenia with tardive dyskinesia: A linear mixed model approach. Sci. Rep. 2018, 8, 17466. [Google Scholar] [CrossRef] [PubMed]
- Schrott, R.; Acharya, K.; Itchon-Ramos, N.; Hawkey, A.B.; Pippen, E.; Mitchell, J.T.; Kollins, S.H.; Levin, E.D.; Murphy, S.K. Cannabis use is associated with potentially heritable widespread changes in autism candidate gene DLGAP2 DNA methylation in sperm. Epigenetics 2020, 15, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Kajimoto, Y.; Shirakawa, O.; Lin, X.H.; Hashimoto, T.; Kitamura, N.; Murakami, N.; Takumi, T.; Maeda, K. Synapse-associated protein 90/postsynaptic density-95-associated protein (SAPAP) is expressed differentially in phencyclidine-treated rats and is increased in the nucleus accumbens of patients with schizophrenia. Neuropsychopharmacology 2003, 28, 1831–1839. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, J.; Yan, Y.; Gu, Y.; Ma, Y.; Wang, M.; Zhang, H.; Tao, K.; Lu, Y.; Yu, W.; et al. SAPAP3 regulates epileptic seizures involving GluN2A in post-synaptic densities. Cell Death Dis. 2022, 13, 437. [Google Scholar] [CrossRef]
- Schutt, J.; Falley, K.; Richter, D.; Kreienkamp, H.J.; Kindler, S. Fragile X mental retardation protein regulates the levels of scaffold proteins and glutamate receptors in postsynaptic densities. J. Biol. Chem. 2009, 284, 25479–25487. [Google Scholar] [CrossRef]
- Krueger, D.D.; Osterweil, E.K.; Chen, S.P.; Tye, L.D.; Bear, M.F. Cognitive dysfunction and prefrontal synaptic abnormalities in a mouse model of fragile X syndrome. Proc. Natl. Acad. Sci. USA 2011, 108, 2587–2592. [Google Scholar] [CrossRef]
- Been, L.E.; Moore, K.M.; Kennedy, B.C.; Meisel, R.L. Metabotropic glutamate receptor and fragile X signaling in a female model of escalated aggression. Biol. Psychiatry 2016, 79, 685–692. [Google Scholar] [CrossRef][Green Version]
- Brown, V.; Jin, P.; Ceman, S.; Darnell, J.C.; O’Donnell, W.T.; Tenenbaum, S.A.; Jin, X.; Feng, Y.; Wilkinson, K.D.; Keene, J.D.; et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 2001, 107, 477–487. [Google Scholar] [CrossRef]
- Darnell, J.C.; Klann, E. The translation of translational control by FMRP: Therapeutic targets for FXS. Nat. Neurosci. 2013, 16, 1530–1536. [Google Scholar] [CrossRef] [PubMed]
- Bassell, G.J.; Warren, S.T. Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron 2008, 60, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Dictenberg, J.B.; Swanger, S.A.; Antar, L.N.; Singer, R.H.; Bassell, G.J. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell 2008, 14, 926–939. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, M.; Nalavadi, V.; Epstein, M.P.; Narayanan, U.; Bassell, G.J.; Warren, S.T. Fragile X mental retardation protein deficiency leads to excessive mGluR5-dependent internalization of AMPA receptors. Proc. Natl. Acad. Sci. USA 2007, 104, 15537–15542. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bey, A.L.; Katz, B.M.; Badea, A.; Kim, N.; David, L.K.; Duffney, L.J.; Kumar, S.; Mague, S.D.; Hulbert, S.W.; et al. Altered mGluR5-Homer scaffolds and corticostriatal connectivity in a Shank3 complete knockout model of autism. Nat. Commun. 2016, 7, 11459. [Google Scholar] [CrossRef]
- Elkins, R.L.; Orr, T.E.; Rausch, J.L.; Fei, Y.J.; Carl, G.F.; Hobbs, S.H.; Buccafusco, J.J.; Edwards, G.L. Cocaine-induced expression differences in PSD-95/SAP-90-associated protein 4 and in Ca2+/calmodulin-dependent protein kinase subunits in amygdalae of taste aversion-prone and taste aversion-resistant rats. Ann. N. Y. Acad. Sci. 2003, 1003, 386–390. [Google Scholar] [CrossRef]
- Kim, Y.; Zhang, Y.; Pang, K.; Kang, H.; Park, H.; Lee, Y.; Lee, B.; Lee, H.J.; Kim, W.K.; Geum, D.; et al. Bipolar disorder associated microRNA, miR-1908-5p, regulates the expression of genes functioning in neuronal glutamatergic synapses. Exp. Neurobiol. 2016, 25, 296–306. [Google Scholar] [CrossRef]
- Mahgoub, M.; Adachi, M.; Suzuki, K.; Liu, X.; Kavalali, E.T.; Chahrour, M.H.; Monteggia, L.M. MeCP2 and histone deacetylases 1 and 2 in dorsal striatum collectively suppress repetitive behaviors. Nat. Neurosci. 2016, 19, 1506–1512. [Google Scholar] [CrossRef]
- Saadatmand, F.; Gurdziel, K.; Jackson, L.; Kwabi-Addo, B.; Ruden, D.M. DNA methylation and exposure to violence among African American young adult males. Brain Behav. Immun. Health 2021, 14, 100247. [Google Scholar] [CrossRef]
- Minocherhomji, S.; Seemann, S.; Mang, Y.; El-Schich, Z.; Bak, M.; Hansen, C.; Papadopoulos, N.; Josefsen, K.; Nielsen, H.; Gorodkin, J.; et al. Sequence and expression analysis of gaps in human chromosome 20. Nucleic Acids Res. 2012, 40, 6660–6672. [Google Scholar] [CrossRef]
- Aoyama, S.; Shirakawa, O.; Ono, H.; Hashimoto, T.; Kajimoto, Y.; Maeda, K. Mutation and association analysis of the DAP-1 gene with schizophrenia. Psychiatry Clin. Neurosci. 2003, 57, 545–547. [Google Scholar] [CrossRef] [PubMed]
- Li, J.M.; Lu, C.L.; Cheng, M.C.; Luu, S.U.; Hsu, S.H.; Chen, C.H. Genetic analysis of the DLGAP1 gene as a candidate gene for schizophrenia. Psychiatry Res. 2013, 205, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shi, M.; Ma, Z.; Zhao, S.; Euskirchen, G.; Ziskin, J.; Urban, A.; Hallmayer, J.; Snyder, M. Integrated systems analysis reveals a molecular network underlying autism spectrum disorders. Mol. Syst. Biol. 2014, 10, 774. [Google Scholar] [CrossRef] [PubMed]
- Mathias, S.R.; Knowles, E.E.; Kent, J.W., Jr.; McKay, D.R.; Curran, J.E.; de Almeida, M.A.; Dyer, T.D.; Goring, H.H.; Olvera, R.L.; Duggirala, R.; et al. Recurrent major depression and right hippocampal volume: A bivariate linkage and association study. Hum. Brain Mapp. 2016, 37, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, O.; Meera, P.; Ghosh, S.; Kubendran, S.; Kiran, K.; Manjunath, K.R.; Subhash, M.N.; Benegal, V.; Brahmachari, S.K.; Majumder, P.P.; et al. Evidence of linkage and association on 18p11.2 for psychosis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006, 141B, 868–873. [Google Scholar] [CrossRef]
- Schwab, S.G.; Hallmayer, J.; Lerer, B.; Albus, M.; Borrmann, M.; Honig, S.; Strauss, M.; Segman, R.; Lichtermann, D.; Knapp, M.; et al. Support for a chromosome 18p locus conferring susceptibility to functional psychoses in families with schizophrenia, by association and linkage analysis. Am. J. Hum. Genet. 1998, 63, 1139–1152. [Google Scholar] [CrossRef][Green Version]
- Chien, W.H.; Gau, S.S.; Liao, H.M.; Chiu, Y.N.; Wu, Y.Y.; Huang, Y.S.; Tsai, W.C.; Tsai, H.M.; Chen, C.H. Deep exon resequencing of DLGAP2 as a candidate gene of autism spectrum disorders. Mol. Autism 2013, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Delaby, E.; Merico, D.; Barbosa, M.; Merikangas, A.; Klei, L.; Thiruvahindrapuram, B.; Xu, X.; Ziman, R.; Wang, Z.; et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 2014, 94, 677–694. [Google Scholar] [CrossRef]
- Autism Genome Project, C.; Szatmari, P.; Paterson, A.D.; Zwaigenbaum, L.; Roberts, W.; Brian, J.; Liu, X.Q.; Vincent, J.B.; Skaug, J.L.; Thompson, A.P.; et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 2007, 39, 319–328. [Google Scholar] [CrossRef]
- Marshall, C.R.; Noor, A.; Vincent, J.B.; Lionel, A.C.; Feuk, L.; Skaug, J.; Shago, M.; Moessner, R.; Pinto, D.; Ren, Y.; et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008, 82, 477–488. [Google Scholar] [CrossRef]
- Woodbury-Smith, M.; Zarrei, M.; Wei, J.; Thiruvahindrapuram, B.; O’Connor, I.; Paterson, A.D.; Yuen, R.K.C.; Dastan, J.; Stavropoulos, D.J.; Howe, J.L.; et al. Segregating patterns of copy number variations in extended autism spectrum disorder (ASD) pedigrees. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2020, 183, 268–276. [Google Scholar] [CrossRef]
- Catusi, I.; Garzo, M.; Capra, A.P.; Briuglia, S.; Baldo, C.; Canevini, M.P.; Cantone, R.; Elia, F.; Forzano, F.; Galesi, O.; et al. 8p23.2-pter microdeletions: Seven new cases narrowing the candidate region and review of the literature. Genes 2021, 12, 652. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Lin, S.; Chen, B.; Zhou, Y. Isolated chromosome 8p23.2pter deletion: Novel evidence for developmental delay, intellectual disability, microcephaly and neurobehavioral disorders. Mol. Med. Rep. 2017, 16, 6837–6845. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Hanna, G.L.; Easter, P.; Kennedy, J.L.; Rosenberg, D.R.; Arnold, P.D. Glutamate system genes and brain volume alterations in pediatric obsessive-compulsive disorder: A preliminary study. Psychiatry Res. 2013, 211, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ji, T.; Wang, J.; Xiao, J.; Wang, H.; Li, J.; Gao, Z.; Yang, Y.; Cai, B.; Wang, L.; et al. Submicroscopic subtelomeric aberrations in Chinese patients with unexplained developmental delay/mental retardation. BMC Med. Genet. 2010, 11, 72. [Google Scholar] [CrossRef]
- Guilmatre, A.; Dubourg, C.; Mosca, A.L.; Legallic, S.; Goldenberg, A.; Drouin-Garraud, V.; Layet, V.; Rosier, A.; Briault, S.; Bonnet-Brilhault, F.; et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch. Gen. Psychiatry 2009, 66, 947–956. [Google Scholar] [CrossRef]
- Purcell, S.M.; Moran, J.L.; Fromer, M.; Ruderfer, D.; Solovieff, N.; Roussos, P.; O’Dushlaine, C.; Chambert, K.; Bergen, S.E.; Kahler, A.; et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014, 506, 185–190. [Google Scholar] [CrossRef]
- Costain, G.; Lionel, A.C.; Merico, D.; Forsythe, P.; Russell, K.; Lowther, C.; Yuen, T.; Husted, J.; Stavropoulos, D.J.; Speevak, M.; et al. Pathogenic rare copy number variants in community-based schizophrenia suggest a potential role for clinical microarrays. Hum. Mol. Genet. 2013, 22, 4485–4501. [Google Scholar] [CrossRef]
- White, C.C.; Yang, H.S.; Yu, L.; Chibnik, L.B.; Dawe, R.J.; Yang, J.; Klein, H.U.; Felsky, D.; Ramos-Miguel, A.; Arfanakis, K.; et al. Identification of genes associated with dissociation of cognitive performance and neuropathological burden: Multistep analysis of genetic, epigenetic, and transcriptional data. PLoS Med. 2017, 14, e1002287. [Google Scholar] [CrossRef]
- Chaudhry, M.; Wang, X.; Bamne, M.N.; Hasnain, S.; Demirci, F.Y.; Lopez, O.L.; Kamboh, M.I. Genetic variation in imprinted genes is associated with risk of late-onset Alzheimer’s disease. J. Alzheimers Dis. 2015, 44, 989–994. [Google Scholar] [CrossRef]
- Chien, W.H.; Gau, S.S.; Wu, Y.Y.; Huang, Y.S.; Fang, J.S.; Chen, Y.J.; Soong, W.T.; Chiu, Y.N.; Chen, C.H. Identification and molecular characterization of two novel chromosomal deletions associated with autism. Clin. Genet. 2010, 78, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Zuchner, S.; Wendland, J.R.; Ashley-Koch, A.E.; Collins, A.L.; Tran-Viet, K.N.; Quinn, K.; Timpano, K.C.; Cuccaro, M.L.; Pericak-Vance, M.A.; Steffens, D.C.; et al. Multiple rare SAPAP3 missense variants in trichotillomania and OCD. Mol. Psychiatry 2009, 14, 6–9. [Google Scholar] [CrossRef]
- Allen-Brady, K.; Miller, J.; Matsunami, N.; Stevens, J.; Block, H.; Farley, M.; Krasny, L.; Pingree, C.; Lainhart, J.; Leppert, M.; et al. A high-density SNP genome-wide linkage scan in a large autism extended pedigree. Mol. Psychiatry 2009, 14, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Pauls, D.L.; Abramovitch, A.; Rauch, S.L.; Geller, D.A. Obsessive-compulsive disorder: An integrative genetic and neurobiological perspective. Nat. Rev. Neurosci. 2014, 15, 410–424. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Holder, J.L., Jr.; Schaaf, C.P.; Lu, H.; Chen, H.; Kang, H.; Tang, J.; Wu, Z.; Hao, S.; Cheung, S.W.; et al. SHANK3 overexpression causes manic-like behaviour with unique pharmacogenetic properties. Nature 2013, 503, 72–77. [Google Scholar] [CrossRef]
- Horner, A.E.; Norris, R.H.; McLaren-Jones, R.; Alexander, L.; Komiyama, N.H.; Grant, S.G.N.; Nithianantharajah, J.; Kopanitsa, M.V. Learning and reaction times in mouse touchscreen tests are differentially impacted by mutations in genes encoding postsynaptic interacting proteins SYNGAP1, NLGN3, DLGAP1, DLGAP2 and SHANK2. Genes Brain Behav. 2021, 20, e12723. [Google Scholar] [CrossRef]
- van den Boom, B.J.G.; Mooij, A.H.; Miseviciute, I.; Denys, D.; Willuhn, I. Behavioral flexibility in a mouse model for obsessive-compulsive disorder: Impaired Pavlovian reversal learning in SAPAP3 mutants. Genes Brain Behav. 2019, 18, e12557. [Google Scholar] [CrossRef]
- Manning, E.E.; Dombrovski, A.Y.; Torregrossa, M.M.; Ahmari, S.E. Impaired instrumental reversal learning is associated with increased medial prefrontal cortex activity in Sapap3 knockout mouse model of compulsive behavior. Neuropsychopharmacology 2019, 44, 1494–1504. [Google Scholar] [CrossRef]
- Hadjas, L.C.; Luscher, C.; Simmler, L.D. Aberrant habit formation in the Sapap3-knockout mouse model of obsessive-compulsive disorder. Sci. Rep. 2019, 9, 12061. [Google Scholar] [CrossRef]
- Ehmer, I.; Crown, L.; van Leeuwen, W.; Feenstra, M.; Willuhn, I.; Denys, D. Evidence for distinct forms of compulsivity in the SAPAP3 mutant-mouse model for obsessive-compulsive disorder. eNeuro 2020, 7. [Google Scholar] [CrossRef]
- Ehmer, I.; Feenstra, M.; Willuhn, I.; Denys, D. Instrumental learning in a mouse model for obsessive-compulsive disorder: Impaired habit formation in Sapap3 mutants. Neurobiol. Learn. Mem. 2020, 168, 107162. [Google Scholar] [CrossRef] [PubMed]
- Benzina, N.; N’Diaye, K.; Pelissolo, A.; Mallet, L.; Burguiere, E. A cross-species assessment of behavioral flexibility in compulsive disorders. Commun. Biol. 2021, 4, 96. [Google Scholar] [CrossRef] [PubMed]
- Manning, E.E.; Geramita, M.A.; Piantadosi, S.C.; Pierson, J.L.; Ahmari, S.E. Distinct patterns of abnormal lateral orbitofrontal cortex activity during compulsive grooming and reversal learning normalize after fluoxetine. Biol. Psychiatry 2021. [Google Scholar] [CrossRef] [PubMed]
- Kajs, B.L.; van Roessel, P.J.; Davis, G.L.; Williams, L.M.; Rodriguez, C.I.; Gunaydin, L.A. Valence processing alterations in SAPAP3 knockout mice and human OCD. J. Psychiatr. Res. 2022, 151, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Pinhal, C.M.; van den Boom, B.J.G.; Santana-Kragelund, F.; Fellinger, L.; Bech, P.; Hamelink, R.; Feng, G.; Willuhn, I.; Feenstra, M.G.P.; Denys, D. Differential effects of deep brain stimulation of the internal capsule and the striatum on excessive grooming in Sapap3 mutant mice. Biol. Psychiatry 2018, 84, 917–925. [Google Scholar] [CrossRef]
- Glorie, D.; Verhaeghe, J.; Miranda, A.; Kertesz, I.; Wyffels, L.; Stroobants, S.; Staelens, S. Progression of obsessive compulsive disorder-like grooming in Sapap3 knockout mice: A longitudinal [(11)C]ABP688 PET study. Neuropharmacology 2020, 177, 108160. [Google Scholar] [CrossRef]
- Mintzopoulos, D.; Gillis, T.E.; Robertson, H.R.; Dalia, T.; Feng, G.; Rauch, S.L.; Kaufman, M.J. Striatal magnetic resonance spectroscopy abnormalities in young adult SAPAP3 knockout mice. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2016, 1, 39–48. [Google Scholar] [CrossRef]
- Glorie, D.; Verhaeghe, J.; Miranda, A.; De Lombaerde, S.; Stroobants, S.; Staelens, S. Sapap3 deletion causes dynamic synaptic density abnormalities: A longitudinal [(11)C]UCB-J PET study in a model of obsessive-compulsive disorder-like behaviour. EJNMMI Res. 2020, 10, 140. [Google Scholar] [CrossRef]
- Xu, P.; Grueter, B.A.; Britt, J.K.; McDaniel, L.; Huntington, P.J.; Hodge, R.; Tran, S.; Mason, B.L.; Lee, C.; Vong, L.; et al. Double deletion of melanocortin 4 receptors and SAPAP3 corrects compulsive behavior and obesity in mice. Proc. Natl. Acad. Sci. USA 2013, 110, 10759–10764. [Google Scholar] [CrossRef]
- Lei, H.; Lai, J.; Sun, X.; Xu, Q.; Feng, G. Lateral orbitofrontal dysfunction in the Sapap3 knockout mouse model of obsessive-compulsive disorder. J. Psychiatry Neurosci. 2019, 44, 120–131. [Google Scholar] [CrossRef]
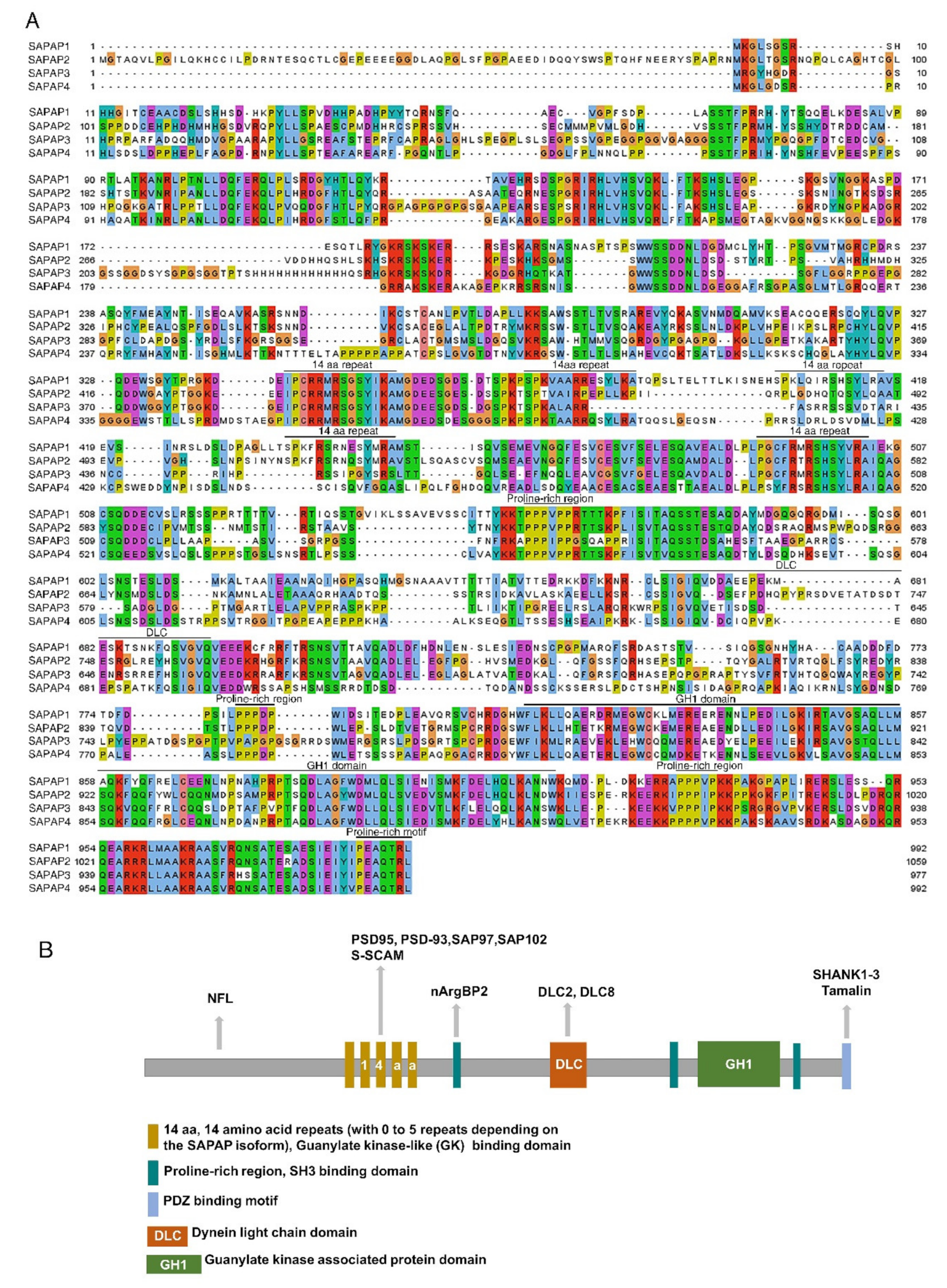
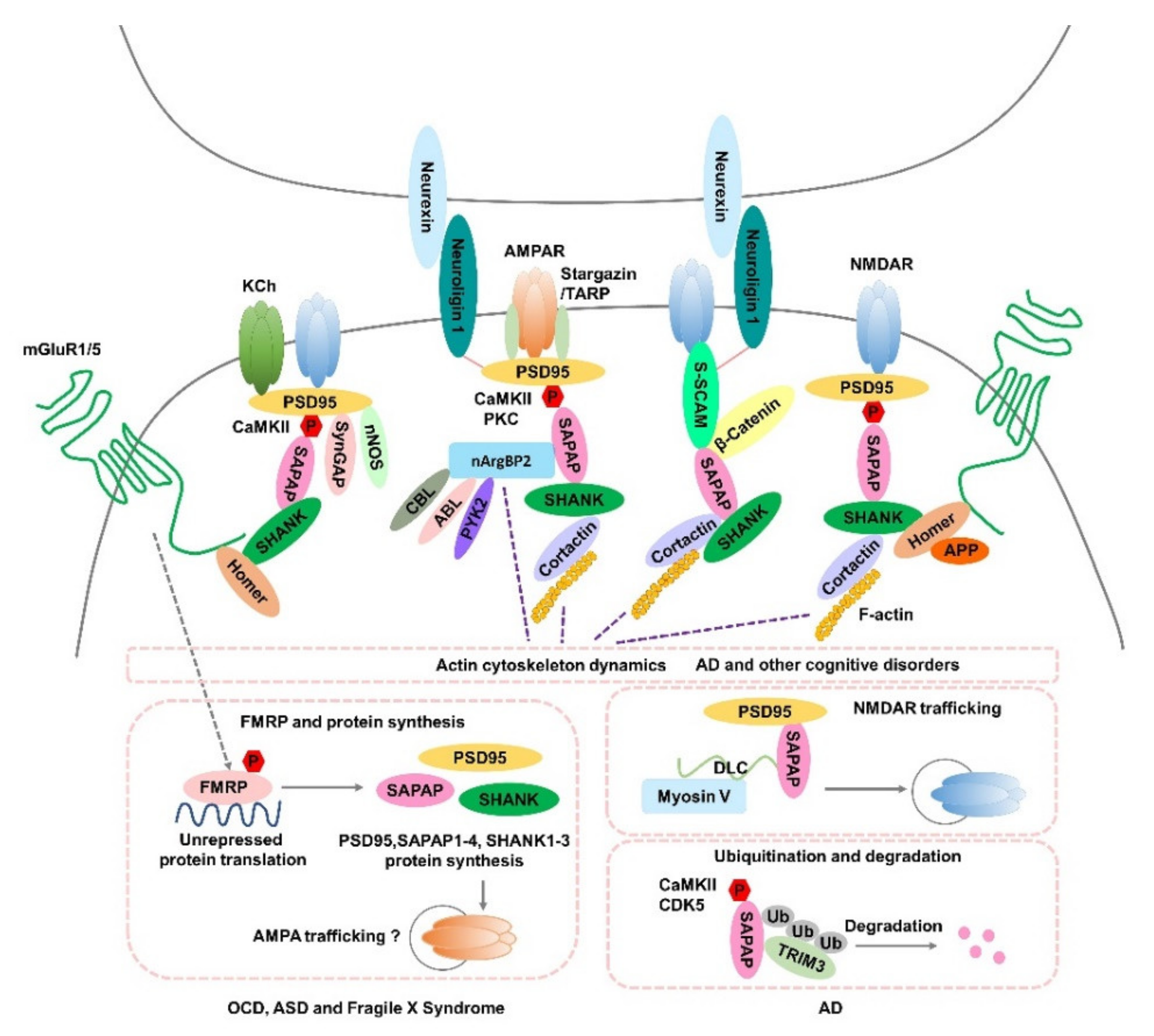
| Gene | Regulation | Expression | Brain Regions | Animal Models/Patients | References |
|---|---|---|---|---|---|
| SAPAP1 | Upregulation | Protein | Cortex or hippocampus | Fmr1-KO mice | [117] |
| Upregulation | mRNA or protein | Rats: NAc and hippocampus; patients: NAc | Schizophrenia rat model and patients | [115] | |
| SAPAP2 | Upregulation | Protein | Cortex or hippocampus | Fmr1-KO mice | [117] |
| SAPAP3 | Upregulation | Protein | Cortex or hippocampus | Fmr1-KO mice | [117] |
| Upregulation | mRNAor Protein | NAc | Syrian hamsters after aggressive experience | [119] | |
| Downregulation | Protein | OFC and mPFC | Fmr1-KO mice | [118] | |
| Upregulation | Protein | Mice: cortex and hippocampus; patients: cortex | Epilepsy patients and murine models | [116] | |
| SAPAP4 | Upregulation in the hippocampus | mRNA | Hippocampus | Fmr1-KO mice | [123] |
| Downregulation | mRNA | Amygdala | Taste-aversion-resistant rats after cocaine exposure | [126] |
| Gene | Regulation | Expression | Brain Regions/Cells | Animal Models/Patients | References |
|---|---|---|---|---|---|
| SAPAP2 | Upregulation (with lower DNA methylation) | mRNA | Hippocampus | PTSD rat model | [112] |
| Upregulation (with lower DNA methylation) | mRNA | dlPFC or NAc | Patients with alcohol dependence | [21] | |
| Downregulation (related to DNA methylation) | mRNA protein | dlPFC | AD patients | [22] | |
| SAPAP3 | Downregulation (by absence of histone deacetylases) | mRNA | Frontal cortex and striatum | Mice deficient in histone deacetylases 1 and 2 | [128] |
| SAPAP4 | Disruption of SAPAP4 by the chromosome translocation results in monoallelic hypermethylation of the truncated SAPAP4 promoter CpG island | N/A | Cerebellum | Early-onset cerebellar ataxia patients | [23] |
| Identified as a target of the BD-associated microRNA miR-1908-5p | N/A | Neural progenitor cells | BD patients | [127] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, Y.; Wang, H.; Li, C. SAPAP Scaffold Proteins: From Synaptic Function to Neuropsychiatric Disorders. Cells 2022, 11, 3815. https://doi.org/10.3390/cells11233815
Bai Y, Wang H, Li C. SAPAP Scaffold Proteins: From Synaptic Function to Neuropsychiatric Disorders. Cells. 2022; 11(23):3815. https://doi.org/10.3390/cells11233815
Chicago/Turabian StyleBai, Yunxia, Huimin Wang, and Chunxia Li. 2022. "SAPAP Scaffold Proteins: From Synaptic Function to Neuropsychiatric Disorders" Cells 11, no. 23: 3815. https://doi.org/10.3390/cells11233815
APA StyleBai, Y., Wang, H., & Li, C. (2022). SAPAP Scaffold Proteins: From Synaptic Function to Neuropsychiatric Disorders. Cells, 11(23), 3815. https://doi.org/10.3390/cells11233815






