The Role of 8-oxoG Repair Systems in Tumorigenesis and Cancer Therapy
Abstract
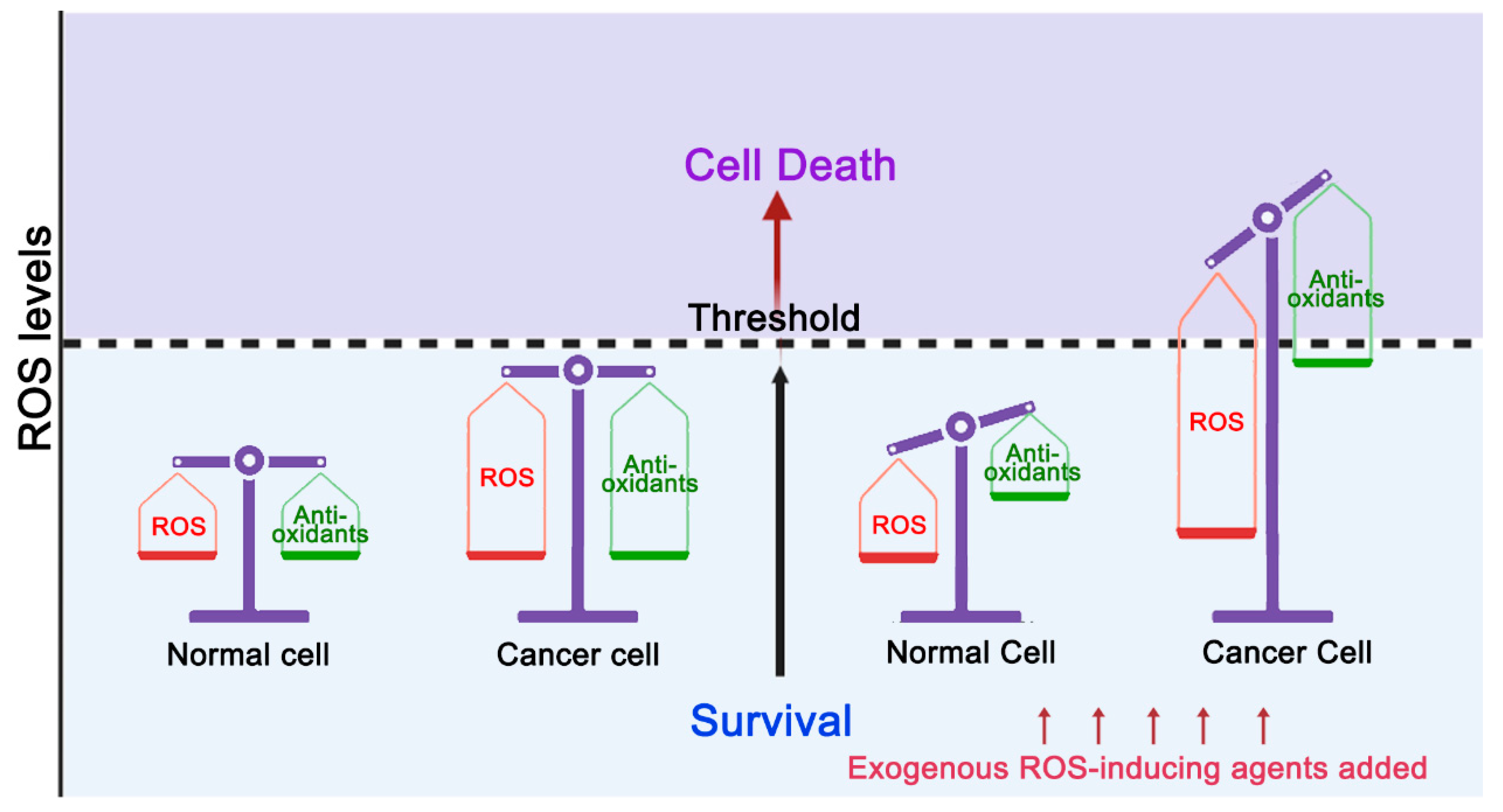
1. Introduction
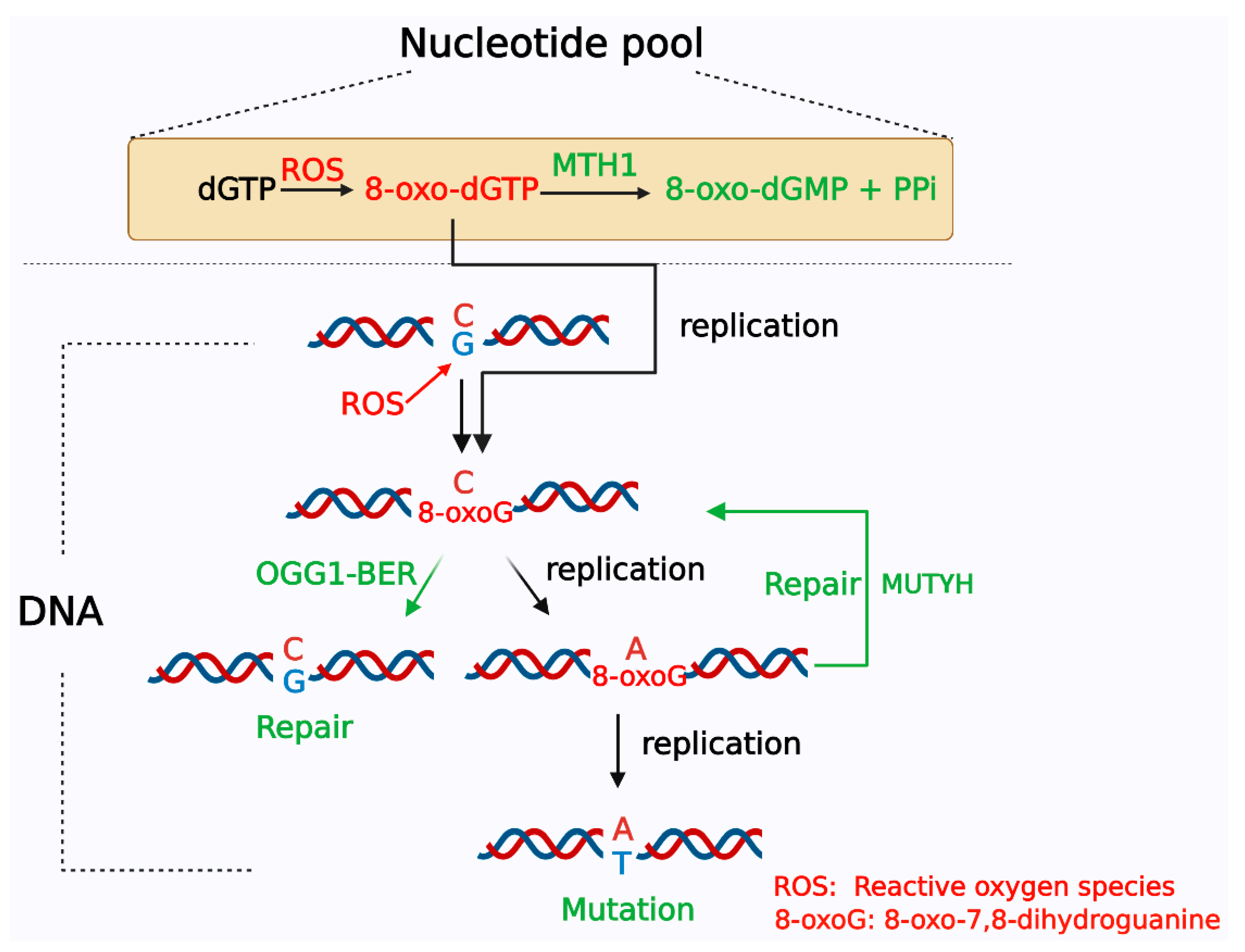
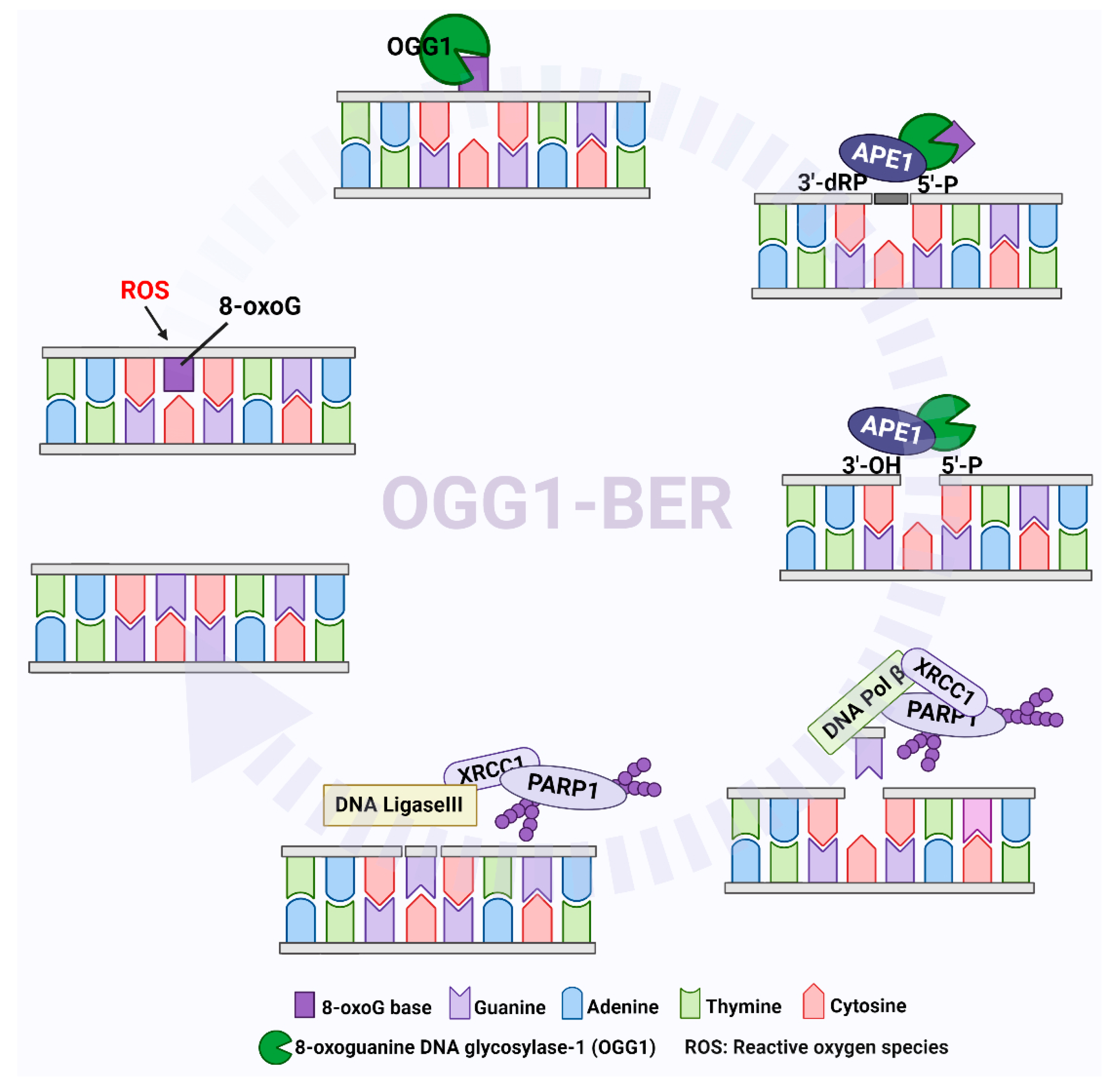
2. Systems for Formation and Elimination of 8-oxoG
3. The Effect of Accumulation of 8-oxoG on Tumorigenesis
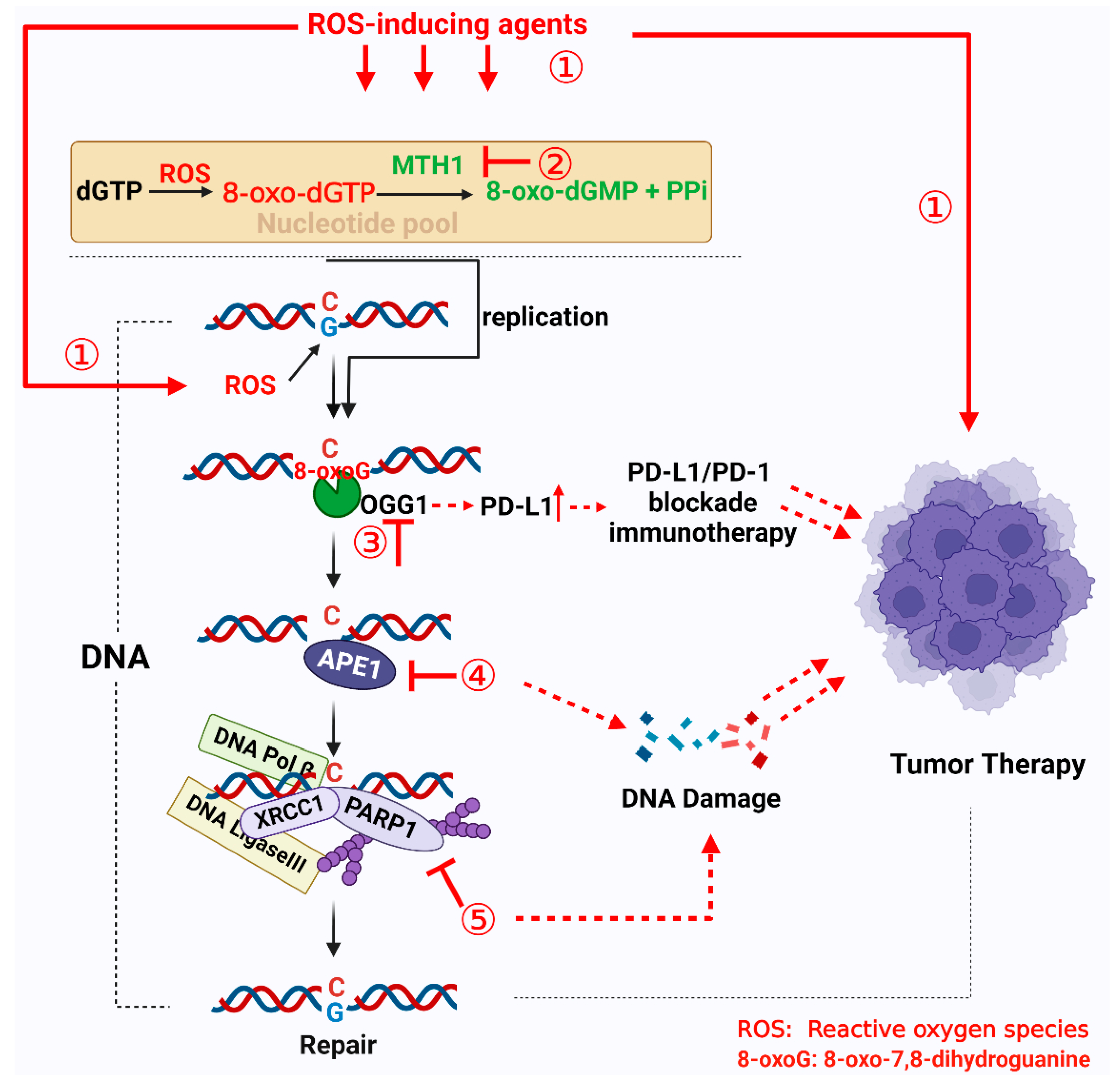
4. Targeting 8-oxoG Repair Systems as a Tumor Therapy Strategy
4.1. Targeting MTH1
4.2. Targeting OGG1
4.3. Targeting APE1
4.4. Targeting PARP1
5. Conclusions and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Cairns, R.A.; Harris, I.S.; Mak, T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 2011, 11, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Cheung, E.C.; Vousden, K.H. The role of ROS in tumour development and progression. Nat. Rev. Cancer 2022, 22, 280–297. [Google Scholar] [CrossRef] [PubMed]
- Handy, D.E.; Loscalzo, J. Redox regulation of mitochondrial function. Antioxid. Redox. Signal 2012, 16, 1323–1367. [Google Scholar] [CrossRef]
- Finkel, T. Signal transduction by reactive oxygen species. J. Cell Biol. 2011, 194, 7–15. [Google Scholar] [CrossRef]
- Verbon, E.H.; Post, J.A.; Boonstra, J. The influence of reactive oxygen species on cell cycle progression in mammalian cells. Gene 2012, 511, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Radak, Z.; Zhao, Z.; Koltai, E.; Ohno, H.; Atalay, M. Oxygen consumption and usage during physical exercise: The balance between oxidative stress and ROS-dependent adaptive signaling. Antioxid. Redox. Signal 2013, 18, 1208–1246. [Google Scholar] [CrossRef] [PubMed]
- Radak, Z.; Boldogh, I. 8-Oxo-7,8-dihydroguanine: Links to gene expression, aging, and defense against oxidative stress. Free Radic. Biol. Med. 2010, 49, 587–596. [Google Scholar] [CrossRef]
- Barnes, D.E.; Lindahl, T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004, 38, 445–476. [Google Scholar] [CrossRef]
- Burrows, C.J.; Muller, J.G. Oxidative Nucleobase Modifications Leading to Strand Scission. Chem. Rev. 1998, 98, 1109–1152. [Google Scholar] [CrossRef]
- Candeias, L.P.; Steenken, S. Reaction of HO. with Guanine Derivatives in Aqueous Solution: Formation of Two Different Redox-Active OH-Adduct Radicals and Their Unimolecular Transformation Reactions. Properties of G(-H). Chem.-A Eur. J. 2000, 6, 475–484. [Google Scholar] [CrossRef]
- Boiteux, S.; Coste, F.; Castaing, B. Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: Properties and biological roles of the Fpg and OGG1 DNA N-glycosylases. Free Radic. Biol. Med. 2017, 107, 179–201. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, B.; Jan, K.Y.; Chen, C.H.; Hour, T.C.; Yu, H.J.; Pu, Y.S. Resistance to paclitaxel is proportional to cellular total antioxidant capacity. Cancer Res. 2005, 65, 8455–8460. [Google Scholar] [CrossRef] [PubMed]
- Attia, S.; Kolesar, J.; Mahoney, M.R.; Pitot, H.C.; Laheru, D.; Heun, J.; Huang, W.; Eickhoff, J.; Erlichman, C.; Holen, K.D. A phase 2 consortium (P2C) trial of 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP) for advanced adenocarcinoma of the pancreas. Investig. New Drugs 2008, 26, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Gerber, D.E.; Beg, M.S.; Fattah, F.; Frankel, A.E.; Fatunde, O.; Arriaga, Y.; Dowell, J.E.; Bisen, A.; Leff, R.D.; Meek, C.C.; et al. Phase 1 study of ARQ 761, a beta-lapachone analogue that promotes NQO1-mediated programmed cancer cell necrosis. Br. J. Cancer 2018, 119, 928–936. [Google Scholar] [CrossRef]
- Pink, J.J.; Planchon, S.M.; Tagliarino, C.; Varnes, M.E.; Siegel, D.; Boothman, D.A. NAD(P)H.; Quinone oxidoreductase activity is the principal determinant of beta-lapachone cytotoxicity. J. Biol. Chem. 2000, 275, 5416–5424. [Google Scholar] [CrossRef] [PubMed]
- Kasai, H.; Nishimura, S. Hydroxylation of deoxyguanosine at the C-8 position by ascorbic acid and other reducing agents. Nucleic. Acids Res. 1984, 12, 2137–2145. [Google Scholar] [CrossRef]
- Chao, M.R.; Evans, M.D.; Hu, C.W.; Ji, Y.; Moller, P.; Rossner, P.; Cooke, M.S. Biomarkers of nucleic acid oxidation—A summary state-of-the-art. Redox. Biol. 2021, 42, 101872. [Google Scholar] [CrossRef]
- Dominissini, D.; He, C. Cancer: Damage prevention targeted. Nature 2014, 508, 191–192. [Google Scholar] [CrossRef] [PubMed]
- Koag, M.C.; Jung, H.; Lee, S. Mutagenesis mechanism of the major oxidative adenine lesion 7,8-dihydro-8-oxoadenine. Nucleic. Acids Res. 2020, 48, 5119–5134. [Google Scholar] [CrossRef]
- Katafuchi, A.; Nohmi, T. DNA polymerases involved in the incorporation of oxidized nucleotides into DNA: Their efficiency and template base preference. Mutat. Res. 2010, 703, 24–31. [Google Scholar] [CrossRef]
- Wang, Y.; Reddy, S.; Beard, W.A.; Wilson, S.H.; Schlick, T. Differing conformational pathways before and after chemistry for insertion of dATP versus dCTP opposite 8-oxoG in DNA polymerase beta. Biophys. J. 2007, 92, 3063–3070. [Google Scholar] [CrossRef] [PubMed]
- Furuichi, M.; Yoshida, M.C.; Oda, H.; Tajiri, T.; Nakabeppu, Y.; Tsuzuki, T.; Sekiguchi, M. Genomic structure and chromosome location of the human mutT homologue gene MTH1 encoding 8-oxo-dGTPase for prevention of A:T to C:G transversion. Genomics 1994, 24, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Rai, P.; Sobol, R.W. Mechanisms of MTH1 inhibition-induced DNA strand breaks: The slippery slope from the oxidized nucleotide pool to genotoxic damage. DNA Repair (Amst) 2019, 77, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Mishima, M.; Sakai, Y.; Itoh, N.; Kamiya, H.; Furuichi, M.; Takahashi, M.; Yamagata, Y.; Iwai, S.; Nakabeppu, Y.; Shirakawa, M. Structure of human MTH1, a Nudix family hydrolase that selectively degrades oxidized purine nucleoside triphosphates. J. Biol. Chem. 2004, 279, 33806–33815. [Google Scholar] [CrossRef] [PubMed]
- Svensson, L.M.; Jemth, A.S.; Desroses, M.; Loseva, O.; Helleday, T.; Hogbom, M.; Stenmark, P. Crystal structure of human MTH1 and the 8-oxo-dGMP product complex. FEBS Lett. 2011, 585, 2617–2621. [Google Scholar] [CrossRef]
- Nissink, J.W.; Bista, M.; Breed, J.; Carter, N.; Embrey, K.; Read, J.; Winter-Holt, J.J. MTH1 Substrate Recognition—An Example of Specific Promiscuity. PLoS ONE 2016, 11, e0151154. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M.; Kirkali, G.; Jaruga, P. Formamidopyrimidines in DNA: Mechanisms of formation, repair, and biological effects. Free Radic. Biol. Med. 2008, 45, 1610–1621. [Google Scholar] [CrossRef]
- Hazra, T.K.; Das, A.; Das, S.; Choudhury, S.; Kow, Y.W.; Roy, R. Oxidative DNA damage repair in mammalian cells: A new perspective. DNA Repair (Amst) 2007, 6, 470–480. [Google Scholar] [CrossRef]
- Michaels, M.L.; Pham, L.; Cruz, C.; Miller, J.H. MutM, a protein that prevents G.C—T.A transversions, is formamidopyrimidine-DNA glycosylase. Nucleic. Acids Res. 1991, 19, 3629–3632. [Google Scholar] [CrossRef] [PubMed]
- Nakabeppu, Y.; Ohta, E.; Abolhassani, N. MTH1 as a nucleotide pool sanitizing enzyme: Friend or foe? Free Radic. Biol. Med. 2017, 107, 151–158. [Google Scholar] [CrossRef]
- Hashimoto, K.; Tominaga, Y.; Nakabeppu, Y.; Moriya, M. Futile short-patch DNA base excision repair of adenine: 8-oxoguanine mispair. Nucleic. Acids Res. 2004, 32, 5928–5934. [Google Scholar] [CrossRef] [PubMed]
- Dupuy, P.; Howlader, M.; Glickman, M.S. A multilayered repair system protects the mycobacterial chromosome from endogenous and antibiotic-induced oxidative damage. Proc. Natl. Acad. Sci. USA 2020, 117, 19517–19527. [Google Scholar] [CrossRef]
- Bruner, S.D.; Norman, D.P.; Verdine, G.L. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 2000, 403, 859–866. [Google Scholar] [CrossRef]
- Izumi, T.; Wiederhold, L.R.; Roy, G.; Roy, R.; Jaiswal, A.; Bhakat, K.K.; Mitra, S.; Hazra, T.K. Mammalian DNA base excision repair proteins: Their interactions and role in repair of oxidative DNA damage. Toxicology 2003, 193, 43–65. [Google Scholar] [CrossRef] [PubMed]
- Krokan, H.E.; Nilsen, H.; Skorpen, F.; Otterlei, M.; Slupphaug, G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000, 476, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Ba, X.; Aguilera-Aguirre, L.; Rashid, Q.T.; Bacsi, A.; Radak, Z.; Sur, S.; Hosoki, K.; Hegde, M.L.; Boldogh, I. The role of 8-oxoguanine DNA glycosylase-1 in inflammation. Int. J. Mol. Sci. 2014, 15, 16975–16997. [Google Scholar] [CrossRef]
- Wallace, S.S. Base excision repair: A critical player in many games. DNA Repair (Amst) 2014, 19, 14–26. [Google Scholar] [CrossRef]
- Khodyreva, S.N.; Prasad, R.; Ilina, E.S.; Sukhanova, M.V.; Kutuzov, M.M.; Liu, Y.; Hou, E.W.; Wilson, S.H.; Lavrik, O.I. Apurinic/apyrimidinic (AP) site recognition by the 5′-dRP/AP lyase in poly(ADP-ribose) polymerase-1 (PARP-1). Proc. Natl. Acad. Sci. USA 2010, 107, 22090–22095. [Google Scholar] [CrossRef]
- El-Khamisy, S.F.; Masutani, M.; Suzuki, H.; Caldecott, K.W. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic. Acids Res. 2003, 31, 5526–5533. [Google Scholar] [CrossRef]
- Reynolds, P.; Cooper, S.; Lomax, M.; O’Neill, P. Disruption of PARP1 function inhibits base excision repair of a sub-set of DNA lesions. Nucleic. Acids Res. 2015, 43, 4028–4038. [Google Scholar] [CrossRef]
- Oka, S.; Hsu, C.P.; Sadoshima, J. Regulation of cell survival and death by pyridine nucleotides. Circ. Res. 2012, 111, 611–627. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.L.; Ren, E.C. Functional Aspects of PARP1 in DNA Repair and Transcription. Biomolecules 2012, 2, 524–548. [Google Scholar] [CrossRef] [PubMed]
- Polo, L.M.; Xu, Y.; Hornyak, P.; Garces, F.; Zeng, Z.; Hailstone, R.; Matthews, S.J.; Caldecott, K.W.; Oliver, A.W.; Pearl, L.H. Efficient Single-Strand Break Repair Requires Binding to Both Poly(ADP-Ribose) and DNA by the Central BRCT Domain of XRCC1. Cell Rep. 2019, 26, 573–581.e5. [Google Scholar] [CrossRef]
- Parsons, J.L.; Dianova, I.I.; Allinson, S.L.; Dianov, G.L. Poly(ADP-ribose) polymerase-1 protects excessive DNA strand breaks from deterioration during repair in human cell extracts. FEBS J. 2005, 272, 2012–2021. [Google Scholar] [CrossRef]
- Ray Chaudhuri, A.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef]
- Sharma, R.A.; Dianov, G.L. Targeting base excision repair to improve cancer therapies. Mol. Aspects Med. 2007, 28, 345–374. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chakrabarti, G.; Silvers, M.A.; Ilcheva, M.; Liu, Y.; Moore, Z.R.; Luo, X.; Gao, J.; Anderson, G.; Liu, L.; Sarode, V.; et al. Tumor-selective use of DNA base excision repair inhibition in pancreatic cancer using the NQO1 bioactivatable drug, beta-lapachone. Sci. Rep. 2015, 5, 17066. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, H.; Guo, Y.; Chen, Y.; Chen, H.; Liu, Y. X-ray repair cross-complementing protein 1 (XRCC1) loss promotes beta-lapachone -induced apoptosis in pancreatic cancer cells. BMC Cancer 2021, 21, 1234. [Google Scholar] [CrossRef]
- Strom, C.E.; Johansson, F.; Uhlen, M.; Szigyarto, C.A.; Erixon, K.; Helleday, T. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic. Acids Res. 2011, 39, 3166–3175. [Google Scholar] [CrossRef]
- Bohr, V.A.; Stevnsner, T.; de Souza-Pinto, N.C. Mitochondrial DNA repair of oxidative damage in mammalian cells. Gene 2002, 286, 127–134. [Google Scholar] [CrossRef]
- Hudson, E.K.; Hogue, B.A.; Souza-Pinto, N.C.; Croteau, D.L.; Anson, R.M.; Bohr, V.A.; Hansford, R.G. Age-associated change in mitochondrial DNA damage. Free Radic. Res. 1998, 29, 573–579. [Google Scholar] [CrossRef] [PubMed]
- de Souza-Pinto, N.C.; Bohr, V.A. The mitochondrial theory of aging: Involvement of mitochondrial DNA damage and repair. Int. Rev. Neurobiol. 2002, 53, 519–534. [Google Scholar] [PubMed]
- Shigenaga, M.K.; Hagen, T.M.; Ames, B.N. Oxidative damage and mitochondrial decay in aging. Proc. Natl. Acad. Sci. USA 1994, 91, 10771–10778. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, K.; Stuart, J.A.; de Souza-Pinto, N.C.; Bohr, V.A. The C-terminal alphaO helix of human Ogg1 is essential for 8-oxoguanine DNA glycosylase activity: The mitochondrial beta-Ogg1 lacks this domain and does not have glycosylase activity. Nucleic. Acids Res. 2004, 32, 5596–5608. [Google Scholar] [CrossRef] [PubMed]
- Lia, D.; Reyes, A.; de Melo Campos, J.T.A.; Piolot, T.; Baijer, J.; Radicella, J.P.; Campalans, A. Mitochondrial maintenance under oxidative stress depends on mitochondrially localised alpha-OGG1. J. Cell Sci. 2018, 131, jcs213538. [Google Scholar] [CrossRef] [PubMed]
- Kazak, L.; Reyes, A.; Holt, I.J. Minimizing the damage: Repair pathways keep mitochondrial DNA intact. Nat. Rev. Mol. Cell Biol. 2012, 13, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Fontana, G.A.; Gahlon, H.L. Mechanisms of replication and repair in mitochondrial DNA deletion formation. Nucleic. Acids Res. 2020, 48, 11244–11258. [Google Scholar] [CrossRef]
- Boiteux, S.; Radicella, J.P. The human OGG1 gene: Structure, functions, and its implication in the process of Carcinogenesis. Arch. Biochem. Biophys. 2000, 377, 1–8. [Google Scholar] [CrossRef]
- Kim, S.J.; Cheresh, P.; Jablonski, R.P.; Williams, D.B.; Kamp, D.W. The Role of Mitochondrial DNA in Mediating Alveolar Epithelial Cell Apoptosis and Pulmonary Fibrosis. Int. J. Mol. Sci. 2015, 16, 21486–21519. [Google Scholar] [CrossRef]
- Loeb, L.A. Mutator Phenotype May Be Required for Multistage Carcinogenesis. Cancer Res. 1991, 51, 3075–3079. [Google Scholar] [PubMed]
- Koh, G.; Degasperi, A.; Zou, X.; Momen, S.; Nik-Zainal, S. Mutational signatures: Emerging concepts, caveats and clinical applications. Nat. Rev. Cancer 2021, 21, 619–637. [Google Scholar] [CrossRef] [PubMed]
- Nakabeppu, Y. Cellular levels of 8-oxoguanine in either DNA or the nucleotide pool play pivotal roles in carcinogenesis and survival of cancer cells. Int. J. Mol. Sci. 2014, 15, 12543–12557. [Google Scholar] [CrossRef] [PubMed]
- Nakabeppu, Y.; Sakumi, K.; Sakamoto, K.; Tsuchimoto, D.; Tsuzuki, T.; Nakatsu, Y. Mutagenesis and carcinogenesis caused by the oxidation of nucleic acids. Biol. Chem. 2006, 387, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Tsuzuki, T.; Egashira, A.; Igarashi, H.; Iwakuma, T.; Nakatsuru, Y.; Tominaga, Y.; Kawate, H.; Nakao, K.; Nakamura, K.; Ide, F.; et al. Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase. Proc. Natl. Acad. Sci. USA 2001, 98, 11456–11461. [Google Scholar] [CrossRef]
- Sakumi, K.; Tominaga, Y.; Furuichi, M.; Xu, P.; Tsuzuki, T.; Sekiguchi, M.; Nakabeppu, Y. Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Cancer Res. 2003, 63, 902–905. [Google Scholar]
- Arai, T.; Kelly, V.P.; Minowa, O.; Noda, T.; Nishimura, S. High accumulation of oxidative DNA damage, 8-hydroxyguanine, in Mmh/Ogg1 deficient mice by chronic oxidative stress. Carcinogenesis 2002, 23, 2005–2010. [Google Scholar] [CrossRef] [PubMed]
- Kunisada, M.; Sakumi, K.; Tominaga, Y.; Budiyanto, A.; Ueda, M.; Ichihashi, M.; Nakabeppu, Y.; Nishigori, C. 8-Oxoguanine formation induced by chronic UVB exposure makes Ogg1 knockout mice susceptible to skin Carcinogenesis. Cancer Res. 2005, 65, 6006–6010. [Google Scholar] [CrossRef] [PubMed]
- Kakehashi, A.; Ishii, N.; Okuno, T.; Fujioka, M.; Gi, M.; Wanibuchi, H. Enhanced Susceptibility of Ogg1 Mutant Mice to Multiorgan Carcinogenesis. Int. J. Mol. Sci. 2017, 18, 1801. [Google Scholar] [CrossRef]
- Banda, D.M.; Nunez, N.N.; Burnside, M.A.; Bradshaw, K.M.; David, S.S. Repair of 8-oxoG.;A mismatches by the MUTYH glycosylase: Mechanism, metals and medicine. Free Radic. Biol. Med. 2017, 107, 202–215. [Google Scholar] [CrossRef]
- Oka, S.; Nakabeppu, Y. DNA glycosylase encoded by MUTYH functions as a molecular switch for programmed cell death under oxidative stress to suppress tumorigenesis. Cancer Sci. 2011, 102, 677–682. [Google Scholar] [CrossRef]
- Sakamoto, K.; Tominaga, Y.; Yamauchi, K.; Nakatsu, Y.; Sakumi, K.; Yoshiyama, K.; Egashira, A.; Kura, S.; Yao, T.; Tsuneyoshi, M.; et al. MUTYH-null mice are susceptible to spontaneous and oxidative stress induced intestinal tumorigenesis. Cancer Res. 2007, 67, 6599–6604. [Google Scholar] [CrossRef]
- Xie, Y.; Yang, H.; Cunanan, C.; Okamoto, K.; Shibata, D.; Pan, J.; Barnes, D.E.; Lindahl, T.; McIlhatton, M.; Fishel, R.; et al. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004, 64, 3096–30102. [Google Scholar] [CrossRef]
- Ohno, M.; Sakumi, K.; Fukumura, R.; Furuichi, M.; Iwasaki, Y.; Hokama, M.; Ikemura, T.; Tsuzuki, T.; Gondo, Y.; Nakabeppu, Y. 8-oxoguanine causes spontaneous de novo germline mutations in mice. Sci. Rep. 2014, 4, 4689. [Google Scholar] [CrossRef] [PubMed]
- Bravard, A.; Vacher, M.; Moritz, E.; Vaslin, L.; Hall, J.; Epe, B.; Radicella, J.P. Oxidation status of human OGG1-S326C polymorphic variant determines cellular DNA repair capacity. Cancer Res. 2009, 69, 3642–3649. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.W.; Evans, M.K. Dimerization and opposite base-dependent catalytic impairment of polymorphic S326C OGG1 glycosylase. Nucleic. Acids Res. 2006, 34, 1620–1632. [Google Scholar] [CrossRef]
- Yamane, A.; Kohno, T.; Ito, K.; Sunaga, N.; Aoki, K.; Yoshimura, K.; Murakami, H.; Nojima, Y.; Yokota, J. Differential ability of polymorphic OGG1 proteins to suppress mutagenesis induced by 8-hydroxyguanine in human cell in vivo. Carcinogenesis 2004, 25, 1689–1694. [Google Scholar] [CrossRef] [PubMed]
- Janik, J.; Swoboda, M.; Janowska, B.; Ciesla, J.M.; Gackowski, D.; Kowalewski, J.; Olinski, R.; Tudek, B.; Speina, E. 8-Oxoguanine incision activity is impaired in lung tissues of NSCLC patients with the polymorphism of OGG1 and XRCC1 genes. Mutat. Res. 2011, 709–710, 21–31. [Google Scholar] [CrossRef]
- Wei, B.; Zhou, Y.; Xu, Z.; Xi, B.; Cheng, H.; Ruan, J.; Zhu, M.; Hu, Q.; Wang, Q.; Wang, Z.; et al. The effect of hOGG1 Ser326Cys polymorphism on cancer risk: Evidence from a meta-analysis. PLoS ONE 2011, 6, e27545. [Google Scholar] [CrossRef]
- Duan, W.X.; Hua, R.X.; Yi, W.; Shen, L.J.; Jin, Z.X.; Zhao, Y.H.; Yi, D.H.; Chen, W.S.; Yu, S.Q. The association between OGG1 Ser326Cys polymorphism and lung cancer susceptibility: A meta-analysis of 27 studies. PLoS ONE 2012, 7, e35970. [Google Scholar] [CrossRef]
- Zhang, M.; Mo, R. Association of hOGG1 Ser326Cys polymorphism with colorectal cancer risk: An updated meta-analysis including 5235 cases and 8438 controls. Tumour. Biol. 2014, 35, 12627–12633. [Google Scholar] [CrossRef]
- Zhou, P.T.; Li, B.; Ji, J.; Wang, M.M.; Gao, C.F. A systematic review and meta-analysis of the association between OGG1 Ser326Cys polymorphism and cancers. Med. Oncol. 2015, 32, 472. [Google Scholar] [CrossRef]
- Wang, R.; Hao, W.; Pan, L.; Boldogh, I.; Ba, X. The roles of base excision repair enzyme OGG1 in gene expression. Cell Mol. Life Sci. 2018, 75, 3741–3750. [Google Scholar] [CrossRef]
- Mabley, J.G.; Pacher, P.; Deb, A.; Wallace, R.; Elder, R.H.; Szabo, C. Potential role for 8-oxoguanine DNA glycosylase in regulating inflammation. FASEB J. 2005, 19, 290–292. [Google Scholar] [CrossRef]
- Touati, E.; Michel, V.; Thiberge, J.M.; Ave, P.; Huerre, M.; Bourgade, F.; Klungland, A.; Labigne, A. Deficiency in OGG1 protects against inflammation and mutagenic effects associated with H. pylori infection in mouse. Helicobacter 2006, 11, 494–505. [Google Scholar] [CrossRef]
- Li, G.; Yuan, K.; Yan, C.; Fox, J., III.; Gaid, M.; Breitwieser, W.; Bansal, A.K.; Zeng, H.; Gao, H.; Wu, M. 8-Oxoguanine-DNA glycosylase 1 deficiency modifies allergic airway inflammation by regulating STAT6 and IL-4 in cells and in mice. Free Radic. Biol. Med. 2012, 52, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Reeve, A.K.; Krishnan, K.J.; Turnbull, D. Mitochondrial DNA mutations in disease, aging, and neurodegeneration. Ann. N. Y. Acad. Sci. 2008, 1147, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Torres-Gonzalez, M.; Gawlowski, T.; Kocalis, H.; Scott, B.T.; Dillmann, W.H. Mitochondrial 8-oxoguanine glycosylase decreases mitochondrial fragmentation and improves mitochondrial function in H9C2 cells under oxidative stress conditions. Am. J. Physiol. Cell Physiol. 2014, 306, C221–C229. [Google Scholar] [CrossRef] [PubMed]
- Yuzefovych, L.V.; Kahn, A.G.; Schuler, M.A.; Eide, L.; Arora, R.; Wilson, G.L.; Tan, M.; Rachek, L.I. Mitochondrial DNA Repair through OGG1 Activity Attenuates Breast Cancer Progression and Metastasis. Cancer Res. 2016, 76, 30–34. [Google Scholar] [CrossRef]
- de Souza-Pinto, N.C.; Eide, L.; Hogue, B.A.; Thybo, T.; Stevnsner, T.; Seeberg, E.; Klungland, A.; Bohr, V.A. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001, 61, 5378–5381. [Google Scholar]
- Leon, J.; Sakumi, K.; Castillo, E.; Sheng, Z.; Oka, S.; Nakabeppu, Y. 8-Oxoguanine accumulation in mitochondrial DNA causes mitochondrial dysfunction and impairs neuritogenesis in cultured adult mouse cortical neurons under oxidative conditions. Sci. Rep. 2016, 6, 22086. [Google Scholar] [CrossRef]
- Kim, S.J.; Cheresh, P.; Jablonski, R.P.; Rachek, L.; Yeldandi, A.; Piseaux-Aillon, R.; Ciesielski, M.J.; Ridge, K.; Gottardi, C.; Lam, A.P.; et al. Mitochondrial 8-oxoguanine DNA glycosylase mitigates alveolar epithelial cell PINK1 deficiency, mitochondrial DNA damage, apoptosis, and lung fibrosis. Am. J. Physiol. Lung. Cell Mol. Physiol. 2020, 318, L1084–L1096. [Google Scholar] [CrossRef]
- Grundy, G.J.; Parsons, J.L. Base excision repair and its implications to cancer therapy. Essays. Biochem. 2020, 64, 831–843. [Google Scholar] [PubMed]
- Kennedy, C.H.; Cueto, R.; Belinsky, S.A.; Lechner, J.F.; Pryor, W.A. Overexpression of hMTH1 mRNA: A molecular marker of oxidative stress in lung cancer cells. FEBS Lett. 1998, 429, 17–20. [Google Scholar] [CrossRef]
- Coskun, E.; Jaruga, P.; Jemth, A.S.; Loseva, O.; Scanlan, L.D.; Tona, A.; Lowenthal, M.S.; Helleday, T.; Dizdaroglu, M. Addiction to MTH1 protein results in intense expression in human breast cancer tissue as measured by liquid chromatography-isotope-dilution tandem mass spectrometry. DNA Repair (Amst) 2015, 33, 101–110. [Google Scholar] [CrossRef]
- Obtulowicz, T.; Swoboda, M.; Speina, E.; Gackowski, D.; Rozalski, R.; Siomek, A.; Janik, J.; Janowska, B.; Ciesla, J.M.; Jawien, A.; et al. Oxidative stress and 8-oxoguanine repair are enhanced in colon adenoma and carcinoma patients. Mutagenesis 2010, 25, 463–471. [Google Scholar] [CrossRef] [PubMed]
- McPherson, L.A.; Troccoli, C.I.; Ji, D.; Bowles, A.E.; Gardiner, M.L.; Mohsen, M.G.; Nagathihalli, N.S.; Nguyen, D.M.; Robbins, D.J.; Merchant, N.B.; et al. Increased MTH1-specific 8-oxodGTPase activity is a hallmark of cancer in colon, lung and pancreatic tissue. DNA Repair (Amst) 2019, 83, 102644. [Google Scholar] [CrossRef] [PubMed]
- Gad, H.; Koolmeister, T.; Jemth, A.-S.; Eshtad, S.; Jacques, S.A.; Ström, C.E.; Svensson, L.M.; Schultz, N.; Lundbäck, T.; Einarsdottir, B.O.; et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature 2014, 508, 215–221. [Google Scholar] [CrossRef]
- Huber, K.V.; Salah, E.; Radic, B.; Gridling, M.; Elkins, J.M.; Stukalov, A.; Jemth, A.S.; Gokturk, C.; Sanjiv, K.; Stromberg, K.; et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature 2014, 508, 222–227. [Google Scholar] [CrossRef]
- Wang, R.; Li, C.; Qiao, P.; Xue, Y.; Zheng, X.; Chen, H.; Zeng, X.; Liu, W.; Boldogh, I.; Ba, X. OGG1-initiated base excision repair exacerbates oxidative stress-induced parthanatos. Cell Death Disease 2018, 9, 628. [Google Scholar] [CrossRef]
- Petrocchi, A.; Leo, E.; Reyna, N.J.; Hamilton, M.M.; Shi, X.; Parker, C.A.; Mseeh, F.; Bardenhagen, J.P.; Leonard, P.; Cross, J.B.; et al. Identification of potent and selective MTH1 inhibitors. Bioorg. Med. Chem. Lett. 2016, 26, 1503–1507. [Google Scholar] [CrossRef]
- Warpman Berglund, U.; Sanjiv, K.; Gad, H.; Kalderén, C.; Koolmeister, T.; Pham, T.; Gokturk, C.; Jafari, R.; Maddalo, G.; Seashore-Ludlow, B.; et al. Validation and development of MTH1 inhibitors for treatment of cancer. Ann. Oncol. 2016, 27, 2275–2283. [Google Scholar] [CrossRef] [PubMed]
- Moukengue, B.; Brown, H.K.; Charrier, C.; Battaglia, S.; Baud’huin, M.; Quillard, T.; Pham, T.M.; Pateras, I.S.; Gorgoulis, V.G.; Helleday, T.; et al. TH1579, MTH1 inhibitor, delays tumour growth and inhibits metastases development in osteosarcoma model. EBioMedicine 2020, 53, 102704. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Sanjiv, K.; Gad, H.; Pham, T.; Gokturk, C.; Rasti, A.; Zhao, Z.; He, K.; Feng, M.; Zang, Y.; et al. Karonudib is a promising anticancer therapy in hepatocellular carcinoma. Ther. Adv. Med. Oncol. 2019, 11, 1758835919866960. [Google Scholar] [CrossRef]
- Oksvold, M.P.; Berglund, U.W.; Gad, H.; Bai, B.; Stokke, T.; Rein, I.D.; Pham, T.; Sanjiv, K.; Oy, G.F.; Norum, J.H.; et al. Karonudib has potent anti-tumor effects in preclinical models of B-cell lymphoma. Sci. Rep. 2021, 11, 6317. [Google Scholar] [CrossRef]
- Zhou, W.; Ma, L.; Yang, J.; Qiao, H.; Li, L.; Guo, Q.; Ma, J.; Zhao, L.; Wang, J.; Jiang, G.; et al. Potent and specific MTH1 inhibitors targeting gastric cancer. Cell Death Dis. 2019, 10, 434. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Jin, L.; Yan, X.G.; Sherwin, S.; Farrelly, M.; Zhang, Y.Y.; Liu, F.; Wang, C.Y.; Guo, S.T.; Yari, H.; et al. Reactive Oxygen Species Dictate the Apoptotic Response of Melanoma Cells to TH588. J. Investig. Dermatol. 2016, 136, 2277–2286. [Google Scholar] [CrossRef] [PubMed]
- Kettle, J.G.; Alwan, H.; Bista, M.; Breed, J.; Davies, N.L.; Eckersley, K.; Fillery, S.; Foote, K.M.; Goodwin, L.; Jones, D.R.; et al. Potent and Selective Inhibitors of MTH1 Probe Its Role in Cancer Cell Survival. J. Med. Chem. 2016, 59, 2346–2361. [Google Scholar] [CrossRef]
- Coppede, F.; Migliore, L. DNA damage in neurodegenerative diseases. Mutat. Res. 2015, 776, 84–97. [Google Scholar] [CrossRef]
- Abolhassani, N.; Leon, J.; Sheng, Z.; Oka, S.; Hamasaki, H.; Iwaki, T.; Nakabeppu, Y. Molecular pathophysiology of impaired glucose metabolism, mitochondrial dysfunction, and oxidative DNA damage in Alzheimer’s disease brain. Mech. Ageing. Dev. 2017, 161, 95–104. [Google Scholar] [CrossRef]
- Ventura, I.; Russo, M.T.; De Nuccio, C.; De Luca, G.; Degan, P.; Bernardo, A.; Visentin, S.; Minghetti, L.; Bignami, M. hMTH1 expression protects mitochondria from Huntington’s disease-like impairment. Neurobiol. Dis. 2013, 49, 148–158. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Kajitani, K.; Dan, Y.; Furuichi, M.; Ohno, M.; Sakumi, K.; Kang, D.; Nakabeppu, Y. MTH1, an oxidized purine nucleoside triphosphatase, protects the dopamine neurons from oxidative damage in nucleic acids caused by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Cell Death Differ. 2006, 13, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, S.; Song, L.; Qu, M.; Zou, Z. Synergistic lethality between PARP-trapping and alantolactone-induced oxidative DNA damage in homologous recombination-proficient cancer cells. Oncogene 2020, 39, 2905–2920. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, J.; Su, Y.; Yang, L.; Chen, L.; Qiang, L.; Wang, Y.; Xiang, H.; Tham, H.P.; Peng, J.; et al. MTH1 inhibitor amplifies the lethality of reactive oxygen species to tumor in photodynamic therapy. Sci. Adv. 2020, 6, eaaz0575. [Google Scholar] [CrossRef] [PubMed]
- Centio, A.; Estruch, M.; Reckzeh, K.; Sanjiv, K.; Vittori, C.; Engelhard, S.; Warpman Berglund, U.; Helleday, T.; Theilgaard-Mönch, K. Inhibition of oxidized nucleotide sanitation by TH1579 and conventional chemotherapy cooperatively enhance oxidative DNA-damage and survival in AML. Mol. Cancer Ther. 2022, 21, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Visnes, T.; Cazares-Korner, A.; Hao, W.; Wallner, O.; Masuyer, G.; Loseva, O.; Mortusewicz, O.; Wiita, E.; Sarno, A.; Manoilov, A.; et al. Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation. Science 2018, 362, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Mezzadra, R.; Schumacher, T.N. Regulation and Function of the PD-L1 Checkpoint. Immunity 2018, 48, 434–452. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.H.; Chan, L.C.; Li, C.W.; Hsu, J.L.; Hung, M.C. Mechanisms Controlling PD-L1 Expression in Cancer. Mol. Cell 2019, 76, 359–370. [Google Scholar] [CrossRef]
- Glorieux, C.; Xia, X.; Huang, P. The Role of Oncogenes and Redox Signaling in the Regulation of PD-L1 in Cancer. Cancers 2021, 13, 4426. [Google Scholar] [CrossRef]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef]
- Brahmer, J.R.; Tykodi, S.S.; Chow, L.Q.; Hwu, W.J.; Topalian, S.L.; Hwu, P.; Drake, C.G.; Camacho, L.H.; Kauh, J.; Odunsi, K.; et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 2012, 366, 2455–2465. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. Regulation of PD-L1 expression on cancer cells with ROS-modulating drugs. Life Sci. 2020, 246, 117403. [Google Scholar] [CrossRef] [PubMed]
- Permata, T.B.M.; Hagiwara, Y.; Sato, H.; Yasuhara, T.; Oike, T.; Gondhowiardjo, S.; Held, K.D.; Nakano, T.; Shibata, A. Base excision repair regulates PD-L1 expression in cancer cells. Oncogene 2019, 38, 4452–4466. [Google Scholar] [CrossRef]
- Glorieux, C.; Xia, X.; He, Y.Q.; Hu, Y.; Cremer, K.; Robert, A.; Liu, J.; Wang, F.; Ling, J.; Chiao, P.J.; et al. Regulation of PD-L1 expression in K-ras-driven cancers through ROS-mediated FGFR1 signaling. Redox. Biol. 2021, 38, 101780. [Google Scholar] [CrossRef]
- Wang, X.; Li, J.; Dong, K.; Lin, F.; Long, M.; Ouyang, Y.; Wei, J.; Chen, X.; Weng, Y.; He, T.; et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal 2015, 27, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Z.; Zhang, A.; Han, C.; Shen, A.; Jiang, L.; Boothman, D.A.; Qiao, J.; Wang, Y.; Huang, X.; et al. NQO1 targeting prodrug triggers innate sensing to overcome checkpoint blockade resistance. Nat. Commun. 2019, 10, 3251. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wilson, D.M., III. Human apurinic/apyrimidinic endonuclease 1. Antioxid. Redox. Signal 2014, 20, 678–707. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Herman, T.; Demple, B. Two distinct human DNA diesterases that hydrolyze 3′-blocking deoxyribose fragments from oxidized DNA. Nucleic. Acids Res. 1991, 19, 5907–5914. [Google Scholar] [CrossRef]
- Weaver, T.M.; Hoitsma, N.M.; Spencer, J.J.; Gakhar, L.; Schnicker, N.J.; Freudenthal, B.D. Structural basis for APE1 processing DNA damage in the nucleosome. Nat. Commun. 2022, 13, 5390. [Google Scholar] [CrossRef]
- McNeill, D.R.; Lam, W.; DeWeese, T.L.; Cheng, Y.C.; Wilson, D.M., III. Impairment of APE1 function enhances cellular sensitivity to clinically relevant alkylators and antimetabolites. Mol. Cancer Res. 2009, 7, 897–906. [Google Scholar] [CrossRef]
- Wang, D.; Xiang, D.B.; Yang, X.Q.; Chen, L.S.; Li, M.X.; Zhong, Z.Y.; Zhang, Y.S. APE1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and targeted inhibition of APE1 enhances the activity of cisplatin in A549 cells. Lung Cancer 2009, 66, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Poletto, M.; Legrand, A.J.; Dianov, G.L. DNA Base Excision Repair: The Achilles’ Heel of Tumour Cells and their Microenvironment? Curr. Pharm. Des. 2017, 23, 4758–4772. [Google Scholar] [CrossRef] [PubMed]
- Liuzzi, M.; Weinfeld, M.; Paterson, M.C. Selective inhibition by methoxyamine of the apurinic/apyrimidinic endonuclease activity associated with pyrimidine dimer-DNA glycosylases from Micrococcus luteus and bacteriophage T4. Biochemistry 1987, 26, 3315–3321. [Google Scholar] [CrossRef]
- Montaldi, A.P.; Sakamoto-Hojo, E.T. Methoxyamine sensitizes the resistant glioblastoma T98G cell line to the alkylating agent temozolomide. Clin. Exp. Med. 2013, 13, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.S.; Rosen, L.S.; Mendelson, D.; Ramanathan, R.K.; Goldman, J.; Liu, L.; Xu, Y.; Gerson, S.L.; Anthony, S.P.; Figg, W.D.; et al. A phase 1 study of TRC102, an inhibitor of base excision repair, and pemetrexed in patients with advanced solid tumors. Investig. New Drugs 2013, 31, 714–723. [Google Scholar] [CrossRef]
- Liu, L.; Nakatsuru, Y.; Gerson, S.L. Base excision repair as a therapeutic target in colon cancer. Clin. Cancer Res. 2002, 8, 2985–2991. [Google Scholar]
- Eads, J.R.; Krishnamurthi, S.S.; Saltzman, J.; Bokar, J.A.; Savvides, P.; Meropol, N.J.; Gibbons, J.; Koon, H.; Sharma, N.; Rogers, L.; et al. Phase I clinical trial of temozolomide and methoxyamine (TRC-102), an inhibitor of base excision repair, in patients with advanced solid tumors. Investig. New Drugs 2021, 39, 142–151. [Google Scholar] [CrossRef]
- Madhusudan, S.; Smart, F.; Shrimpton, P.; Parsons, J.L.; Gardiner, L.; Houlbrook, S.; Talbot, D.C.; Hammonds, T.; Freemont, P.A.; Sternberg, M.J.; et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic. Acids Res. 2005, 33, 4711–4724. [Google Scholar] [CrossRef]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef]
- Helleday, T. The underlying mechanism for the PARP and BRCA synthetic lethality: Clearing up the misunderstandings. Mol. Oncol. 2011, 5, 387–393. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Wang, P.Y.; Wang, Y.T.; Yang, G.F.; Zhang, A.; Miao, Z.H. An Update on Poly(ADP-ribose)polymerase-1 (PARP-1) Inhibitors: Opportunities and Challenges in Cancer Therapy. J. Med. Chem. 2016, 59, 9575–9598. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, S.; Weller, M.C.; Repmann, S.; Moch, H.; Jiricny, J. Synthetic lethality between BRCA1 deficiency and poly(ADP-ribose) polymerase inhibition is modulated by processing of endogenous oxidative DNA damage. Nucleic. Acids Res. 2019, 47, 9132–9143. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Wang, M.; Sang, X.; Liu, P.; Gao, J.; Jiang, K.; Cheng, H. Phenethyl Isothiocyanate Enhances the Cytotoxic Effects of PARP Inhibitors in High-Grade Serous Ovarian Cancer Cells. Front. Oncol. 2021, 11, 812264. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Wang, H.; Niu, J.; Song, Y.; Zou, Z. Alkannin-Induced Oxidative DNA Damage Synergizes With PARP Inhibition to Cause Cancer-Specific Cytotoxicity. Front. Pharmacol. 2020, 11, 610205. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Motea, E.A.; Moore, Z.R.; Yao, J.; Dong, Y.; Chakrabarti, G.; Kilgore, J.A.; Silvers, M.A.; Patidar, P.L.; Cholka, A.; et al. Leveraging an NQO1 Bioactivatable Drug for Tumor-Selective Use of Poly(ADP-ribose) Polymerase Inhibitors. Cancer Cell 2016, 30, 940–952. [Google Scholar] [CrossRef] [PubMed]
| Target | Inhibitor | Validation | Current Status * |
|---|---|---|---|
| MTH1 | Karonudib (TH1579) | In vitro, Cell lines, Xenografts | Phase I |
| TH588, TH287 | In vitro, Cell lines, Xenografts | ||
| (S)-crizotinib | In vitro, Cell lines, Xenografts | ||
| IACS-4759, IACS-4619 | In vitro, Cell lines | ||
| APE1 | TRC102 (Methoxyamine) | In vitro, Cell lines, Xenografts | Phase I/II |
| Gossypol | In vitro, Cell lines, Xenografts | Phase III | |
| CRT0044876 | In vitro, Cell lines | ||
| AR03 | In vitro, Cell lines | ||
| PARP1/2 | Olaparib | In vitro, Cell lines, Xenografts | FDA-approved |
| Talazoparib | In vitro, Cell lines, Xenografts | FDA-approved | |
| Niraparib | In vitro, Cell lines, Xenografts | FDA-approved | |
| Rucaparib | In vitro, Cell lines, Xenografts | FDA-approved | |
| Veliparib | In vitro, Cell lines, Xenografts | Phase III |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Xue, Y.; Ba, X.; Wang, R. The Role of 8-oxoG Repair Systems in Tumorigenesis and Cancer Therapy. Cells 2022, 11, 3798. https://doi.org/10.3390/cells11233798
Li C, Xue Y, Ba X, Wang R. The Role of 8-oxoG Repair Systems in Tumorigenesis and Cancer Therapy. Cells. 2022; 11(23):3798. https://doi.org/10.3390/cells11233798
Chicago/Turabian StyleLi, Chunshuang, Yaoyao Xue, Xueqing Ba, and Ruoxi Wang. 2022. "The Role of 8-oxoG Repair Systems in Tumorigenesis and Cancer Therapy" Cells 11, no. 23: 3798. https://doi.org/10.3390/cells11233798
APA StyleLi, C., Xue, Y., Ba, X., & Wang, R. (2022). The Role of 8-oxoG Repair Systems in Tumorigenesis and Cancer Therapy. Cells, 11(23), 3798. https://doi.org/10.3390/cells11233798






