CircZXDC Promotes Vascular Smooth Muscle Cell Transdifferentiation via Regulating miRNA-125a-3p/ABCC6 in Moyamoya Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Specimen Collection and Ethics Statement
2.2. Network Establishing
2.3. Reverse Transcription-Quantitative PCR (RT-qPCR)
2.4. Cell Culture
2.5. ROC Curve
2.6. Cell TRANSFECTION
2.7. Dual-Luciferase Reporter Assay
2.8. OGD /RP Model
2.9. Immunohistochemistry (IHC)
2.10. Hematoxylin–Eosin (H&E) Staining
2.11. Immunofluorescence
2.12. Western Blotting
2.13. Cell Counting Kit-8 (CCK-8) Assay
2.14. Transwell Assay
2.15. Wound-Healing Assay
3. Result
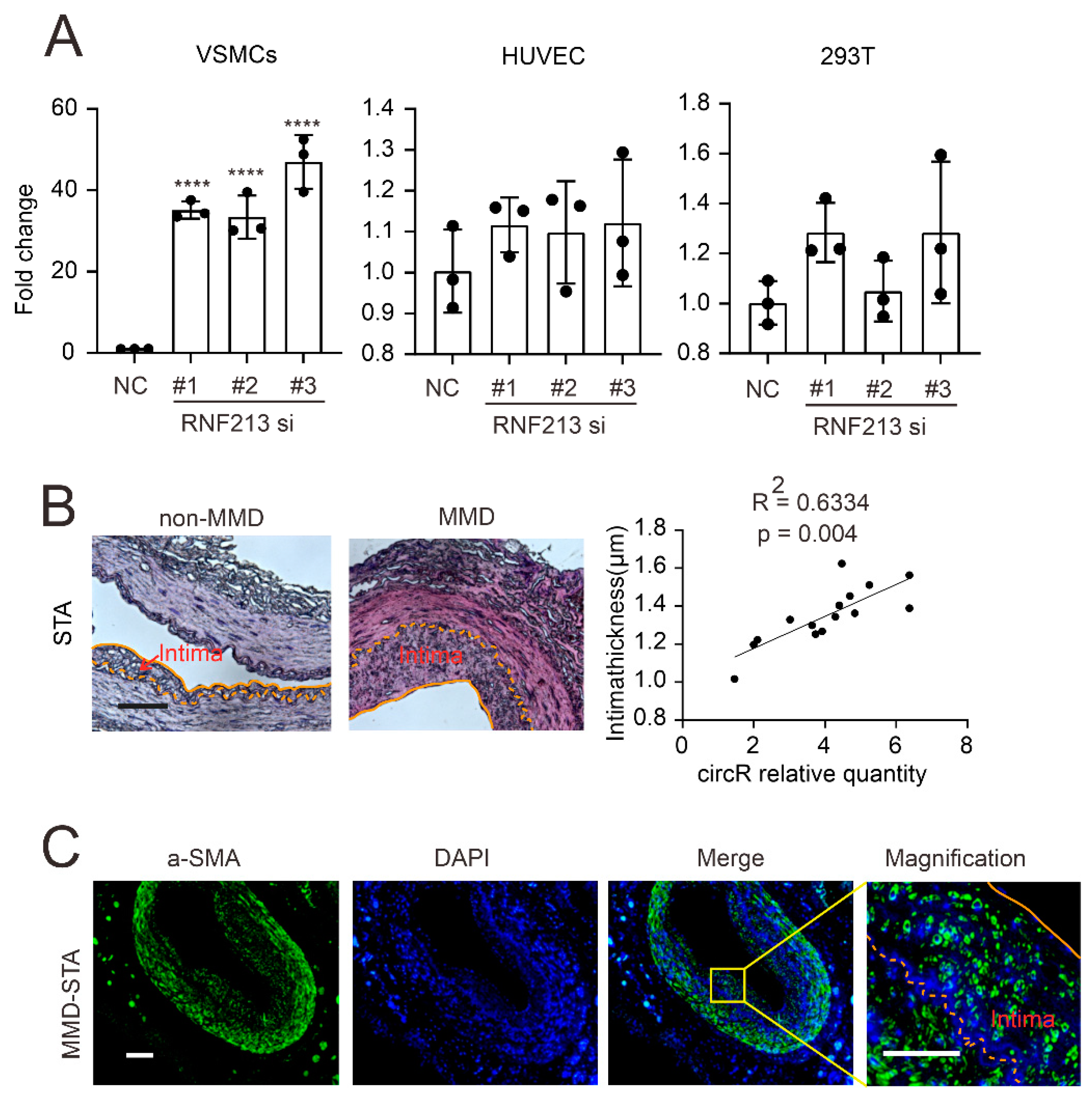
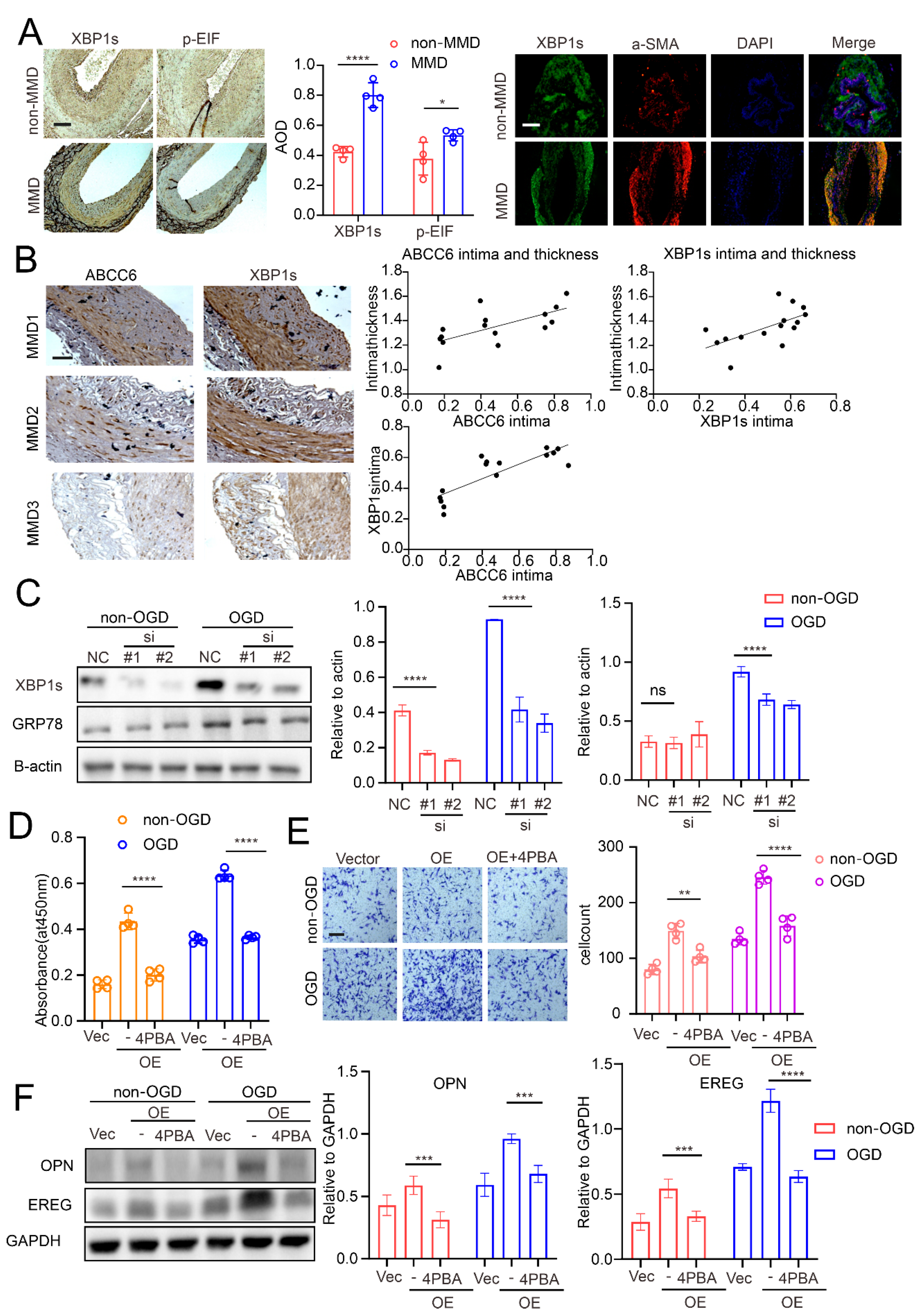
3.1. A CircZXDC–miR-125a-3p–ABCC6 Axis Was Identified in the MMD Vessels, and the Plasma Level of CircZXDC Could Be Used as A Diagnostic Biomarker of MMD
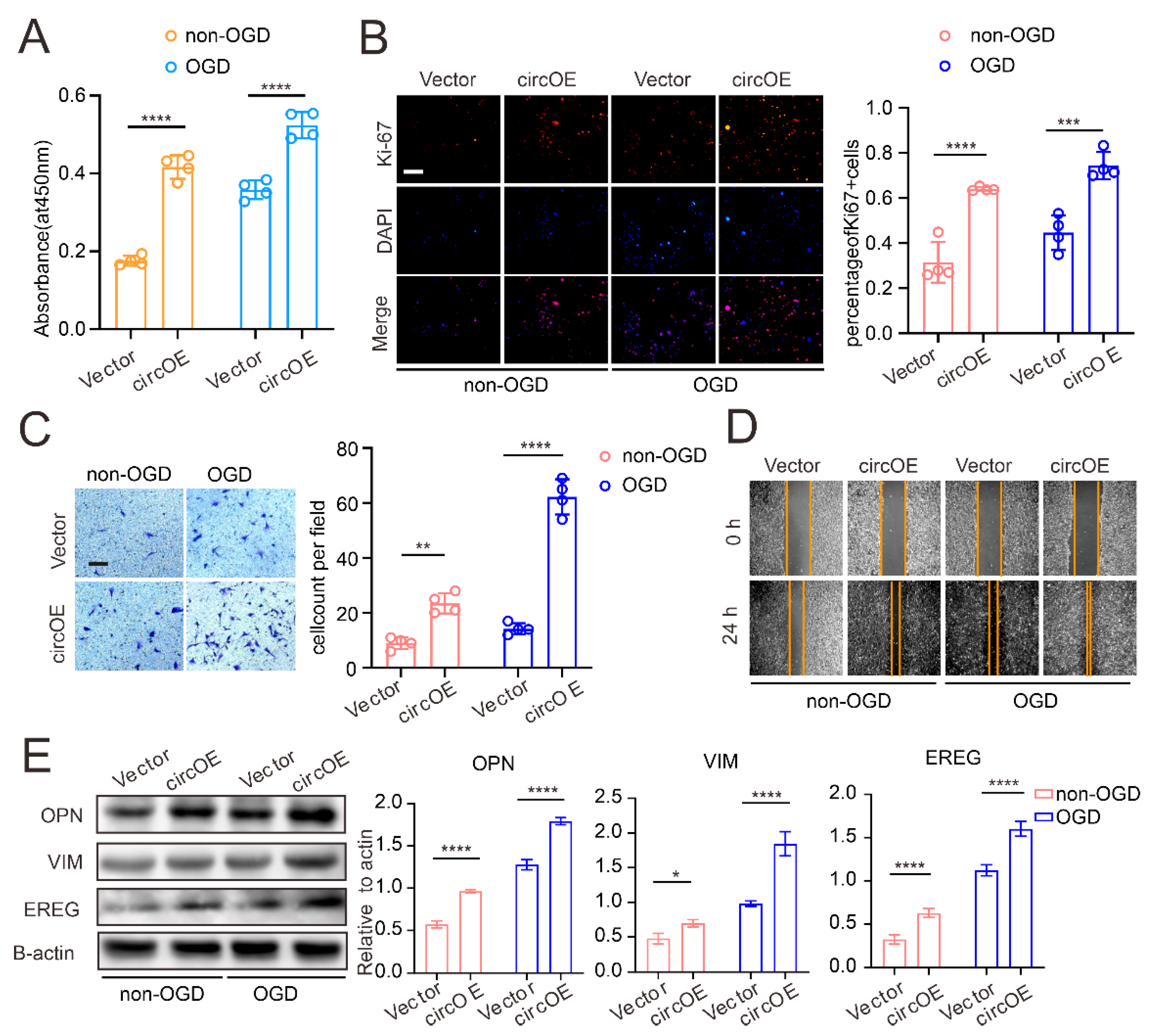
3.2. CircZXDC–miR-125a-3p Axis Regulates VSMC Transdifferentiation towards the Synthetic Phenotype
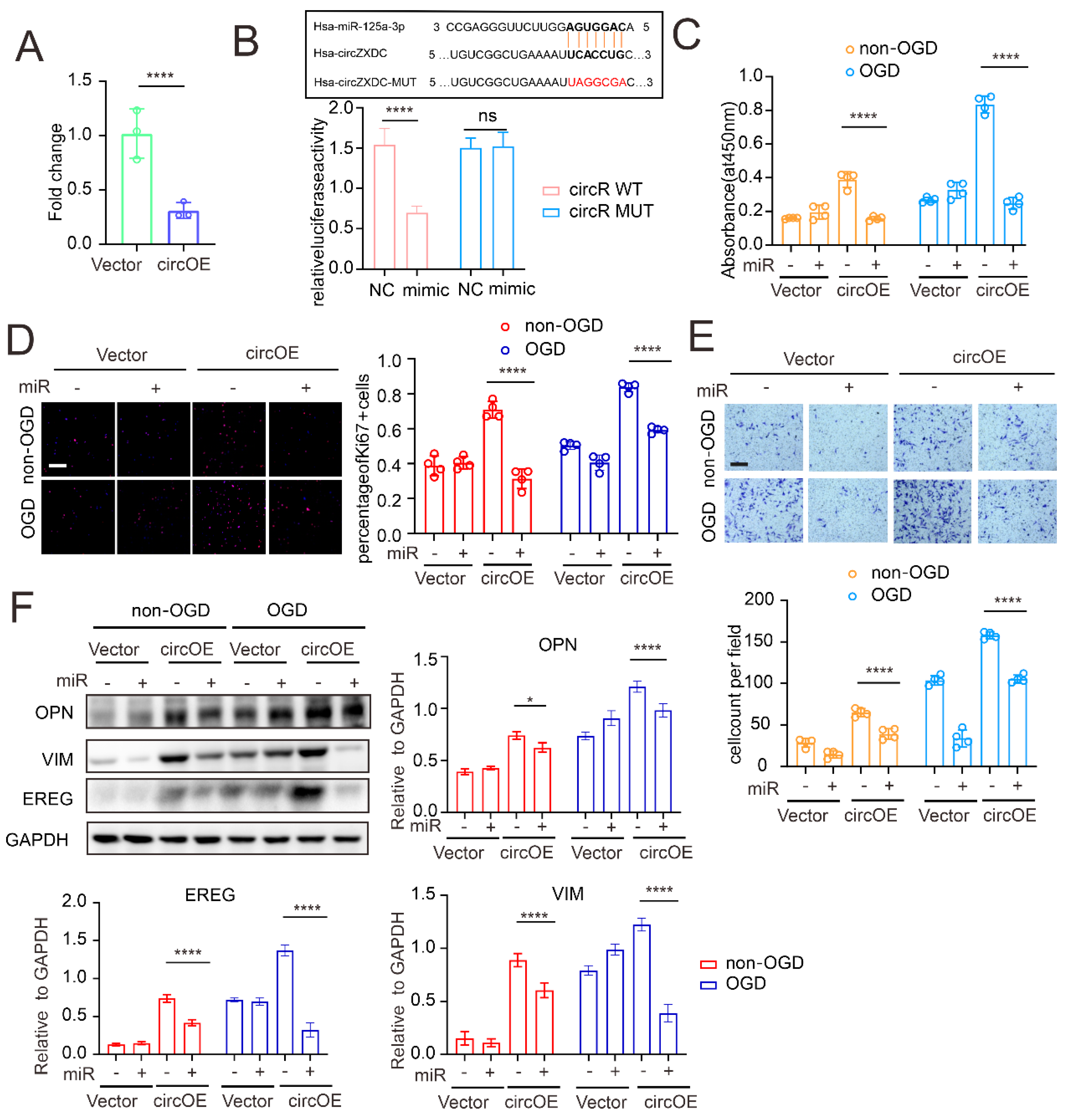
3.3. miR-125a-3p Regulates VSMC Transdifferentiation via ABCC6 mRNA Sponging
3.4. ABCC6 Affects VSMC Transdifferentiation via Regulating Endoplasmic Reticulum Stress
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, J.S. Moyamoya Disease: Epidemiology, Clinical Features, and Diagnosis. J. Stroke 2016, 18, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhou, G.; Feng, J.; Chen, L.; Liu, G.; Wang, J.; Wang, Q.; Yu, J.; Yang, X.; Yang, Z.; et al. Incidence and prevalence of moyamoya disease in urban China: A nation wide retrospective cohort study. Stroke Vasc. Neurol. 2021, 6, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, Y.; Zhang, D.; Wang, R.; Zhang, Y.; Wang, S.; Yu, L.; Lu, C.; Liu, F.; Zhou, J.; et al. RNF213 as the major susceptibility gene for Chinese patients with moyamoya disease and its clinical relevance. J. Neurosurg. 2017, 126, 1106–1113. [Google Scholar] [CrossRef] [PubMed]
- Kamada, F.; Aoki, Y.; Narisawa, A.; Abe, Y.; Komatsuzaki, S.; Kikuchi, A.; Kanno, J.; Niihori, T.; Ono, M.; Ishii, N.; et al. A genome-wide association study identifies RNF213 as the first Moyamoya disease gene. J. Hum. Genet. 2011, 56, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Hitomi, T.; Kobayashi, H.; Harada, K.H.; Koizumi, A. Distribution of moyamoya disease susceptibility polymorphism p.R4810K in RNF213 in East and Southeast Asian populations. Neurol. Med.-Chir. 2012, 52, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Zhang, H.; Liu, Q.; Zhang, S.; Hu, L.; Deng, F. Clinical and experimental pathology of Moyamoya disease. Chin. Med. J. 2013, 116, 1845–1849. [Google Scholar]
- Achrol, A.S.; Guzman, R.; Lee, M.; Steinberg, G.K. Pathophysiology and genetic factors in moyamoya disease. Neurosurg. Focus 2009, 26, E4. [Google Scholar] [CrossRef]
- Masuda, J.; Ogata, J.; Yutani, C. Smooth muscle cell proliferation and localization of macrophages and T cells in the occlusive intracranial major arteries in moyamoya disease. Stroke 1993, 24, 1960–1967. [Google Scholar] [CrossRef]
- Tokairin, K.; Hamauchi, S.; Ito, M.; Kazumata, K.; Sugiyama, T.; Nakayama, N.; Kawabori, M.; Osanai, T.; Houkin, K. Vascular Smooth Muscle Cell Derived from IPS Cell of Moyamoya Disease—Comparative Characterization with Endothelial Cell Transcriptome. J. Stroke Cerebrovasc. Dis. 2020, 29, 105305. [Google Scholar] [CrossRef]
- Blecharz-Lang, K.G.; Prinz, V.; Burek, M.; Frey, D.; Schenkel, T.; Krug, S.M.; Fromm, M.; Vajkoczy, P. Gelatinolytic activity of autocrine matrix metalloproteinase-9 leads to endothelial de-arrangement in Moyamoya disease. J. Cereb. Blood Flow Metab. 2018, 38, 1940–1953. [Google Scholar] [CrossRef]
- Kobayashi, H.; Kabata, R.; Kinoshita, H.; Morimoto, T.; Ono, K.; Takeda, M.; Choi, J.; Okuda, H.; Liu, W.; Harada, K.H.; et al. Rare variants in RNF213, a susceptibility gene for moyamoya disease, are found in patients with pulmonary hypertension and aggravate hypoxia-induced pulmonary hypertension in mice. Pulm. Circ. 2018, 8, 2045894018778155. [Google Scholar]
- Koizumi, A.; Kobayashi, H.; Hitomi, T.; Harada, K.H.; Habu, T.; Youssefian, S. A new horizon of moyamoya disease and associated health risks explored through RNF213. Environ. Health Prev. Med. 2016, 21, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Morito, D.; Takashima, S.; Mineharu, Y.; Kobayashi, H.; Hitomi, T.; Hashikata, H.; Matsuura, N.; Yamazaki, S.; Toyoda, A.; et al. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS ONE 2011, 6, e22542. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.S. Single Nucleotide Polymorphism in Patients with Moyamoya Disease. J. Korean Neurosurg. Soc. 2015, 57, 422–427. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Z.; Liu, W.; Xiong, Y.; Sun, W.; Huang, X.; Jiang, Y.; Ni, G.; Sun, W.; Zhou, L.; et al. Impacts and interactions of PDGFRB, MMP-3, TIMP-2, and RNF213 polymorphisms on the risk of Moyamoya disease in Han Chinese human subjects. Gene 2013, 526, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Lu, Q.; Huang, Q.; Yang, P.; Hong, B.; Xu, Y.; Zhao, W.; Liu, J.; Li, Q. Serum miRNA signature in Moyamoya disease. PLoS ONE 2014, 9, e102382. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Gao, F.; Zhang, D.; Wang, S.; Zhang, Y.; Wang, R.; Zhao, J. Altered expression of circular RNAs in Moyamoya disease. J. Neurol. Sci. 2017, 381, 25–31. [Google Scholar] [CrossRef]
- Wang, G.; Wen, Y.; Faleti, O.D.; Zhao, Q.; Liu, J.; Zhang, G.; Li, M.; Qi, S.; Feng, W.; Lyu, X. A Panel of Exosome-Derived miRNAs of Cerebrospinal Fluid for the Diagnosis of Moyamoya Disease. Front. Neurosci. 2020, 14, 548278. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Ma, Q.; Li, L.; Yu, B.; Jiao, L.; Han, Z.; Zhao, H.; Li, G.; Ma, Y.; Luo, Y. Circular RNA profiling of neutrophil transcriptome provides insights into asymptomatic Moyamoya disease. Brain Res. 2019, 1719, 104–112. [Google Scholar] [CrossRef]
- Issah, M.A.; Wu, D.; Zhang, F.; Zheng, W.; Liu, Y.; Fu, H.; Zhou, H.; Chen, R.; Shen, J. Epigenetic modifications in acute myeloid leukemia: The emerging role of circular RNAs (Review). Int. J. Oncol. 2021, 59, 107. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Kanamori, F.; Yokoyama, K.; Ota, A.; Yoshikawa, K.; Karnan, S.; Maruwaka, M.; Shimizu, K.; Ota, S.; Uda, K.; Araki, Y.; et al. Transcriptome-wide analysis of intracranial artery in patients with moyamoya disease showing upregulation of immune response, and down regulation of oxidative phosphorylation and DNA repair. Neurosurg. Focus 2021, 51, E3. [Google Scholar] [CrossRef] [PubMed]
- Frid, M.G.; Dempsey, E.C.; Durmowicz, A.G.; Stenmark, K.R. Smooth muscle cell heterogeneity in pulmonary and systemic vessels. Importance in vascular disease. Arterioscler. Thromb. Vasc. Biol. 1997, 17, 1203–1209. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-J.; Huang, C.-C.; Chen, P.-R.; Lai, Y.-J. Remodeling Matrix Synthesis in a Rat Model of Aortocaval Fistula and the Cyclic Stretch: Impaction in Pulmonary Arterial Hypertension-Congenital Heart Disease. Int. J. Mol. Sci. 2020, 21, 4676. [Google Scholar] [CrossRef]
- Ma, X.; Huang, Y.; He, X.; Zhang, X.; Liu, Y.; Yang, Y.; Yue, P.; Liu, Y.; Gan, C.; Shu, K.; et al. Endothelial Cell-Derived Let-7c-Induced TLR7 Activation on Smooth Muscle Cell Mediate Vascular Wall Remodeling in Moyamoya Disease. Transl. Stroke Res. 2022. [Google Scholar] [CrossRef]
- Armentano, M.F.; Caterino, M.; Miglionico, R.; Ostuni, A.; Pace, M.C.; Cozzolino, F.; Monti, M.; Milella, L.; Carmosino, M.; Pucci, P.; et al. New insights on the functional role of URG7 in the cellular response to ER stress. Biol. Cell 2018, 110, 147–158. [Google Scholar] [CrossRef]
- Hosen, M.J.; Coucke, P.J.; Le Saux, O.; De Paepe, A.; Vanakker, O.M. Perturbation of specific pro-mineralizing signalling pathways in human and murine pseudoxanthoma elasticum. Orphanet J. Rare. Dis. 2014, 9, 66. [Google Scholar] [CrossRef]
- Pan, H.; Zhao, F.; Yang, Y.; Chang, N. Overexpression of long non-coding RNA SNHG16 against cerebral ischemia-reperfusion injury through miR-106b-5p/LIMK1 axis. Life Sci. 2020, 254, 117778. [Google Scholar] [CrossRef]
- Guo, M.-M.; Qu, S.-B.; Lu, H.-L.; Wang, W.-B.; He, M.-L.; Su, J.-L.; Chen, J.; Wang, Y. Biochanin A Alleviates Cerebral Ischemia/Reperfusion Injury by Suppressing Endoplasmic Reticulum Stress-Induced Apoptosis and p38MAPK Signaling Pathway In Vivo and In Vitro. Front. Endocrinol. 2021, 12, 646720. [Google Scholar] [CrossRef]
- Zhang, L.; Rashad, S.; Zhou, Y.; Niizuma, K.; Tominaga, T. RNF213 loss of function reshapes vascular transcriptome and spliceosome leading to disrupted angiogenesis and aggravated vascular inflammatory responses. J. Cereb. Blood Flow Metab. 2022, 42, 2107–2122. [Google Scholar] [CrossRef]
- Mertens, R.; Graupera, M.; Gerhardt, H.; Bersano, A.; Tournier-Lasserve, E.; Mensah, M.A.; Mundlos, S.; Vajkoczy, P. The Genetic Basis of Moyamoya Disease. Transl. Stroke Res. 2022, 13, 25–45. [Google Scholar] [CrossRef] [PubMed]
- Al-Kandari, W.; Jambunathan, S.; Navalgund, V.; Koneni, R.; Freer, M.; Parimi, N.; Mudhasani, R.; Fontes, J.D. ZXDC, a novel zinc finger protein that binds CIITA and activates MHC gene transcription. Mol. Immunol. 2007, 44, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, L.D.; Clark, M.J.; Patwardhan, A.; Chandratillake, G.; Garcia, S.; Chen, R.; Morgan, A.A.; Leng, N.; Kirk, S.; Chen, R.; et al. Disease Variant Landscape of a Large Multiethnic Population of Moyamoy a Patients by Exome Sequencing. G3 Genes|Genomes|Genet. 2015, 6, 41–49. [Google Scholar] [CrossRef]
- Meng, E.; Deng, J.; Jiang, R.; Wu, H. CircRNA-Encoded Peptides or Proteins as New Players in Digestive System Neoplasms. Front. Oncol. 2022, 12, 944159. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Jia, L.; Luo, L.; Xu, X.; Xiang, Y.; Ren, Y.; Ren, D.; Shen, L.; Liang, T. Critical Roles of Circular RNA in Tumor Metastasis via Acting as a Sponge of miRNA/isomiR. Int. J. Mol. Sci. 2022, 23, 7024. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E. A 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip. Rev. RNA 2018, 9, e1478. [Google Scholar] [CrossRef]
- Xiao, M.-S.; Wilusz, J.E. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3’ ends. Nucleic Acids. Res. 2019, 47, 8755–8769. [Google Scholar] [CrossRef]
- Chen, L.-L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef]
- Ahmed, S.P.; Castresana, J.S.; Shahi, M.H. Role of Circular RNA in Brain Tumor Development. Cells 2022, 11, 2130. [Google Scholar] [CrossRef]
- Shoda, K.; Kuwano, Y.; Ichikawa, D.; Masuda, K. circRNA: A New Biomarker and Therapeutic Target for Esophageal Cancer. Biomedicines 2022, 10, 1643. [Google Scholar] [CrossRef]
- Thery, F.; Martina, L.; Asselman, C.; Zhang, Y.; Vessely, M.; Repo, H.; Sedeyn, K.; Moschonas, G.D.; Bredow, C.; Teo, Q.W.; et al. Ring finger protein 213 assembles into a sensor for ISGylated proteins with antimicrobial activity. Nat. Commun. 2021, 12, 5772. [Google Scholar] [CrossRef] [PubMed]
- Prosdocimo, D.A.; Sabeh, M.K.; Jain, M.K. Kruppel-like factors in muscle health and disease. Trends Cardiovasc. Med. 2015, 25, 278–287. [Google Scholar] [CrossRef]
- Miano, J.M.; Fisher, E.A.; Majesky, M.W. Fate and State of Vascular Smooth Muscle Cells in Atherosclerosis. Circulation 2021, 143, 2110–2116. [Google Scholar] [CrossRef] [PubMed]
- Furmanik, M.; Chatrou, M.; van Gorp, R.; Akbulut, A.; Willems, B.; Schmidt, H.; van Eys, G.; Bochaton-Piallat, M.-L.; Proudfoot, D.; Biessen, E.; et al. Reactive Oxygen-Forming Nox5 Links Vascular Smooth Muscle Cell Phenotypic Switching and Extracellular Vesicle-Mediated Vascular Calcification. Circ. Res. 2020, 127, 911–927. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huang, J.; Wang, Y.; Shen, W. Transcriptome analysis revealed a two-step transformation of vascular smooth muscle cells to macrophage-like cells. Atherosclerosis 2022, 346, 26–35. [Google Scholar] [CrossRef]
- Lauf, P.K.; Sharma, N.; Adragna, N.C. Kinetic studies of K-Cl cotransport in cultured rat vascular smooth muscle cells. Am. J. Physiol.-Cell Physiol. 2019, 316, C274–C284. [Google Scholar] [CrossRef]
- Kang, H.-S.; Kim, J.H.; Phi, J.H.; Kim, Y.-Y.; Kim, J.E.; Wang, K.-C.; Cho, B.-K.; Kim, S.-K. Plasma matrix metalloproteinases, cytokines and angiogenic factors in moyamoya disease. J. Neurol. Neurosurg. Psychiatry 2010, 81, 673–678. [Google Scholar] [CrossRef]
- Chen, Y.; Tang, M.; Li, H.; Liu, H.; Wang, J.; Huang, J. TGFβ1 as a Predictive Biomarker for Collateral Formation within Ischemic Moyamoya Disease. Front. Neurol. 2022, 13, 899470. [Google Scholar] [CrossRef]
- Fang, Y.-C.; Wei, L.-F.; Hu, C.-J.; Tu, Y.-K. Pathological Circulating Factors in Moyamoya Disease. Int. J. Mol. Sci. 2021, 22, 1696. [Google Scholar] [CrossRef]
- Wang, C.; Luo, Y.; Tang, H.; Yan, Y.; Chang, X.; Zhao, R.; Li, Q.; Yang, P.; Hong, B.; Xu, Y.; et al. Hsa_circ_0031608: A Potential Modulator of VSMC Phenotype in the Rupture of Intracranial Aneurysms. Front. Mol. Neurosci. 2022, 15, 842865. [Google Scholar] [CrossRef]
- Zhou, W.; Feng, Q.; Cheng, M.; Zhang, D.; Jin, J.; Zhang, S.; Bai, Y.; Xu, J. LncRNA H19 sponges miR-103-3p to promote the high phosphorus-induced osteoblast phenotypic transition of vascular smooth muscle cells by upregulating Runx2. Cell Signal. 2022, 91, 110220. [Google Scholar] [CrossRef]
- Wei, W.; Zhou, Y.J.; Shen, J.L.; Lu, L.; Lv, X.R.; Lu, T.T.; Xu, P.T.; Xue, X.H. The Compatibility of Alisma and Atractylodes Affects the Biological Behaviours of VSMCs by Inhibiting the miR-128-5p/p21 Gene. Evid.-Based Complement. Altern. Med. 2022, 2022, 7617258. [Google Scholar] [CrossRef]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar] [CrossRef]
- Szeri, F.; Niaziorimi, F.; Donnelly, S.; Orndorff, J.; van de Wetering, K. Generation of fully functional fluorescent fusion proteins to gain insights into ABCC6 biology. FEBS Lett. 2021, 595, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Ostuni, A.; Carmosino, M.; Miglionico, R.; Abruzzese, V.; Martinelli, F.; Russo, D.; Laurenzana, I.; Petillo, A.; Bisaccia, F. Inhibition of ABCC6 Transporter Modifies Cytoskeleton and Reduces Motility of HepG2 Cells via Purinergic Pathway. Cells 2020, 9, 1410. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhou, H.; Zhou, X.; Yang, L.; Xiong, Y.; Zhang, L. Comprehensive Analysis of Endoplasmic Reticulum Stress in Intracranial Aneurysm. Front. Cell. Neurosci. 2022, 16, 865005. [Google Scholar] [CrossRef]
- Efentakis, P.; Molitor, M.; Kossmann, S.; Bochenek, M.L.; Wild, J.; Lagrange, J.; Finger, S.; Jung, R.; Karbach, S.; Schäfer, K.; et al. Tubulin-folding cofactor E deficiency promotes vascular dysfunction by increased endoplasmic reticulum stress. Eur. Heart J. 2022, 43, 488–500. [Google Scholar] [CrossRef]
- Zhang, Y.; He, L.; Tu, M.; Huang, M.; Chen, Y.; Pan, D.; Peng, J.; Shen, X. The ameliorative effect of terpinen-4-ol on ER stress-induced vascular calcification depends on SIRT1-mediated regulation of PERK acetylation. Pharmacol. Res. 2021, 170, 105629. [Google Scholar] [CrossRef]
- Ahmed, S.; Habu, T.; Kim, J.; Okuda, H.; Oikawa, S.; Murata, M.; Koizumi, A.; Kobayashi, H. Suppression of RNF213, a susceptibility gene for moyamoya disease, inhibits endoplasmic reticulum stress through SEL1L upregulation. Biochem. Biophys. Res. Commun. 2022, 609, 62–68. [Google Scholar] [CrossRef]
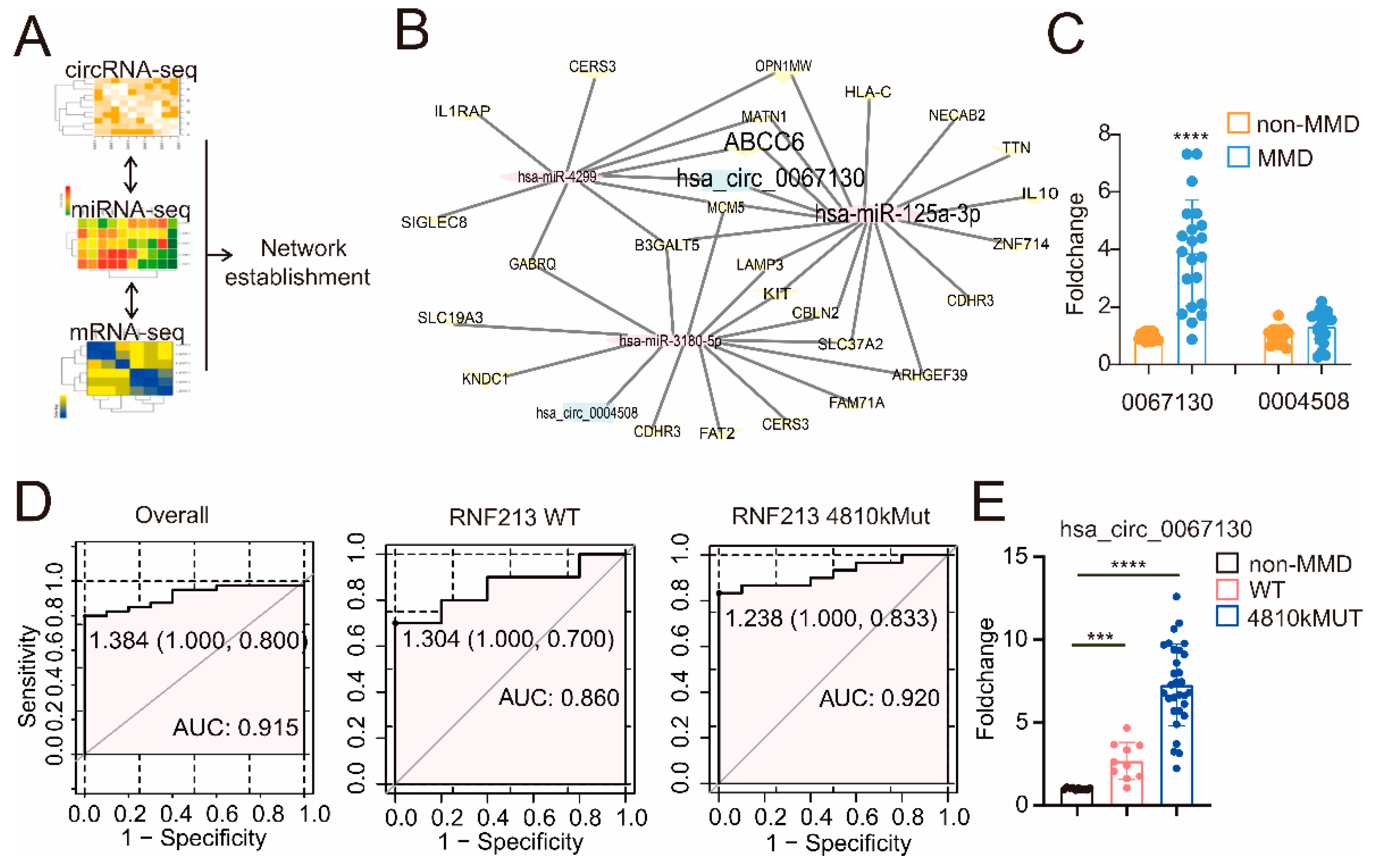

| CircRNA | miRNA | mRNA |
|---|---|---|
| hsa_circ_0004508 | hsa-miR-4299 | ABCC6 |
| hsa_circ_0004508 | hsa-miR-4299 | SIGLEC8 |
| hsa_circ_0004508 | hsa-miR-4299 | B3GALT5 |
| hsa_circ_0004508 | hsa-miR-4299 | GABRQ |
| hsa_circ_0004508 | hsa-miR-4299 | OPN1MW |
| hsa_circ_0004508 | hsa-miR-4299 | MCM5 |
| hsa_circ_0004508 | hsa-miR-4299 | CERS3 |
| hsa_circ_0004508 | hsa-miR-4299 | IL1RAP |
| hsa_circ_0004508 | hsa-miR-4299 | MATN1 |
| hsa_circ_0067130 | hsa-miR-125a-3p | ABCC6 |
| hsa_circ_0067130 | hsa-miR-125a-3p | B3GALT5 |
| hsa_circ_0067130 | hsa-miR-125a-3p | NECAB2 |
| hsa_circ_0067130 | hsa-miR-125a-3p | MCM5 |
| hsa_circ_0067130 | hsa-miR-125a-3p | CBLN2 |
| hsa_circ_0067130 | hsa-miR-125a-3p | IL10 |
| hsa_circ_0067130 | hsa-miR-125a-3p | HLA-C |
| hsa_circ_0067130 | hsa-miR-125a-3p | ARHGEF39 |
| hsa_circ_0067130 | hsa-miR-125a-3p | MATN1 |
| hsa_circ_0067130 | hsa-miR-125a-3p | ZNF714 |
| hsa_circ_0067130 | hsa-miR-125a-3p | LAMP3 |
| hsa_circ_0067130 | hsa-miR-125a-3p | OPN1MW |
| hsa_circ_0067130 | hsa-miR-125a-3p | KIT |
| hsa_circ_0067130 | hsa-miR-125a-3p | SLC37A2 |
| hsa_circ_0067130 | hsa-miR-125a-3p | SLC19A3 |
| hsa_circ_0067130 | hsa-miR-125a-3p | CDHR3 |
| hsa_circ_0067130 | hsa-miR-125a-3p | TTN |
| hsa_circ_0067130 | hsa-miR-3180-5p | KNDC1 |
| hsa_circ_0067130 | hsa-miR-3180-5p | B3GALT5 |
| hsa_circ_0067130 | hsa-miR-3180-5p | FAM71A |
| hsa_circ_0067130 | hsa-miR-3180-5p | MCM5 |
| hsa_circ_0067130 | hsa-miR-3180-5p | CBLN2 |
| hsa_circ_0067130 | hsa-miR-3180-5p | FAT2 |
| hsa_circ_0067130 | hsa-miR-3180-5p | ARHGEF39 |
| hsa_circ_0067130 | hsa-miR-3180-5p | LAMP3 |
| hsa_circ_0067130 | hsa-miR-3180-5p | GABRQ |
| hsa_circ_0067130 | hsa-miR-3180-5p | KIT |
| hsa_circ_0067130 | hsa-miR-3180-5p | CERS3 |
| hsa_circ_0067130 | hsa-miR-3180-5p | SLC37A2 |
| hsa_circ_0067130 | hsa-miR-3180-5p | SLC19A3 |
| hsa_circ_0067130 | hsa-miR-3180-5p | CDHR3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Huang, Y.; Zhang, X.; Ma, X.; He, X.; Gan, C.; Zou, X.; Wang, S.; Shu, K.; Lei, T.; et al. CircZXDC Promotes Vascular Smooth Muscle Cell Transdifferentiation via Regulating miRNA-125a-3p/ABCC6 in Moyamoya Disease. Cells 2022, 11, 3792. https://doi.org/10.3390/cells11233792
Liu Y, Huang Y, Zhang X, Ma X, He X, Gan C, Zou X, Wang S, Shu K, Lei T, et al. CircZXDC Promotes Vascular Smooth Muscle Cell Transdifferentiation via Regulating miRNA-125a-3p/ABCC6 in Moyamoya Disease. Cells. 2022; 11(23):3792. https://doi.org/10.3390/cells11233792
Chicago/Turabian StyleLiu, Yuan, Yimin Huang, Xincheng Zhang, Xiaopeng Ma, Xuejun He, Chao Gan, Xin Zou, Sheng Wang, Kai Shu, Ting Lei, and et al. 2022. "CircZXDC Promotes Vascular Smooth Muscle Cell Transdifferentiation via Regulating miRNA-125a-3p/ABCC6 in Moyamoya Disease" Cells 11, no. 23: 3792. https://doi.org/10.3390/cells11233792
APA StyleLiu, Y., Huang, Y., Zhang, X., Ma, X., He, X., Gan, C., Zou, X., Wang, S., Shu, K., Lei, T., & Zhang, H. (2022). CircZXDC Promotes Vascular Smooth Muscle Cell Transdifferentiation via Regulating miRNA-125a-3p/ABCC6 in Moyamoya Disease. Cells, 11(23), 3792. https://doi.org/10.3390/cells11233792







