Abstract
The metabolites produced by the gut microbiota have been reported as crucial agents against obesity; however, their key targets have not been revealed completely in complex microbiome systems. Hence, the aim of this study was to decipher promising prebiotics, probiotics, postbiotics, and more importantly, key target(s) via a network pharmacology approach. First, we retrieved the metabolites related to gut microbes from the gutMGene database. Then, we performed a meta-analysis to identify metabolite-related targets via the similarity ensemble approach (SEA) and SwissTargetPrediction (STP), and obesity-related targets were identified by DisGeNET and OMIM databases. After selecting the overlapping targets, we adopted topological analysis to identify core targets against obesity. Furthermore, we employed the integrated networks to microbiota–substrate–metabolite–target (MSMT) via R Package. Finally, we performed a molecular docking test (MDT) to verify the binding affinity between metabolite(s) and target(s) with the Autodock 1.5.6 tool. Based on holistic viewpoints, we performed a filtering step to discover the core targets through topological analysis. Then, we implemented protein–protein interaction (PPI) networks with 342 overlapping target, another subnetwork was constructed with the top 30% degree centrality (DC), and the final core networks were obtained after screening the top 30% betweenness centrality (BC). The final core targets were IL6, AKT1, and ALB. We showed that the three core targets interacted with three other components via the MSMT network in alleviating obesity, i.e., four microbiota, two substrates, and six metabolites. The MDT confirmed that equol (postbiotics) converted from isoflavone (prebiotics) via Lactobacillus paracasei JS1 (probiotics) can bind the most stably on IL6 (target) compared with the other four metabolites (3-indolepropionic acid, trimethylamine oxide, butyrate, and acetate). In this study, we demonstrated that the promising substate (prebiotics), microbe (probiotics), metabolite (postbiotics), and target are suitable for obsesity treatment, providing a microbiome basis for further research.
1. Introduction
Obesity is an serious health issue globally because it is related to diverse diseases, such as diabetes, atherosclerosis, hypertension, heart attack, and even cancers [1]. The main cause of obesity a persistent between consumed energy and expended energy for a long period of time [2]. A criterion used to assess the severity of obesity is body mass index (BMI), with BMI values of 30.00 or more indicating obesity [3]. At present, obesity is prevalent in all ages, with the worldwide prevalence of obsesity expected to increase to 573 million by 2030 [4].
Common medications administered for short-term weight management include phentermine and diethylpropion as appetite suppressants [5]. Orlistat is approved for long-term oral administration, acting as a pancreatic lipase inhibitor to interrupt dietary fat absorption [6]. However, these drugs are associated with negative side effects, such as headache, nausea, dry mouth, constipation, diarrhea, and even anxiety [7].
Recently, a report demonstrated that metabolites from the gut microbiota can exert favorable efficacy to ameliorate metabolic disorders, including obesity [8]. Another report suggested that metabolites produced by the gut microbiota act as regulators to maintain energy balance in host system [9]. The gut microbiota is related to the etiology of obesity and its associated metabolic disorders, for instance, by regulating the fermentation of dietary polysaccharides, fat consumption, and even obesity [10]. In addition, a study showed that utilization of prebiotics, probiotics, and synbiotics (the mixture of prebiotics and probiotics) may affect the production of chemical messengers (hormones and neurotransmitters) and inflammatory elements, interrupting diet intake stimulators that result in obesity [11]. Therefore, favorable dietary food (prebiotics) and gut microbes (probiotics) may exert positive effects on obesity. As mentioned above, prebiotics (a precursor of postbiotics) converted into postbiotics (defined as metabolites) via probiotics (known as gut microbiota) have been documented as critical constituents for the treatment of obesity; however, their core targets have not been completely elucidated in highly complicated microbiome systems. Thus, we pioneered potential prebiotics, probiotics, postbiotics, and more importantly, key targets to establish the four key components against obesity.
Flavonoid-abundant foods, such as fruits and vegetables, exert therapeutic effects, including anti-inflammation, antioxidant, hypertension, and anti-obesity actions, a relationship that is expounded by characteristics of the gut microbiome to a certain extent [12]. Flavonoid are considered significant nutritional constituents in therapeutics due to their favorable physicochemical properties, providing an improved absorption rate, increased therapeutic capacity, and fewer adverse effects relative to other compounds [13]. The gut–liver axis is a cross junction between the gut, its microbial colonization, and the liver, regulating the signaling pathways tuned by nutritional, genetic, and environmental variables [14]. Therefore, flavonoids with advantageous pharmacokinetic characteristics for use as agents against some diseases, including obesity, might be promising additives produced by the gut microbiota.
Additionally, the construction of multiple biological networks offers prospective insight to elucidate complex pharmacological information, such as microbial interactions, protein–protein interaction (PPI) network analysis, and even topological analysis [15,16]. Specifically, biological network models can serve as a paradigm to uncover the underlying causes of complex diseases [17]. Network pharmacology is an integrated analytical methodology to pioneer significant components (bioactives, proteins, diseases, and genes) [18]. With the development of bioinformatics, network pharmacology can decode the mechanism of action in complex biological systems through interdisciplinary studies, suggesting that network pharmacology is a convergent approach to shift from “one target, one compound” to “multiple targets, multiple compounds” [18,19,20].
We constructed a microbiota–substrate–metabolite–target (MSMT) network to reveal the underlying therapeutic values of the interactions. A previous report suggested that Lactobacillus paracasei JS1, isoflavone, and equol might be significant components in the treatment of skin and intestinal disorders [21]. Another significant factor related to obesity is IL6. This target is a key element exerting pharmacological efficacy against obesity and is better known as a targeted to inhibit obesity [22].
Our description shows that important components, such as prebiotics, probiotics, and postbiotics, can dampen obesity in the microbiome and bed targeted via network pharmacology analysis. Targets with high betweenness centrality (BC) (the route value of the shortest path between nodes) can be used to identify significant nodes (or targets) in the network [23]. Key targets with highest BC values are noteworthy therapeutically relevant candidates that can be used to treat diverse diseases [24]. Hence, we generated PPI networks based on the BC values of each target, and targets with high connectivity values in the networks were considered therapeutic targets against obesity.
Despite insufficient data to elucidate a crucial therapeutic underpinning of obesity etiology, our findings contribute to unraveling the potentiality of an antiobesity effect in complex microbiome systems.
2. Hypothesis
We postulated that targets with a high degree of betweenness centrality (BC) can serve as therapeutic candidates against obesity. Targets with a high correlation degree value in PPI networks based on a high BC value are potential targets for the treatment of obesity. The most stably bound metabolites on a key target in the molecular docking test (MDT) were considered key postbiotics, and microbes produced by postbiotics were defined as key probiotics.
3. Methods and Materials
Biologically substantial datasets with abundant data on the association between ligands and targets enable researchers to employ network pharmacology as an efficient method for drug discovery or development (Table 1). Such web-based datasets are available freely to users to obtain valuable information that can be applied to microbiome and network pharmacology. In this study, we implemented new approach to explore the complex microbiome and its network via public databases and network pharmacology strategies.

Table 1.
List of databases used in the present study.
The metabolites generated by the gut microbiota were identified via gutMGene (http://bio-annotation.cn/gutmgene/) (accessed on 31 May 2022) [25]. We adopted the similarity ensemble approach (SEA) (https://sea.bkslab.org/) (accessed on 31 May 2022) [26] for mining analysis and SwissTargetPrediction (STP) (http://www.swisstargetprediction.ch/) (accessed on 31 May 2022) [27] to search for targets linked to the metabolites. Obesity-responsive targets were identified by DisGeNET (https://www.disgenet.org/) (accessed on 1 June 2022) [28] and OMIM (https://www.omim.org/) (accessed on 1 June 2022) [29]. Crucial targets were utilized to identify the interaction between each node through PPI networks. Then, we constructed MSMT networks for descriptive purposes. Finally, MDT was implemented to evaluate the binding stability between metabolites and targets. The study protocol was conducted as follows.
Step 1: Retrieval of metabolites produced by gut microbiota through gutMGene. The metabolites converted by gut microbes were identified by a subfolder in the downloads section (http://bio-annotation.cn/gutmgene/public/res/gutMGene-human.xlsx) (accessed on 31 May 2022) in gutMGene v1.0, suggesting key metabolites reported to date.
Step 2: Targets associated with the metabolites were mined by SEA and STP databases. The metabolites were selected in simplified molecular input line entry system (SMILES) format to load in the two databases, the format of which was converted by PubChem (https://pubchem.ncbi.nlm.nih.gov/) (accessed on 31 May 2022). Obesity-related targets were identified via DisGeNET (https://www.disgenet.org/) and OMIM (https://www.omim.org/).
Step 3: Selection of intersecting targets between SEA and STP. We selected the intersecting targets to achieve rigor and exactness from the two databases. In detail, an SEA database was constructed by Dr. Shoichet’s group to identify the affinity of compound targets and eventually reveal their binding stability [26]. Additionally, 23 of 30 drug targets suggested by SEA were experimentally confirmed [30]. STP was used as a tool to predict targets for cudraflavone C, a species of flavanols, which were experimentally demonstrated [31]. Thus, we utilized the two databases to enhance the success rate in this study. With the help of VENNY 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/) (accessed on 1 June 2022) Venn diagram, we selected the overlapping targets.
Step 4: Identification of crucial targets among obesity-related targets and the overlapping targets extracted by SEA and STP. We considered intersecting targets to be crucial targets.
Step 5: The crucial targets from Step 4 were analyzed by the String database version 11.5 (https://string-db.org/) (accessed on 1 June 2022) [32], and we adopted PPI networks to identify relationships using the R package.
Step 6: Construction of sub-PPI networks with the highest degree centrality (DC) values in the upper 30% from the PPI networks (Step 5) via R package. DC is determined as the number of edges on each node [33].
Step 7: Construction of a subnetwork with the highest betweenness centrality (BC) values in the top 30% from the sub-PPI networks (Step 6) via R package. BC is a measurement of the influence of node in a network [34]. The highest BC value reflects the relative significance of aspects of biological effect in the networks, i.e., nodes with higher BC values have increased potential therapeutic value against disease [23].
Step 8: Description of MSMT networks via R package. The most important elements against obesity were indicated by the size degree of the circle.
The size of each component (node) describes the number of interactions (edge) in the MSMT networks.
The microbiota, substrate, metabolite, and target were merged to describe their relationships in Microsoft Excel in combination with R package to identify their connectivity. We defined specific compounds of prebiotics as substrates (S) that are pre-metabolites before becoming primary metabolites in the MSMT networks.
Step 9: Initial screening was performed by MDT, with a cutoff less than -6.0 kcal/mol or the lowest Gibbs energy in each complex. The metabolites were downloaded in .sdf format from PubChem (https://pubchem.ncbi.nlm.nih.gov/) and converted to .pdb format via Pymol. The selected .pdb format was converted to .pdbqt format to implement the MDT. The crystal structure of each target was obtained by RCSB PDB (https://www.rcsb.org/) (accessed on 2 June 2022). The MDT was implemented to verify the affinity between metabolites and targets via AutoDockTools-1.5.6 [35].
The docking box size was solidified with x = 40 Å, y = 40 Å, and z = 40 Å. The active site of the crystal structure was formatted with a cubic box in the center: AKT1 (x = 6.313, y = -7.926, z = 17.198) and IL6 (x = 11.213, y = 33.474, z = 11.162). Both hydrophilic and hydrophobic interaction analyses were performed with LigPlot + 2.2. (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/download2.html) (accessed on 3 June 2022) [36].
Step 10: Drug-resemblance and toxicity properties were validated by SwissADME (http://www.swissadme.ch/) (accessed on 4 June 2022) and the ADMETlab web-based tool (https://admetmesh.scbdd.com/) (accessed on 4 June2022) [37]. These two factors are critical elements to facilitate new agents; thus, we assessed their physicochemical values and side effects. The study workflow is represented in Figure 1.
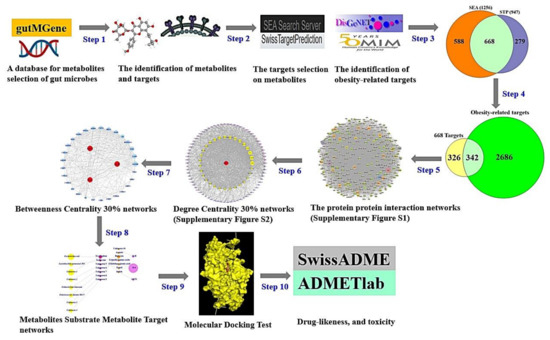
Figure 1.
The workflow of this study.
4. Results
A total of 208 metabolites were obtained via the gutMGene database and identified by the SEA (1256) and STP (947) databases (Supplementary Table S1). A total of 668 targets overlapped between SEA and STP (Figure 2A); a Venn diagram plotter program intersected 342 common targets between the 668 targets and obesity-related targets (3028) (Figure 2B) (Supplementary Table S1).
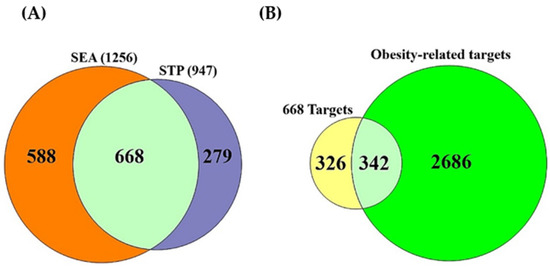
Figure 2.
(A) The common 668 targets between SEA (1256) and STP (947). (B) The common 342 targets between the 668 targets and obesity-related targets (3028).
In the PPI networks, NMUR2, PAM, BRS3, UTS2R, and SSTR4 did not exhibit any interactions with other targets and consisted of 337 nodes and 4492 edges (Supplementary Figure S1). The subnetwork was obtained by selecting the upper top 30% in terms of degree centrality (DC) (Table 2), comprising 106 nodes and 1441 edges (Supplementary Figure S2).

Table 2.
The degree of to 30% DC targets.
After screening the top 30% in terms of betweenness centrality (BC) of the subnetwork, which comprised 32 nodes and 254 edges (Table 3) (Figure 3).

Table 3.
The degree of the top 30% BC targets from Table 1.
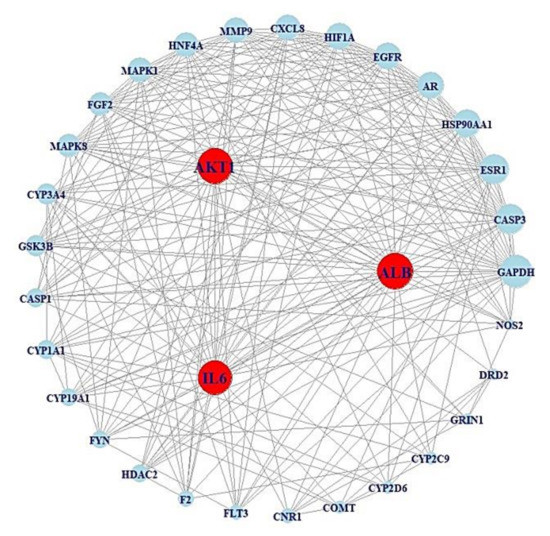
Figure 3.
PPI networks (32 nodes, 254 edges) of the top 30% BC values from Figure 3.
In the BC subnetwork, the targets with the top three BC values were albumin (ALB), interleukin-6 (IL6), and AKT serine/threonine kinase 1 (AKT1), which were considered the core targets connected with microbes to alleviate obesity. The gut microbes directly related to the production of metabolites were identified as Escherichia coli, Lactobacillus paracasei JS1, Eubacterium limosum, and Enterococcus durans M4-5; however, there are additional unknown microbes (Unknown 1, Unknown 2, Unknown 3, and Unknown 4) that produce favorable metabolites against obesity, as indicated in the MSMT networks (Figure 4). Additionally, the beneficial prebiotics to convert into metabolites against obesity were identified as tryptophan and isoflavone, which can produce indole and equol via Escherichia coli and Lactobacillus paracasei JS1, respectively [21,38]. Likewise, there are unknown prebiotics (Unknown 5, Unknown 6, Unknown 7, Unknown 8, and Unknown 9) that may act against obesity. However, the information on prebiotics, probiotics, and postbiotics (Unknown 10) has yet to be confirmed, although ALB was selected as a core antiobesity target.
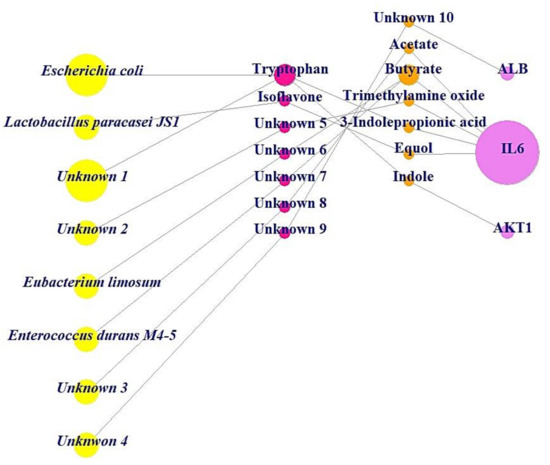
Figure 4.
MSMT networks (25 nodes, 23 edges). Yellow circles: microbiota (probiotics); red circles: substrate (prebiotics); orange circles: metabolites (postbiotics); pink circle: target.
Five metabolites (equol, 3-indolepropionic acid, trimethylamine oxide, butyrate, and acetate) on IL6 (PDB ID: 4NI9) and one metabolite (indole) on AKT1 were selected to perform MDT (Table 4). We observed that the equol–IL6 complex (-7.4 kcal/mol) (Figure 5) docked most stably, indicating its promise as a postbiotic and a target.

Table 4.
Molecular docking test of IL6 (PDB ID: 4NI9) and AKT (PDB ID: 3O96).
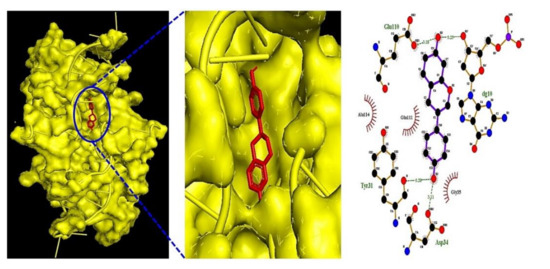
Figure 5.
Equol–IL6 (PDB ID: 4NI9) complex on MDT.
Then, the drug-likeness properties and toxicity of equol were evaluated by the SwissADME and ADMETlab platforms according to Lipinski’s rule of five, including criteria of molecular weight (≤500), H-bond acceptor (≤10), H-bond donor (≤5), MlogP (≤4.15), bioavailability score (>0.1), and topological polar surface area (TPSA) (<140). Our results indicate that equol can be accepted by pharmacokinetics parameters to be assessed as a new agent (Table 5).

Table 5.
Physicochemical properties of equol.
Despite an acceptable therapeutic value, an agent may not be an end product due to unexpected toxicity. Therefore, a drug candidate should exceed the limits of toxicity for further verification. Accordingly, equol was evaluated in terms of hERG blockers, rat oral acute toxicity, eye corrosion, and respiratory toxicity, including LD50 (5.238 mg/kg), via the ADMETlab platform (Table 6). Our observational study suggests that Isoflavone as a prebiotic, Lactobacillus paracasei JS1 as a probiotic, equol as a postbiotic, and IL6 as a target might exert positive effects on obesity.

Table 6.
Toxicity profile of equol.
5. Discussion and Conclusion
Previous studies have demonstrated that the metabolites from the gut microbiota act as significant agents in a wide range of diseases, such as cancer, stroke, irritable bowel syndrome, and even mental disorders, including obesity [39,40]. In this study, we analyzed microbiota–substrate–metabolite–target (MSMT) networks and found that Lactobacillus paracasei JS1, isoflavone, equol, and IL6 exhibited considerable connectivity in the networks, indicating significant antiobesity effects.
Isoflavone is a major compound isolated from soybeans; its pharmacological action has been established against cancers, osteoporosis, metabolic disorders, and neurodegenerative symptoms [41]. Furthermore, equol, as an isoflavone-derived metabolite, has diverse favorable therapeutic effects on human health, such as estrogenic and antioxidant efficacy [42]. An animal test demonstrated that equol can result in a reduction in body weight, white adipose tissue, and depression caused by dietary restriction [43]. More importantly, its therapeutic effects can be confirmed in the context of equol produced by gut microbes. In contrast, in patients who cannot produce equol due to a lack of equol-producing gut microbes, an alternative is to directly administer equol with pharmaceutical forms [44].
A recent report showed that Lactobacillus paracasei JS1 can convert isoflavones into equol via fermentation [45]. Another report demonstrated that equol treatment in collagen-induced arthritis (CIA) inhibited the expression level of IL6 and its receptor at the point of rheumatoid arthritis (RA) [46], suggesting that IL6 is a key regulator of inflammatory levels. The considerable production of IL6 from adipocytes may lead to metabolic disorders, such as obesity; moreover, IL6 cytokine signaling in adipose tissue is associated with hepatic insulin resistance and steatosis [47]. IL6 spontaneously stimulates the secretion of free fatty acid (FFA) by exerting a negative effect on glucose metabolism [47]. Therefore, IL6 inhibition might be an optimal therapeutic target against obesity.
We conducted analysis to identify a key target via topological analysis based on betweenness centrality (BC). In drug network analysis, a drug with a high BC value tends to be associated with several therapeutic applications, with considerable promising for treatment of diverse diseases [36,48]. In particular, a study demonstrated that targets with top 30% BC related to Xiao-Chai-Hu-Tang (Chinese herbal formula) were selected to uncover the mechanism of action against colorectal cancer, providing a theoretical basis for clinical tests [49]. Based on this result, we adopted “top 30% BC” as a threshold in this study.
Network pharmacology research is a powerful tool to monitor targets, pathways, drugs, and diseases in light of the rapid development of databases [50]. We constructed a stepwise workflow to investigate key targets and metabolites to treat obesity by combining public databases. Bioinformatics can be used not only to efficiently mine for drug candidates but also to facilitate drug repurposing [51]. Furthermore, Huangqin decoction was proven an antidiabetic enteritis agent via the combination of network pharmacology and gut microbiota sequencing [52].
Accordingly, we performed a network pharmacology study to evaluate the pharmacological value of the key target identified by a microbiome study. We conducted an observational trial to explicitly elucidate a key target against obesity in complex microbiome networks by reporting up-to-date information. According to the MSTM network results, we selected five potential metabolites and three targets for MDT, with results indicating that equol can bind stably to IL6, which suggests that equol may ameliorate obesity by inhibiting IL6.
Taken together, our results show that isoflavone (prebiotic), Lactobacillus paracasei JS1 (probiotic), equol (postbiotic), and IL6 (Target) are the most crucial components against obesity in current microbiome research. However, accumulation of information concerning the microbiome is subject to some limitations. Due to the limitations of bioinformatics and cheminformatics, we suggest that further preclinical or clinical test should be conducted to specify the four identified elements.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells11182903/s1, Table S1: The 1256 targets identified by SEA; The 947 targets identified by STP; The 668 overlapping targets between SEA and STP; The 3028 obesity-related targets; The 342 final targets. Figure S1: PPI networks (337 nodes and 4492 edges); Figure S2: PPI networks (106 nodes and 1441 edges) and top 30% DC values from Figure S1.
Author Contributions
Conceptualization, K.-T.S. and K.-K.O.; methodology, K.-T.S. and K.-K.O.; software: K.-K.O. and H.G.; validation, K.-T.S., D.J.K., K.-K.O. and H.G.; formal analysis, K.-T.S., K.-K.O. and H.G.; investigation, K.-T.S. and D.J.K.; resources, K.-T.S. and D.J.K.; data curation, K.-K.O. and H.G.; writing—original draft preparation, K.-T.S. and K.-K.O.; writing—review and editing, K.-T.S. and D.J.K.; visualization, R.G., S.-M.W., J.-J.J., S.P.S., S.-B.L., M.-G.C., G.-H.K., B.-H.M., M.-K.J., J.-Y.H., J.-A.E., H.-J.P., S.-J.Y. and M.-R.C.; supervision, K.-T.S. and D.J.K.; project administration, K.-T.S. and D.J.K.; funding acquisition, K.-T.S. and D.J.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Hallym University Research Fund and the Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Education, Science and Technology (NRF-2020R1I1A3073530 and NRF-2020R1A6A1A03043026).
International Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AKT1 | AKT serine/threonine kinase 1 |
| ALB | Albumin |
| BC | Between centrality |
| BMI | Body mass index |
| CIA | Collagen-induced arthritis |
| DC | Degree centrality |
| IL6 | Interleukin-6 |
| MDT | Molecular docking test |
| MSMT | Microbiota–substrate–metabolite–target |
| PPI | Protein–protein interaction |
| RA | Rheumatoid arthritis |
| SEA | Similarity ensemble approach |
| STP | SwissTargetPrediction |
References
- Hruby, A.; Hu, F.B. The Epidemiology of Obesity: A Big Picture. PharmacoEconomics 2015, 33, 673. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Li, H. Obesity: Epidemiology, Pathophysiology, and Therapeutics. Front. Endocrinol. 2021, 12, 1070. [Google Scholar] [CrossRef]
- Weir, C.B.; Jan, A. BMI Classification Percentile And Cut Off Points. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Kelly, T.; Yang, W.; Chen, C.S.; Reynolds, K.; He, J. Global burden of obesity in 2005 and projections to 2030. Int. J. Obes. (2005) 2008, 32, 1431–1437. [Google Scholar] [CrossRef] [PubMed]
- Tchang, B.G.; Aras, M.; Kumar, R.B.; Aronne, L.J. Pharmacologic Treatment of Overweight and Obesity in Adults. In Endotext; MDText.com, Inc.: South Dartmouth, MA, USA, 2021. [Google Scholar]
- Heck, A.M.; Yanovski, J.A.; Calis, K.A. Orlistat, a new lipase inhibitor for the management of obesity. Pharmacotherapy 2000, 20, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Cheung, B.M.Y.; Cheung, T.T.; Samaranayake, N.R. Safety of antiobesity drugs. Ther. Adv. Drug Saf. 2013, 4, 171. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Clément, K.; Sokol, H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 2021, 70, 1174–1182. [Google Scholar] [CrossRef]
- Heiss, C.N.; Olofsson, L.E. Gut Microbiota-Dependent Modulation of Energy Metabolism. J. Innate Immun. 2018, 10, 163–171. [Google Scholar] [CrossRef]
- Muscogiuri, G.; Cantone, E.; Cassarano, S.; Tuccinardi, D.; Barrea, L.; Savastano, S.; Colao, A. Gut microbiota: A new path to treat obesity. Int. J. Obes. Suppl. 2019, 9, 10–19. [Google Scholar] [CrossRef]
- Aoun, A.; Darwish, F.; Hamod, N. The Influence of the Gut Microbiome on Obesity in Adults and the Role of Probiotics, Prebiotics, and Synbiotics for Weight Loss. Prev. Nutr. Food Sci. 2020, 25, 113. [Google Scholar] [CrossRef]
- Tomova, A.; Bukovsky, I.; Rembert, E.; Yonas, W.; Alwarith, J.; Barnard, N.D.; Kahleova, H. The Effects of Vegetarian and Vegan Diets on Gut Microbiota. Front. Nutr. 2019, 6, 47. [Google Scholar] [CrossRef]
- Wang, T.Y.; Li, Q.; Bi, K. shun Bioactive flavonoids in medicinal plants: Structure, activity and biological fate. Asian J. Pharm. Sci. 2018, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef] [PubMed]
- Koh, G.C.K.W.; Porras, P.; Aranda, B.; Hermjakob, H.; Orchard, S.E. Analyzing protein-protein interaction networks. J. Proteome Res. 2012, 11, 2014–2031. [Google Scholar] [CrossRef]
- Janjić, V.; Pržulj, N. Biological function through network topology: A survey of the human diseasome. Brief. Funct. Genom. 2012, 11, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Tian, Y.; Zhang, Z. Network Biology in Medicine and Beyond. Circ. Cardiovasc. Genet. 2014, 7, 536. [Google Scholar] [CrossRef]
- Oh, K.; Adnan, M.; Cho, D. Uncovering mechanisms of Zanthoxylum piperitum fruits for the alleviation of rheumatoid arthritis based on network pharmacology. Biology 2021, 10, 703. [Google Scholar] [CrossRef]
- Li, Y.; Li, R.; Zeng, Z.; Li, S.; Liao, S.; Ma, W.; Zhou, C.; Xu, D. Network pharmacology combined with bioinformatics to investigate the mechanism of Xianlinggubao capsule in the treatment of osteoporosis. Phytomedicine Plus 2021, 1, 100049. [Google Scholar] [CrossRef]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Kwon, J.E.; Lim, J.; Bang, I.; Kim, I.; Kim, D.; Kang, S.C. Fermentation product with new equol-producing Lactobacillus paracasei as a probiotic-like product candidate for prevention of skin and intestinal disorder. J. Sci. Food Agric. 2019, 99, 4200–4210. [Google Scholar] [CrossRef]
- Kang, S.; Tanaka, T.; Narazaki, M.; Kishimoto, T. Targeting Interleukin-6 Signaling in Clinic. Immunity 2019, 50, 1007–1023. [Google Scholar] [CrossRef]
- Somolinos, F.J.; León, C.; Guerrero-Aspizua, S. Drug Repurposing Using Biological Networks. Processes 2021, 9, 1057. [Google Scholar] [CrossRef]
- Oh, K.-K.; Adnan, M.; Cho, D.-H. New Insight into Drugs to Alleviate Atopic March via Network Pharmacology-Based Analysis. Curr. Issues Mol. Biol. 2022, 44, 2257. [Google Scholar] [CrossRef]
- Cheng, L.; Qi, C.; Yang, H.; Lu, M.; Cai, Y.; Fu, T.; Ren, J.; Jin, Q.; Zhang, X. gutMGene: A comprehensive database for target genes of gut microbes and microbial metabolites. Nucleic Acids Res. 2022, 50, D795. [Google Scholar] [CrossRef]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef]
- Gfeller, D.; Michielin, O.; Zoete, V. Shaping the interaction landscape of bioactive molecules. Bioinformatics 2013, 29, 3073–3079. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Saüch, J.; Sanz, F.; Furlong, L.I. The DisGeNET cytoscape app: Exploring and visualizing disease genomics data. Comput. Struct. Biotechnol. J. 2021, 19, 2960–2967. [Google Scholar] [CrossRef] [PubMed]
- Amberger, J.S.; Bocchini, C.A.; Scott, A.F.; Hamosh, A. OMIM.org: Leveraging knowledge across phenotype–gene relationships. Nucleic Acids Res. 2019, 47, D1038–D1043. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Chaput, L.; Villoutreix, B.O. Virtual screening web servers: Designing chemical probes and drug candidates in the cyberspace. Brief. Bioinform. 2021, 22, 1790–1818. [Google Scholar] [CrossRef]
- Soo, H.C.; Chung, F.F.L.; Lim, K.H.; Yap, V.A.; Bradshaw, T.D.; Hii, L.W.; Tan, S.H.; See, S.J.; Tan, Y.F.; Leong, C.O.; et al. Cudraflavone C Induces Tumor-Specific Apoptosis in Colorectal Cancer Cells through Inhibition of the Phosphoinositide 3-Kinase (PI3K)-AKT Pathway. PLoS ONE 2017, 12, e0170551. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Srinivasan, S.; Hyman, J.D.; O’Malley, D.; Karra, S.; Viswanathan, H.S.; Srinivasan, G. Machine learning techniques for fractured media. Adv. Geophys. 2020, 61, 109–150. [Google Scholar] [CrossRef]
- Golbeck, J. Analyzing networks. In Introduction to Social Media Investigation; Elsevier: Amsterdam, The Netherlands, 2015; pp. 221–235. [Google Scholar] [CrossRef]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Modeling 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Wang, N.N.; Yao, Z.J.; Zhang, L.; Cheng, Y.; Ouyang, D.; Lu, A.P.; Cao, D.S. Admetlab: A platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminformatics 2018, 10, 1–11. [Google Scholar] [CrossRef]
- Bansal, T.; Alaniz, R.C.; Wood, T.K.; Jayaraman, A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. USA 2010, 107, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Hills, R.D.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut Microbiome: Profound Implications for Diet and Disease. Nutrients 2019, 11, 1613. [Google Scholar] [CrossRef]
- Asadi, A.; Shadab Mehr, N.; Mohamadi, M.H.; Shokri, F.; Heidary, M.; Sadeghifard, N.; Khoshnood, S. Obesity and gut–microbiota–brain axis: A narrative review. J. Clin. Lab. Anal. 2022, 36, e24420. [Google Scholar] [CrossRef]
- Jung, Y.S.; Rha, C.S.; Baik, M.Y.; Baek, N.I.; Kim, D.O. A brief history and spectroscopic analysis of soy isoflavones. Food Sci. Biotechnol. 2020, 29, 1605–1617. [Google Scholar] [CrossRef]
- Mayo, B.; Vázquez, L.; Flórez, A.B. Equol: A Bacterial Metabolite from The Daidzein Isoflavone and Its Presumed Beneficial Health Effects. Nutrients 2019, 11, E2231. [Google Scholar] [CrossRef]
- Blake, C.; Fabick, K.M.; Setchell, K.D.R.; Lund, T.D.; Lephart, E.D. Neuromodulation by soy diets or equol: Anti-depressive & anti-obesity-like influences, age- & hormone-dependent effects. BMC Neurosci. 2011, 12, 1–13. [Google Scholar] [CrossRef]
- Setchell, K.D.R.; Clerici, C. Equol: Pharmacokinetics and Biological Actions. J. Nutr. 2010, 140, 1363S. [Google Scholar] [CrossRef] [PubMed]
- Ishaq, M.; Khan, A.; Bacha, A.S.; Shah, T.; Hanif, A.; Ahmad, A.A.; Ke, W.; Li, F.; Din, A.U.; Ding, Z.; et al. Microbiota targeted interventions of probiotic lactobacillus as an anti-ageing approach: A review. Antioxidants 2021, 10, 1930. [Google Scholar] [CrossRef] [PubMed]
- Lin, I.C.; Yamashita, S.; Murata, M.; Kumazoe, M.; Tachibana, H. Equol suppresses inflammatory response and bone erosion due to rheumatoid arthritis in mice. J. Nutr. Biochem. 2016, 32, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Wueest, S.; Konrad, D. The role of adipocyte-specific IL-6-type cytokine signaling in FFA and leptin release. Adipocyte 2018, 7, 226. [Google Scholar] [CrossRef]
- Bortoluzzi, S.; Romualdi, C.; Bisognin, A.; Danieli, G.A. Disease genes and intracellular protein networks. Physiol. Genom. 2004, 15, 223–227. [Google Scholar] [CrossRef][Green Version]
- Jin, J.; Chen, B.; Zhan, X.; Zhou, Z.; Liu, H.; Dong, Y. Network pharmacology and molecular docking study on the mechanism of colorectal cancer treatment using Xiao-Chai-Hu-Tang. PLoS ONE 2021, 16, e0252508. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, X.; Bai, H.; Ning, K. Network pharmacology databases for traditional Chinese medicine: Review and assessment. Front. Pharmacol. 2019, 10, 123. [Google Scholar] [CrossRef]
- Xia, X. Bioinformatics and Drug Discovery. Curr. Top. Med. Chem. 2017, 17, 1709. [Google Scholar] [CrossRef]
- Xu, X.; Fang, C.; Lu, F.; Liu, S. Integrated Network Pharmacology and Gut Microbiota Study on the Mechanism of Huangqin Decoction in Treatment Diabetic Enteritis. Appl. Bionics Biomech. 2022, 2022. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).