The Adhesion GPCR VLGR1/ADGRV1 Regulates the Ca2+ Homeostasis at Mitochondria-Associated ER Membranes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Antibodies
2.3. Cell Lines
2.4. Cell Culture
2.5. DNA Constructs
2.6. Tandem Affinity Purification (TAP) and Mass Spectrometry
2.7. Data Processing
2.8. RFP-Trap®/Myc-Trap®
2.9. Immunocytochemistry and Fluorescence Microscopy
2.10. Isolation of Mitochondria-Associated Membrane Fraction
2.11. Co-Localization Analysis
2.12. Transmission Electron Microscopy (TEM)
2.13. Proximity Ligation Assay (PLA)
2.14. Ca2+ Imaging
3. Results
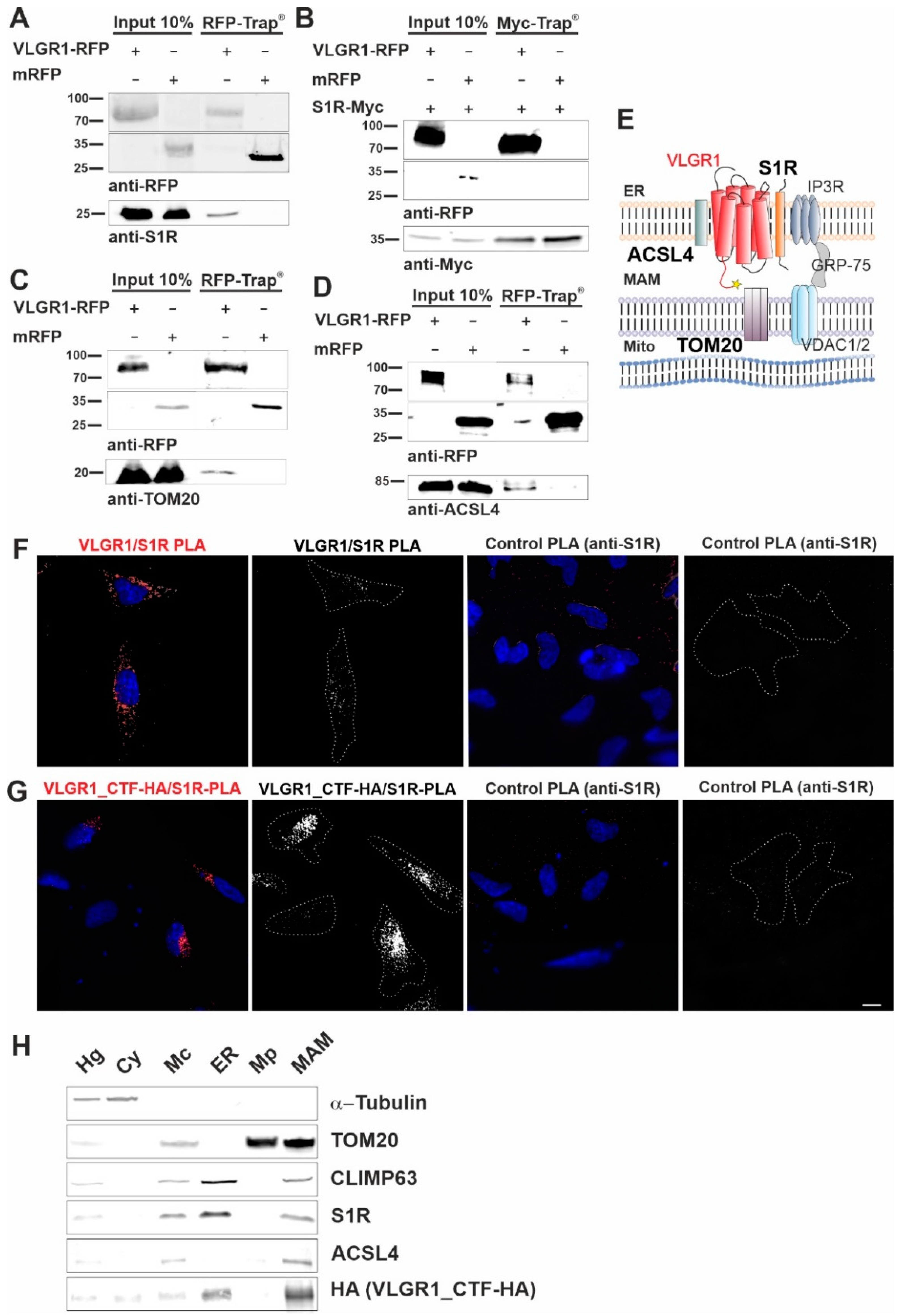
3.1. Affinity Proteomics Identified Interactions of VLGR1 with Proteins of MAMs
3.2. VLGR1 Interacts with MAM-Associated Proteins
3.3. VLGR1 Is Enriched in ER and MAM Fractions
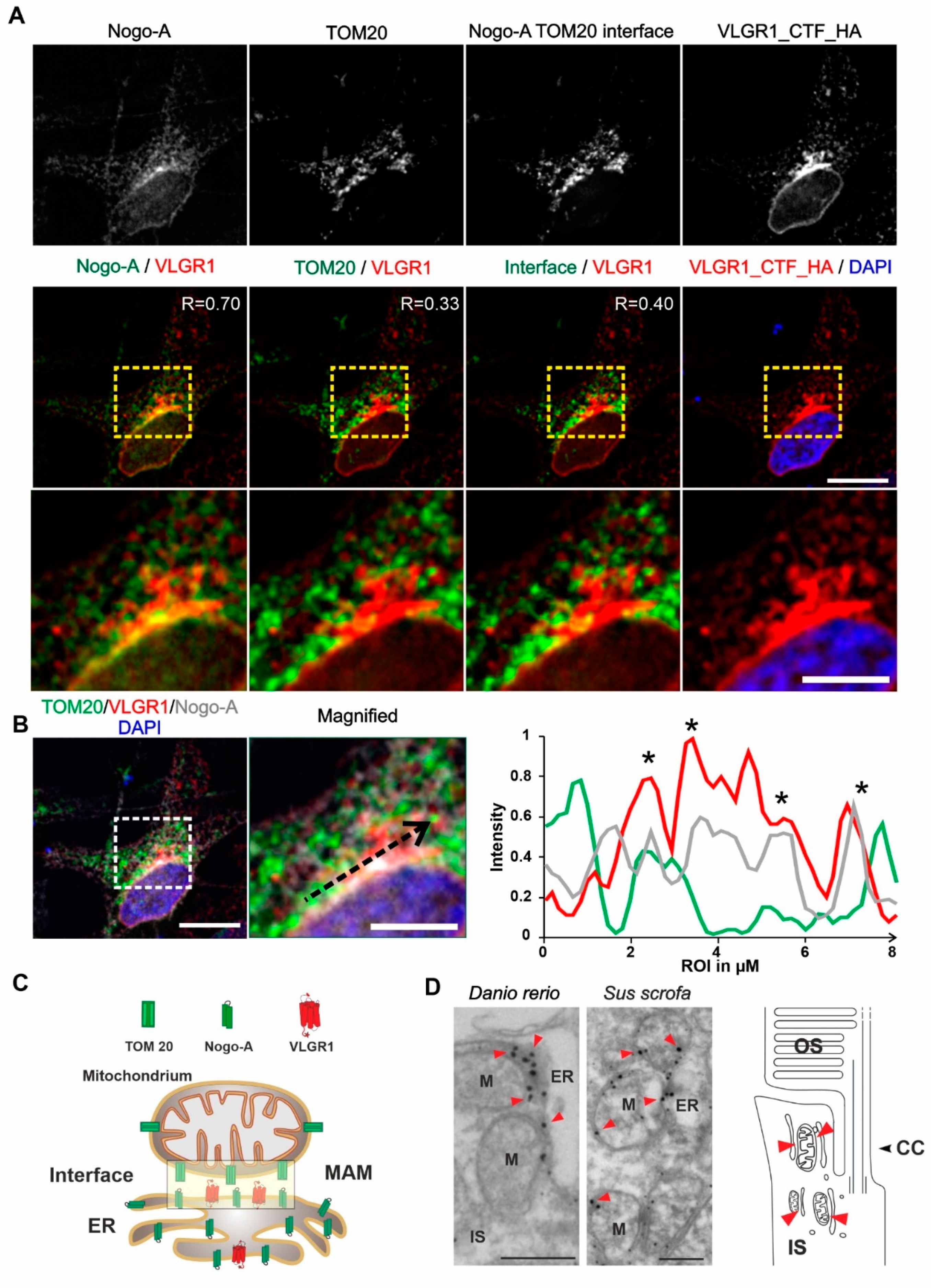
3.4. VLGR1 Is Localized at the ER-Mitochondria Interface
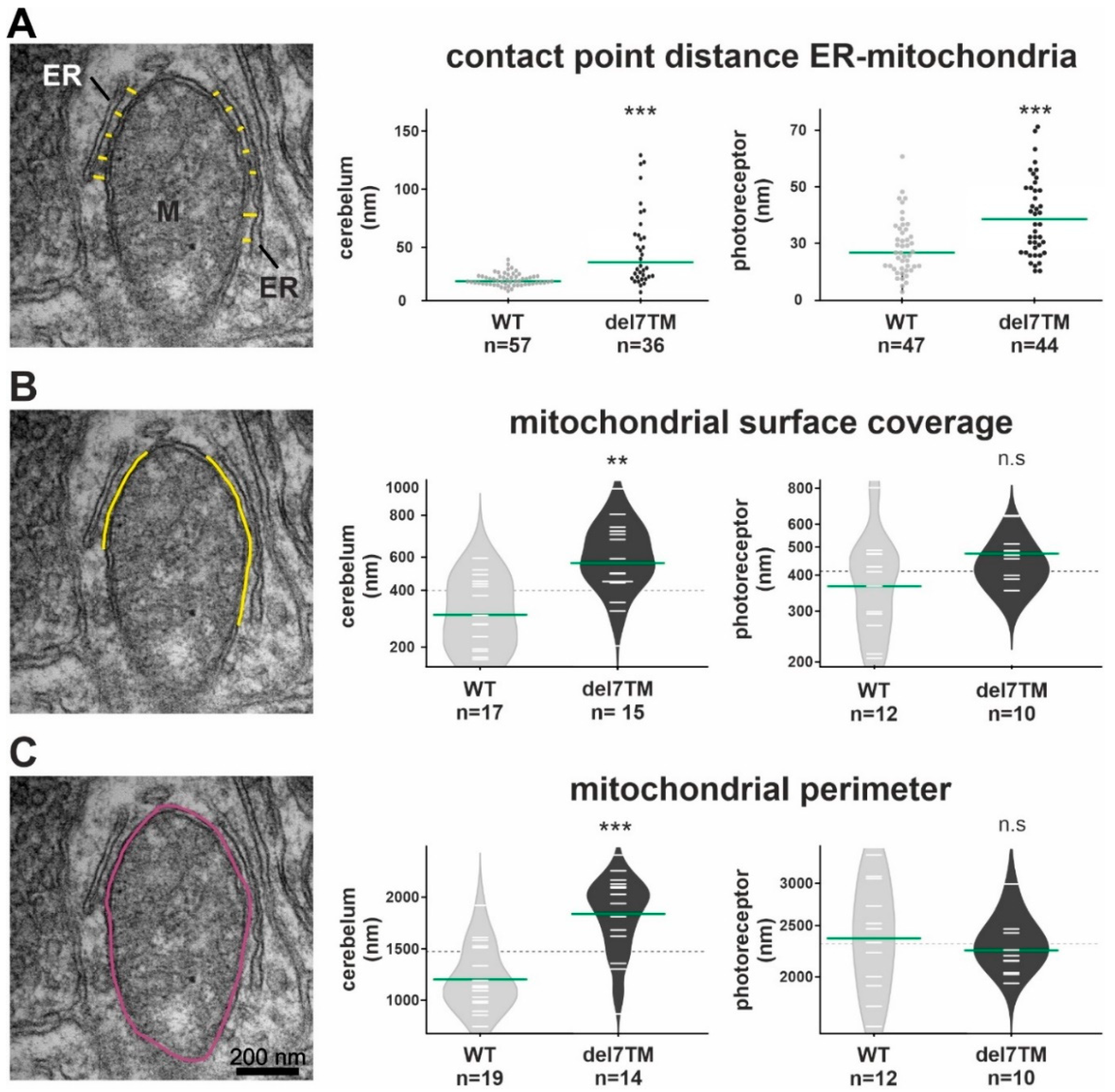
3.5. VLGR1 Deficiency Alters the Structure of the ER-Mitochondria Interface
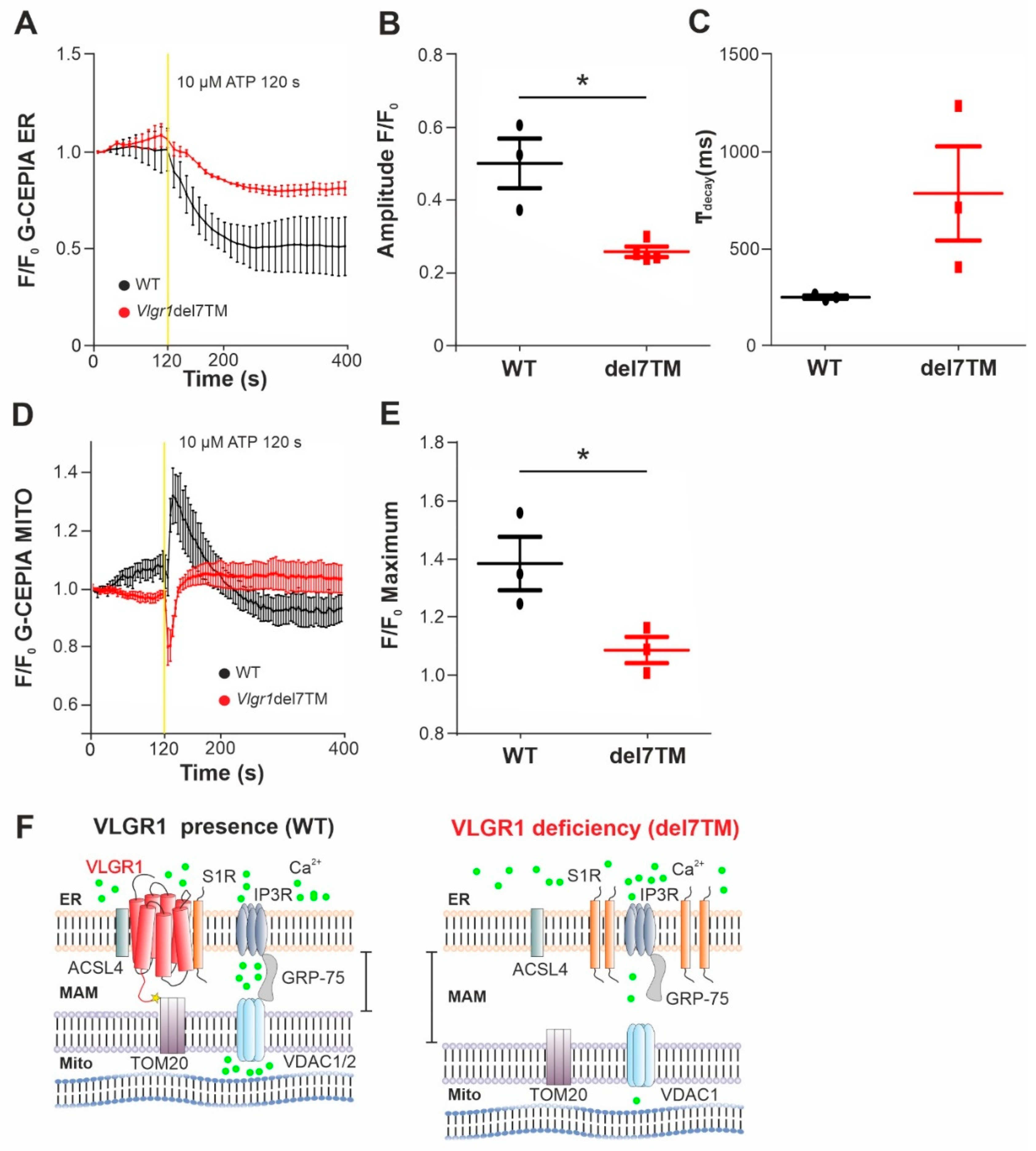
3.6. Ca2+ Transients at ER-Mitochondria Contact Sites Are Impaired by VLGR1 Deficiency
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hamann, J.; Aust, G.; Arac, D.; Engel, F.B.; Formstone, C.; Fredriksson, R.; Hall, R.A.; Harty, B.L.; Kirchhoff, C.; Knapp, B.; et al. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol. Rev. 2015, 67, 338–367. [Google Scholar] [CrossRef]
- Arac, D.; Boucard, A.A.; Bolliger, M.F.; Nguyen, J.; Soltis, S.M.; Sudhof, T.C.; Brunger, A.T. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012, 31, 1364–1378. [Google Scholar] [CrossRef]
- Knapp, B.; Roedig, J.; Roedig, H.; Krzysko, J.; Horn, N.; Guler, B.E.; Kusuluri, D.K.; Yildirim, A.; Boldt, K.; Ueffing, M.; et al. Affinity Proteomics Identifies Interaction Partners and Defines Novel Insights into the Function of the Adhesion GPCR VLGR1/ADGRV1. Molecules 2022, 27, 3108. [Google Scholar] [CrossRef] [PubMed]
- McMillan, D.R.; White, P.C. Studies on the Very Large G Protein-Coupled Receptor: From Initial Discovery to Determining its Role in Sensorineural Deafness in Higher Animals. In Adhesion-GPCRs: Structure to Function; Springer: Boston, MA, USA, 2010; Volume 706, pp. 76–86. [Google Scholar] [CrossRef]
- Reiners, J.; Nagel-Wolfrum, K.; Jurgens, K.; Marker, T.; Wolfrum, U. Molecular basis of human Usher syndrome: Deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp. Eye Res. 2006, 83, 97–119. [Google Scholar] [CrossRef] [PubMed]
- Skradski, S.L.; Clark, A.M.; Jiang, H.; White, H.S.; Fu, Y.H.; Ptacek, L.J. A novel gene causing a mendelian audiogenic mouse epilepsy. Neuron 2001, 31, 537–544. [Google Scholar] [CrossRef]
- Yagi, H.; Noguchi, Y.; Kitamura, K.; Sato, M. Deficiency of Vlgr1 resulted in deafness and susceptibility to audiogenic seizures while the degree of hearing impairment was not correlated with seizure severity in C57BL/6- and 129-backcrossed lines of Vlgr1 knockout mice. Neurosci. Lett. 2009, 461, 190–195. [Google Scholar] [CrossRef]
- Knapp, B.; Roedig, J.; Boldt, K.; Krzysko, J.; Horn, N.; Ueffing, M.; Wolfrum, U. Affinity proteomics identifies novel functional modules related to adhesion GPCRs. Ann. N. Y. Acad. Sci. 2019, 1456, 144–167. [Google Scholar] [CrossRef] [PubMed]
- Kusuluri, D.K.; Guler, B.E.; Knapp, B.; Horn, N.; Boldt, K.; Ueffing, M.; Aust, G.; Wolfrum, U. Adhesion G protein-coupled receptor VLGR1/ADGRV1 regulates cell spreading and migration by mechanosensing at focal adhesions. iScience 2021, 24, 102283. [Google Scholar] [CrossRef] [PubMed]
- Ilacqua, N.; Sanchez-Alvarez, M.; Bachmann, M.; Costiniti, V.; Del Pozo, M.A.; Giacomello, M. Protein Localization at Mitochondria-ER Contact Sites in Basal and Stress Conditions. Front. Cell Dev. Biol. 2017, 5, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedskog, L.; Pinho, C.M.; Filadi, R.; Ronnback, A.; Hertwig, L.; Wiehager, B.; Larssen, P.; Gellhaar, S.; Sandebring, A.; Westerlund, M.; et al. Modulation of the endoplasmic reticulum-mitochondria interface in Alzheimer’s disease and related models. Proc. Natl. Acad. Sci. USA 2013, 110, 7916–7921. [Google Scholar] [CrossRef]
- Lewis, A.; Tsai, S.Y.; Su, T.P. Detection of Isolated Mitochondria-Associated ER Membranes Using the Sigma-1 Receptor. Methods Mol. Biol. 2016, 1376, 133–140. [Google Scholar] [CrossRef] [PubMed]
- McMillan, D.R.; White, P.C. Loss of the transmembrane and cytoplasmic domains of the very large G-protein-coupled receptor-1 (VLGR1 or Mass1) causes audiogenic seizures in mice. Mol. Cell Neurosci. 2004, 26, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Maerker, T.; van Wijk, E.; Overlack, N.; Kersten, F.F.; McGee, J.; Goldmann, T.; Sehn, E.; Roepman, R.; Walsh, E.J.; Kremer, H.; et al. A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum. Mol. Genet. 2008, 17, 71–86. [Google Scholar] [CrossRef]
- Trojan, P.; Krauss, N.; Choe, H.W.; Giessl, A.; Pulvermuller, A.; Wolfrum, U. Centrins in retinal photoreceptor cells: Regulators in the connecting cilium. Prog. Retin. Eye Res. 2008, 27, 237–259. [Google Scholar] [CrossRef]
- Guler, B.E.; Krzysko, J.; Wolfrum, U. Isolation and culturing of primary mouse astrocytes for the analysis of focal adhesion dynamics. STAR Protoc. 2021, 2, 100954. [Google Scholar] [CrossRef]
- Suzuki, J.; Kanemaru, K.; Ishii, K.; Ohkura, M.; Okubo, Y.; Iino, M. Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nat. Commun. 2014, 5, 4153. [Google Scholar] [CrossRef] [PubMed]
- Boldt, K.; van Reeuwijk, J.; Lu, Q.; Koutroumpas, K.; Nguyen, T.M.; Texier, Y.; van Beersum, S.E.; Horn, N.; Willer, J.R.; Mans, D.A.; et al. An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat. Commun. 2016, 7, 11491. [Google Scholar] [CrossRef] [PubMed]
- Wieckowski, M.R.; Giorgi, C.; Lebiedzinska, M.; Duszynski, J.; Pinton, P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 2009, 4, 1582–1590. [Google Scholar] [CrossRef] [PubMed]
- Adler, J.; Parmryd, I. Quantifying colocalization by correlation: The Pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytometry A 2010, 77, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Sutendra, G.; Dromparis, P.; Wright, P.; Bonnet, S.; Haromy, A.; Hao, Z.; McMurtry, M.S.; Michalak, M.; Vance, J.E.; Sessa, W.C.; et al. The Role of Nogo and the Mitochondria-Endoplasmic Reticulum Unit in Pulmonary Hypertension. Sci. Transl. Med. 2011, 3, 88ra55. [Google Scholar] [CrossRef] [PubMed]
- Karlstetter, M.; Sorusch, N.; Caramoy, A.; Dannhausen, K.; Aslanidis, A.; Fauser, S.; Boesl, M.R.; Nagel-Wolfrum, K.; Tamm, E.R.; Jagle, H.; et al. Disruption of the retinitis pigmentosa 28 gene Fam161a in mice affects photoreceptor ciliary structure and leads to progressive retinal degeneration. Hum. Mol. Genet. 2014, 23, 5197–5210. [Google Scholar] [CrossRef] [PubMed]
- Sedmak, T.; Wolfrum, U. Intraflagellar transport molecules in ciliary and nonciliary cells of the retina. J. Cell Biol. 2010, 189, 171–186. [Google Scholar] [CrossRef] [PubMed]
- Gloeckner, C.J.; Boldt, K.; Schumacher, A.; Roepman, R.; Ueffing, M. A novel tandem affinity purification strategy for the efficient isolation and characterisation of native protein complexes. Proteomics 2007, 7, 4228–4234. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, V.N.; Perlman, D.H.; Costello, C.E.; McComb, M.E. Software tool for researching annotations of proteins: Open-source protein annotation software with data visualization. Anal. Chem. 2009, 81, 9819–9823. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Stoica, R.; Paillusson, S.; Gomez-Suaga, P.; Mitchell, J.C.; Lau, D.H.; Gray, E.H.; Sancho, R.M.; Vizcay-Barrena, G.; De Vos, K.J.; Shaw, C.E.; et al. ALS/FTD-associated FUS activates GSK-3beta to disrupt the VAPB-PTPIP51 interaction and ER-mitochondria associations. EMBO Rep. 2016, 17, 1326–1342. [Google Scholar] [CrossRef] [PubMed]
- Shoshan-Barmatz, V.; De, S. Mitochondrial VDAC, the Na+/Ca2+ Exchanger, and the Ca2+ Uniporter in Ca2+ Dynamics and Signaling. Adv. Exp. Med. Biol. 2017, 981, 323–347. [Google Scholar] [CrossRef] [PubMed]
- Lewin, T.M.; Van Horn, C.G.; Krisans, S.K.; Coleman, R.A. Rat liver acyl-CoA synthetase 4 is a peripheral-membrane protein located in two distinct subcellular organelles, peroxisomes, and mitochondrial-associated membrane. Arch. Biochem. Biophys. 2002, 404, 263–270. [Google Scholar] [CrossRef]
- Chiang, S.F.; Huang, C.Y.; Lin, T.Y.; Chiou, S.H.; Chow, K.C. An alternative import pathway of AIF to the mitochondria. Int. J. Mol. Med. 2012, 29, 365–372. [Google Scholar] [CrossRef]
- Goetz, J.G.; Genty, H.; St-Pierre, P.; Dang, T.; Joshi, B.; Sauve, R.; Vogl, W.; Nabi, I.R. Reversible interactions between smooth domains of the endoplasmic reticulum and mitochondria are regulated by physiological cytosolic Ca2+ levels. J. Cell Sci. 2007, 120, 3553–3564. [Google Scholar] [CrossRef] [PubMed]
- Iwasawa, R.; Mahul-Mellier, A.L.; Datler, C.; Pazarentzos, E.; Grimm, S. Fis1 and Bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J. 2011, 30, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Hussien, R.; Brooks, G.A. Colocalization of MCT1, CD147, and LDH in mitochondrial inner membrane of L6 muscle cells: Evidence of a mitochondrial lactate oxidation complex. Am. J. Physiol. Endocrinol. Metab. 2006, 290, E1237–E1244. [Google Scholar] [CrossRef] [PubMed]
- Myhill, N.; Lynes, E.M.; Nanji, J.A.; Blagoveshchenskaya, A.D.; Fei, H.; Carmine Simmen, K.; Cooper, T.J.; Thomas, G.; Simmen, T. The subcellular distribution of calnexin is mediated by PACS-2. Mol. Biol. Cell 2008, 19, 2777–2788. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Chen, Y.F.; Wu, C.Y.; Wu, P.C.; Huang, Y.L.; Kao, C.H.; Lin, C.H.; Kao, L.S.; Tsai, T.F.; Wei, Y.H. Cisd2 modulates the differentiation and functioning of adipocytes by regulating intracellular Ca2+ homeostasis. Hum. Mol. Genet. 2014, 23, 4770–4785. [Google Scholar] [CrossRef] [PubMed]
- Browman, D.T.; Resek, M.E.; Zajchowski, L.D.; Robbins, S.M. Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J. Cell Sci. 2006, 119, 3149–3160. [Google Scholar] [CrossRef]
- Gilady, S.Y.; Bui, M.; Lynes, E.M.; Benson, M.D.; Watts, R.; Vance, J.E.; Simmen, T. Ero1alpha requires oxidizing and normoxic conditions to localize to the mitochondria-associated membrane (MAM). Cell Stress Chaperones 2010, 15, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Bionda, C.; Portoukalian, J.; Schmitt, D.; Rodriguez-Lafrasse, C.; Ardail, D. Subcellular compartmentalization of ceramide metabolism: MAM (mitochondria-associated membrane) and/or mitochondria? Biochem. J. 2004, 382, 527–533. [Google Scholar] [CrossRef]
- Hayashi, T.; Su, T.P. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca2+ signaling and cell survival. Cell 2007, 131, 596–610. [Google Scholar] [CrossRef]
- Szabadkai, G.; Bianchi, K.; Varnai, P.; De Stefani, D.; Wieckowski, M.R.; Cavagna, D.; Nagy, A.I.; Balla, T.; Rizzuto, R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 2006, 175, 901–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinoshita, T.; Inoue, N. Dissecting and manipulating the pathway for glycosylphos-phatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 2000, 4, 632–638. [Google Scholar] [CrossRef]
- Rusinol, A.E.; Cui, Z.; Chen, M.H.; Vance, J.E. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 1994, 269, 27494–27502. [Google Scholar] [CrossRef]
- Horner, S.M.; Liu, H.M.; Park, H.S.; Briley, J.; Gale, M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl. Acad. Sci. USA 2011, 108, 14590–14595. [Google Scholar] [CrossRef] [PubMed]
- Hoffstrom, B.G.; Kaplan, A.; Letso, R.; Schmid, R.S.; Turmel, G.J.; Lo, D.C.; Stockwell, B.R. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat. Chem. Biol. 2010, 6, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Area-Gomez, E.; de Groof, A.J.; Boldogh, I.; Bird, T.D.; Gibson, G.E.; Koehler, C.M.; Yu, W.H.; Duff, K.E.; Yaffe, M.P.; Pon, L.A.; et al. Presenilins are enriched in endoplasmic reticulum membranes associated with mitochondria. Am. J. Pathol. 2009, 175, 1810–1816. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.J.; Vance, J.E. Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated membranes. J. Biol. Chem. 2000, 275, 34534–34540. [Google Scholar] [CrossRef] [PubMed]
- Saotome, M.; Safiulina, D.; Szabadkai, G.; Das, S.; Fransson, A.; Aspenstrom, P.; Rizzuto, R.; Hajnoczky, G. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc. Natl. Acad. Sci. USA 2008, 105, 20728–20733. [Google Scholar] [CrossRef] [PubMed]
- Hubner, C.A.; Kurth, I. Membrane-shaping disorders: A common pathway in axon degeneration. Brain 2014, 137, 3109–3121. [Google Scholar] [CrossRef]
- Man, W.C.; Miyazaki, M.; Chu, K.; Ntambi, J. Colocalization of SCD1 and DGAT2: Implying preference for endogenous monounsaturated fatty acids in triglyceride synthesis. J. Lipid Res. 2006, 47, 1928–1939. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Pei, Z.; Maiguel, D.; Toomer, C.J.; Watkins, P.A. The fatty acid transport protein (FATP) family: Very long chain acyl-CoA synthetases or solute carriers? J. Mol. Neurosci. 2007, 33, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Subedi, K.P.; Kim, J.C.; Kang, M.; Son, M.J.; Kim, Y.S.; Woo, S.H. Voltage-dependent anion channel 2 modulates resting Ca2+ sparks, but not action potential-induced Ca2+ signaling in cardiac myocytes. Cell Calcium 2011, 49, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Delprat, B.; Maurice, T.; Delettre, C. Wolfram syndrome: MAMs’ connection? Cell Death Dis. 2018, 9, 364. [Google Scholar] [CrossRef]
- Smith, M.E.; Saraceno, G.E.; Capani, F.; Castilla, R. Long-chain acyl-CoA synthetase 4 is regulated by phosphorylation. Biochem. Biophys. Res. Commun. 2013, 430, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Meisinger, C.; Ryan, M.T.; Hill, K.; Model, K.; Lim, J.H.; Sickmann, A.; Muller, H.; Meyer, H.E.; Wagner, R.; Pfanner, N. Protein import channel of the outer mitochondrial membrane: A highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small tom proteins, and import receptors. Mol. Cell Biol. 2001, 21, 2337–2348. [Google Scholar] [CrossRef] [PubMed]
- Sala-Vila, A.; Navarro-Lerida, I.; Sanchez-Alvarez, M.; Bosch, M.; Calvo, C.; Lopez, J.A.; Calvo, E.; Ferguson, C.; Giacomello, M.; Serafini, A.; et al. Interplay between hepatic mitochondria-associated membranes, lipid metabolism and caveolin-1 in mice. Sci. Rep. 2016, 6, 27351. [Google Scholar] [CrossRef]
- Klopfenstein, D.R.; Klumperman, J.; Lustig, A.; Kammerer, R.A.; Oorschot, V.; Hauri, H.P. Subdomain-specific localization of CLIMP-63 (p63) in the endoplasmic reticulum is mediated by its luminal alpha-helical segment. J. Cell Biol. 2001, 153, 1287–1300. [Google Scholar] [CrossRef]
- Giacomello, M.; Pellegrini, L. The coming of age of the mitochondria-ER contact: A matter of thickness. Cell Death Differ. 2016, 23, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- De Mario, A.; Quintana-Cabrera, R.; Martinvalet, D.; Giacomello, M. (Neuro)degenerated Mitochondria-ER contacts. Biochem. Biophys. Res. Commun. 2017, 483, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Streng, M.L.; Krook-Magnuson, E. The cerebellum and epilepsy. Epilepsy Behav. 2021, 121, 106909. [Google Scholar] [CrossRef]
- Kuo, I.Y.; Brill, A.L.; Lemos, F.O.; Jiang, J.Y.; Falcone, J.L.; Kimmerling, E.P.; Cai, Y.; Dong, K.; Kaplan, D.L.; Wallace, D.P.; et al. Polycystin 2 regulates mitochondrial Ca2+ signaling, bioenergetics, and dynamics through mitofusin 2. Sci. Signal. 2019, 12, eaat7397. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, Y.; Kwon, S.K.; Paek, H.; Pernice, W.M.; Paul, M.A.; Lee, J.; Erfani, P.; Raczkowski, A.; Petrey, D.S.; Pon, L.A.; et al. ER-mitochondria tethering by PDZD8 regulates Ca2+ dynamics in mammalian neurons. Science 2017, 358, 623–630. [Google Scholar] [CrossRef]
- Salter, M.W.; Hicks, J.L. ATP causes release of intracellular Ca2+ via the phospholipase C beta/IP3 pathway in astrocytes from the dorsal spinal cord. J. Neurosci. 1995, 15, 2961–2971. [Google Scholar] [CrossRef] [PubMed]
- Dubyak, G.R.; El-Moatassim, C. Signal transduction via P2-purinergic receptors for extracellular ATP and other nucleotides. Am. J. Physiol. 1993, 265, C577–C606. [Google Scholar] [CrossRef] [PubMed]
- Lefevre, G.; Michel, V.; Weil, D.; Lepelletier, L.; Bizard, E.; Wolfrum, U.; Hardelin, J.P.; Petit, C. A core cochlear phenotype in USH1 mouse mutants implicates fibrous links of the hair bundle in its cohesion, orientation and differential growth. Development 2008, 135, 1427–1437. [Google Scholar] [CrossRef]
- Wu, H.; Carvalho, P.; Voeltz, G.K. Here, there, and everywhere: The importance of ER membrane contact sites. Science 2018, 361, eaan5835. [Google Scholar] [CrossRef]
- Rizzuto, R.; Pinton, P.; Carrington, W.; Fay, F.S.; Fogarty, K.E.; Lifshitz, L.M.; Tuft, R.A.; Pozzan, T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 1998, 280, 1763–1766. [Google Scholar] [CrossRef]
- Szabadkai, G.; Simoni, A.M.; Bianchi, K.; De Stefani, D.; Leo, S.; Wieckowski, M.R.; Rizzuto, R. Mitochondrial dynamics and Ca2+ signaling. Biochim. Biophys. Acta 2006, 1763, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Bhagawati, M.; Arroum, T.; Webeling, N.; Montoro, A.G.; Mootz, H.D.; Busch, K.B. The receptor subunit Tom20 is dynamically associated with the TOM complex in mitochondria of human cells. Mol. Biol. Cell 2021, 32, br1. [Google Scholar] [CrossRef]
- Lee, K.S.; Huh, S.; Lee, S.; Wu, Z.; Kim, A.K.; Kang, H.Y.; Lu, B. Altered ER-mitochondria contact impacts mitochondria calcium homeostasis and contributes to neurodegeneration in vivo in disease models. Proc. Natl. Acad. Sci. USA 2018, 115, E8844–E8853. [Google Scholar] [CrossRef]
- Paillusson, S.; Stoica, R.; Gomez-Suaga, P.; Lau, D.H.; Mueller, S.; Miller, T.; Miller, C.C. There’s Something Wrong with my MAM; the ER-Mitochondria Axis and Neurodegenerative Diseases. Trends Neurosci. 2016, 39, 146–157. [Google Scholar] [CrossRef] [Green Version]
- Idevall-Hagren, O.; Lu, A.; Xie, B.; De Camilli, P. Triggered Ca2+ influx is required for extended synaptotagmin 1-induced ER-plasma membrane tethering. EMBO J. 2015, 34, 2291–2305. [Google Scholar] [CrossRef] [PubMed]
- Denton, R.M.; Richards, D.A.; Chin, J.G. Calcium ions and the regulation of NAD+-linked isocitrate dehydrogenase from the mitochondria of rat heart and other tissues. Biochem. J. 1978, 176, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, C.; Miller, R.A.; Smith, I.; Bui, T.; Molgo, J.; Muller, M.; Vais, H.; Cheung, K.H.; Yang, J.; Parker, I.; et al. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell 2010, 142, 270–283. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Irwin, R.W.; Zhao, L.; Nilsen, J.; Hamilton, R.T.; Brinton, R.D. Mitochondrial bioenergetic deficit precedes Alzheimer’s pathology in female mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2009, 106, 14670–14675. [Google Scholar] [CrossRef] [PubMed]
- Eckmiller, M.S. Energy depletion hypothesis for retinitis pigmentosa. Adv. Exp. Med. Biol. 2003, 533, 277–285. [Google Scholar] [CrossRef] [PubMed]

| Gene | Protein | Protein Function | Reference | HEK | RPE1 |
|---|---|---|---|---|---|
| ACSL4 | Long-chain-fatty-acid—CoA ligase 4 | lipid biosynthesis & fatty acid degradation | [29] | + | − |
| AIFM1 | Mitochondrial apoptosis-inducing factor 1 | apoptosis, mito. morphology | [30] | + | − |
| AMFR | Autocrine motility factor receptor | ubiquitination | [31] | + | − |
| BCAP31 | B-cell receptor-associated protein 31 | apoptosis | [32] | + | − |
| BSG | Basigin | regulatory component of γ-secretase | [33] | + | + |
| CANX | Calnexin | calcium pump | [34] | + | + |
| CISD2 | CDGSH iron sulfur domain 2 | calcium homeostasis | [35] | + | − |
| ERLIN2 | Erlin-2 | targets IP3Rs for degradation | [36] | + | + |
| ERP44 | ER resident protein 44 | ER protein retention | [37] | + | − |
| G6PC3 | Glucose-6-phosphatase 3 | gluconeogenesis | [38] | + | − |
| HSPA5 | Heat shock protein 5 | chaperone, ER stress | [39] | + | + |
| HSPA9 | Mitochondrial heat shock protein 9 | chaperone, binds VDAC | [40] | + | + |
| LCLAT1 | Lysocardiolipin acyltransferase 1 | chardolipin acyl chain remodeling | [41] | + | − |
| LMAN1 | ERGIC-53 | mannose-specific lectin | [42] | + | − |
| MAVS | Mito. antiviral-signaling protein | activation of NF-kb/IRF3 | [43] | + | + |
| P4HB | Protein disulfide-isomerase | ER protein retention | [44] | + | + |
| PIGN | Phosphatidylinositol glycan anchor biosynthesis, class N | glycolipid, GPI syntheses | [41] | + | − |
| PSEN1 | Presenilin-1 | component of γ-secretase | [45] | + | − |
| PSEN2 | Presenilin-2 | component of γ-secretase | [45] | + | − |
| PTDSS1 | Phosphatidylserine synthase 1 | phospholipid metabolism | [46] | + | − |
| PTDSS2 | Phosphatidylserine synthase 1 | phospholipid metabolism | [46] | + | − |
| RHOT1 | Mitochondrial Rho GTPase 1 | mitochondrial trafficking | [47] | + | − |
| RTN2 | Reticulon-2 | formation of tubular ER | [48] | + | + |
| RTN4 | Reticulon 4, neurite outgrowth inhibitor, Nogo-A | ER-mitochondria tethering, membrane trafficking | [21] | + | + |
| SCD | Stearoyl-CoA desaturase | fatty acid transport | [49] | + | + |
| SIGMAR1 | Sigma-1 receptor, SR1 | chaperone, lipid transport, Ca2+ signaling | [39] | + | + |
| SLC27A4 | Long-chain fatty acid transport protein 4 | fatty acid transport | [50] | + | + |
| SOAT1 | Sterol O-acyltransferase 1 | cholesterol metabolism | [42] | + | + |
| VAPB | VAMP-associated protein | ER-mitochondria tethering | [27] | + | − |
| VDAC1 | Voltage-dependent anion-channel 1 | ion exchange, Ca2+ transport | [40] | + | + |
| VDAC2 | Voltage-dependent anion-channel 2 | ion exchange, Ca2+ transport | [51] | − | + |
| WSF1 | Wolframin, ER, transmembrane | Ca2+ transport, ER stress | [52] | + | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krzysko, J.; Maciag, F.; Mertens, A.; Güler, B.E.; Linnert, J.; Boldt, K.; Ueffing, M.; Nagel-Wolfrum, K.; Heine, M.; Wolfrum, U. The Adhesion GPCR VLGR1/ADGRV1 Regulates the Ca2+ Homeostasis at Mitochondria-Associated ER Membranes. Cells 2022, 11, 2790. https://doi.org/10.3390/cells11182790
Krzysko J, Maciag F, Mertens A, Güler BE, Linnert J, Boldt K, Ueffing M, Nagel-Wolfrum K, Heine M, Wolfrum U. The Adhesion GPCR VLGR1/ADGRV1 Regulates the Ca2+ Homeostasis at Mitochondria-Associated ER Membranes. Cells. 2022; 11(18):2790. https://doi.org/10.3390/cells11182790
Chicago/Turabian StyleKrzysko, Jacek, Filip Maciag, Anna Mertens, Baran Enes Güler, Joshua Linnert, Karsten Boldt, Marius Ueffing, Kerstin Nagel-Wolfrum, Martin Heine, and Uwe Wolfrum. 2022. "The Adhesion GPCR VLGR1/ADGRV1 Regulates the Ca2+ Homeostasis at Mitochondria-Associated ER Membranes" Cells 11, no. 18: 2790. https://doi.org/10.3390/cells11182790
APA StyleKrzysko, J., Maciag, F., Mertens, A., Güler, B. E., Linnert, J., Boldt, K., Ueffing, M., Nagel-Wolfrum, K., Heine, M., & Wolfrum, U. (2022). The Adhesion GPCR VLGR1/ADGRV1 Regulates the Ca2+ Homeostasis at Mitochondria-Associated ER Membranes. Cells, 11(18), 2790. https://doi.org/10.3390/cells11182790







