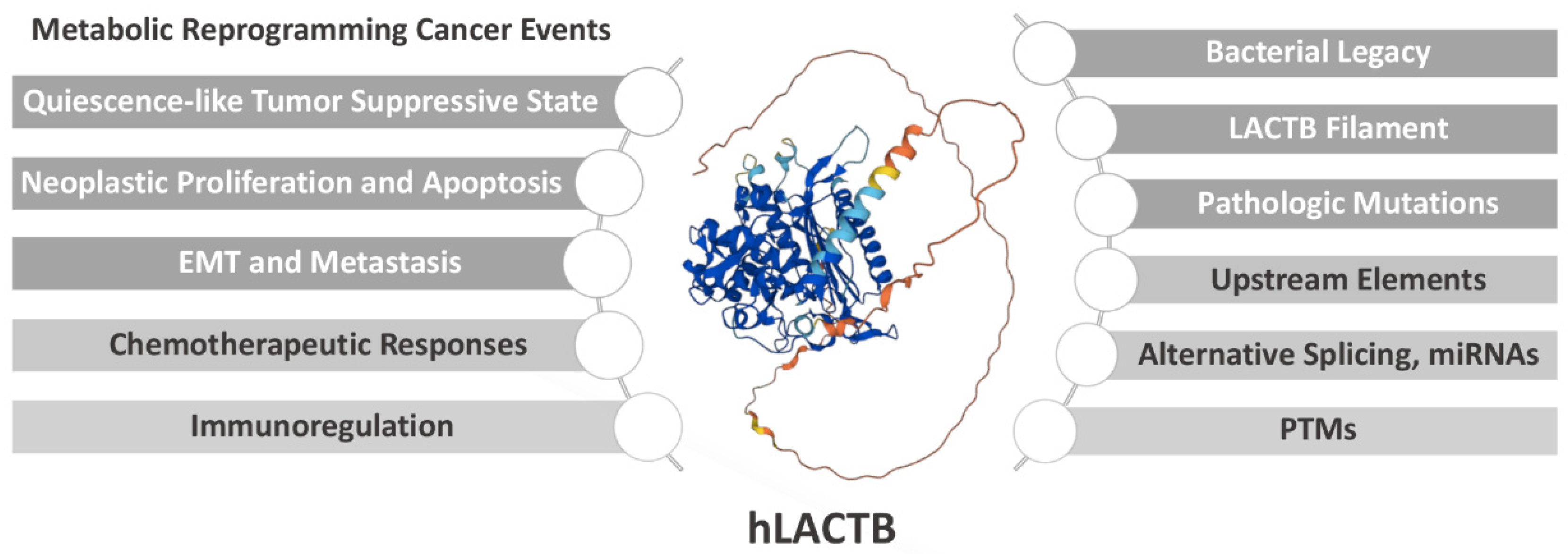
LACTB, a Metabolic Therapeutic Target in Clinical Cancer Application
Abstract
:1. Introduction
2. Potential Prospects of LACTB in Cancer Applications
| Cancer | Phenotype and Effect | Mechanism | Ref |
|---|---|---|---|
| Breast cancer | LACTB expression is downregulated in breast cancer and associated with malignancies; Knockdown of miR-374a suppresses the cell proliferative and colony-formation activity as well as migration and invasion capacity in vitro, and LACTB silencing reverses this change. | LACTB expression is downregulated by miR-374a, promoting cancer progression in breast cancer. | [9] |
| Breast cancer | LACTB expression is downregulated in breast cancer cell lines and tissues; LACTB drives the tumor suppressive mitochondrial state, reduced proliferation, and increased differentiation; Low LACTB, high mutant LACTB expression, or a molecular context with dysfunctional LACTB leads to enhanced proliferation and reduced differentiation. | LACTB inhibits phosphatidylserine decarboxylase and reduces the abundance of phosphatidylethanolamine and lyso-phosphatidylethanolamine, leading to a mitochondrial state compatible with tumor suppression, decreased proliferation, and enhanced differentiation. | [4] |
| Colorectal cancer | LACTB expression is downregulated in colorectal cancer; LACTB is associated with metastasis and advanced clinical stage; Low LACTB expression is associated with poor overall survival; Ectopic LACTB expression suppresses colorectal cancer cells proliferation, migration, invasion, and epithelial-to-mesenchymal transition in vitro, while knockout of LACTB results in an opposite phenotype; Ectopic LACTB expression inhibits colorectal cancer growth and metastasis in vivo, while knockout of LACTB results in an opposite phenotype. | LACTB directly binds to the C-terminus of p53 to inhibit p53 degradation by preventing E3 ubiquitin-protein ligase Mdm2 from interacting with p53; The antitumorigenic effects of LACTB overexpression is counterbalanced by p53 ablation in colorectal cancer; The downregulated expression of LACTB is due to promoter methylation and histone deacetylation. | [10] |
| Colorectal cancer | LACTB expression in colorectal cancer tissue samples is lower than that in nonmalignant tissue samples; LACTB inhibits cell invasion, migration, and proliferation by promoting autophagy in vitro; LACTB modulates tumorigenesis in vivo. | LACTB regulates the activity of phosphoinositide-3-kinase regulatory subunit 3, influences the level of phosphoinositide 3-kinases, and promotes autophagy via the protein kinase B/rapamycin signaling pathway. | [11] |
| Colorectal cancer | LACTB expression is lower in colorectal cancer tissue than that in the adjacent tissue; LACTB is associated with clinical stage in colorectal cancer; LACTB expression is lower in the colorectal cancer patients with lymph node metastasis than those without lymph node metastasis; Low LACTB expression is associated with a lower 5-year survival rate in colorectal cancer patients compared to those with high LACTB expression. | Not mentioned. | [12] |
| Colorectal cancer | Silencing LACTB promotes the viability, migration, and invasiveness in colorectal cancer cells; Silencing LACTB partially abolishes the apoptotic effect of pinocembrin on colorectal cancer cells. | Silencing LACTB enhances the expression of Matrix metalloproteinase-2 and N-cadherin but inhibits that of E-cadherin. | [13] |
| Colon cancer | LACTB mRNA expression is lower in colon cancer than in normal tissue; Overexpressing miR-1276 in colon cancer cells increases proliferation, migration, invasiveness, and epithelial-to-mesenchymal transition and decreases apoptosis, while supplementing LACTB suppresses these effects of miR-1276. | LACTB is inhibited miR-1276 inhibits in colon cancer cells, which inhibits autophagy. | [14] |
| Gastric cancer | LACTB is downregulated in the oxaliplatin-resistant MGC-803 cells; Overexpressing LACTB reduces the resistance of gastric cancer cells to oxaliplatin. | LACTB overexpression induces apoptosis via reducing the mitochondrial membrane potential and accelerating reactive oxygen species accumulation in the oxaliplatin-resistant MGC-803 cells; LACTB overexpression decreases glucose uptake and ATP synthesis, induces mitochondria and DNA damages, and inhibits autophagy of oxaliplatin-resistant MGC-803 cells. | [15] |
| Gastric cancer | No correlation is found between LACTB protein expression and clinicopathological indices of gastric cancer, including sex, age, histological differentiation, tumor location, Borrmman type, and TNM stage; LACTB expression is decreased in the gastric cancer tissues following oxaliplatin plus S-1 neoadjuvant chemotherapy; LACTB expression decreases in 68.6% patients, while LC3 increases in 60.8% following oxaliplatin plus S-1 neoadjuvant chemotherapy; High LC3 expression and low LACTB expression are associated with a poor response of patients with advanced gastric cancer to oxaliplatin plus S-1 neoadjuvant chemotherapy. | The expressions of LACTB and LC3 are negatively correlated following oxaliplatin plus S-1 neoadjuvant chemotherapy. | [16] |
| Glioma | LACTB expression is decreased in glioma; Low LACTB expression is correlated with a poor prognosis of glioma patients; Overexpressing LACTB suppresses the proliferation, invasion, and angiogenesis of glioma cells. | Overexpressing LACTB inhibits the expression of Proliferating cell nuclear antigen, Matrix metalloproteinase-2, Matrix metalloproteinase-9, and vascular endothelial growth factor. | [17] |
| Hepatocellular carcinoma | The mRNA and protein expressions of LACTB are down-regulated in hepatocellular carcinoma; Low LACTB expression is associated with TNM stage, histologic grade, and overall survival of patients; Overexpressing LACTB inhibits proliferation, invasion, and migration in vitro and decreases tumor growth in vivo. | LACTB is markedly correlated with genes involved in the lipid metabolism pathway indicated by online prediction, including fibrillin-1, carnitine O-palmitoyltransferase 1, medium-chain specific acyl-CoA dehydrogenase, phospholipid-transporting ATPase ABCA1, ferrochelatase, peroxisome assembly protein 12, E3 ubiquitin-protein ligase UBR1, and lathosterol oxidase. | [18] |
| Leukemia | Total LACTB is highly expressed in HL60 cells and low expressed in Raji cells; LACTB has multiple splicing isomers in leukemia cell lines, including XR1, V1, V2, and V3; The expression of V1 is high in U937 cells and low in Raji cells, that of V2 is high in HL60 cells and low in Raji cells, and V3 is poorly expressed in THP-1 cells. | Not mentioned. | [19] |
| Lung cancer | LACTB expression is downregulated in lung cancer tissues, while its methylation level is increased; Patients with high LACTB level exhibit improved survival; Overexpressing LACTB inhibits cell migration and invasion and induces apoptosis in H1299 and H1975 cells, while knocking down LACTB has opposite effects; LACTB negatively regulates the epithelial-to-mesenchymal transition process in lung cancer; LACTB strengthens the anticancer role of docetaxel in lung cancer; i.e., LACTB combined with docetaxel leads to a higher apoptotic rate and more potent inhibitory effects on H1299 and H1975 cells compared to docetaxel. | Knockdown of LACTB decreases the expression of E-cadherin (an epithelial cell-derived marker) and increases that of N-cadherin and vimentin (stromal cell-derived markers); Overexpressing LACTB increases the expression of E-cadherin and decreases that of N-cadherin and vimentin. | [20] |
| Melanoma | LACTB expression is low in melanoma tissues and cell lines; Low LACTB expression is correlated with short survival time in melanoma patients; Overexpressing LACTB suppresses the proliferation, migration, and invasion of melanoma cells in vitro; Overexpressing LACTB induces cell apoptosis and cell cycle arrest at the G2/M phase; Overexpressing LACTB suppresses the tumorigenicity and lung metastasis of MUM2B uveal melanoma cells in vivo. | Sex-determining region Y (SRY)-related HMG box-containing factor 10 binds to the promoter of LACTB and negatively regulates its transcription; Overexpressing LACTB prevents Yes-associated protein from translocating to the nucleus; LACTB directly binds to protein phosphatase-1, attenuates the interaction between protein phosphatase-1 and Yes-associated protein, and decreases the consequent dephosphorylation of Yes-associated protein. | [21] |
| Melanoma | LACTB expression is downregulated in melanoma tissues compared to the normal epithelium; Decreased LACTB expression predicts poor prognosis; iDPP/LACTB nanocomplexes promote cell apoptosis and block the cell cycle in vitro; iDPP/LACTB nanocomplexes inhibit tumor proliferation and induce cell apoptosis, consequently inhibiting melanoma growth in the subcutaneous B16-F10 melanoma model. | iDPP/LACTB nanocomplexes upregulates the p53 pathway and increases the mRNA expression of apoptosis- and cell cycle-related genes, including p21, BCL2-associated X, BH3-interacting domain death agonist, p53-induced death domain protein 1, and apoptosis regulatory protein Siva. | [22] |
| Nasopharyngeal carcinoma | Elevated LACTB expression is correlated with malignant behaviors and poorer survival of with multiple types of malignancies in nasopharyngeal carcinoma; LACTB expression is upregulated in high-metastatic nasopharyngeal carcinoma cells with reduced methylation in the promoter region; Overexpressing LACTB in the nasopharyngeal carcinoma cells promotes motility in vitro and metastasis in vivo, and knock-down of LACTB reduces metastasis of nasopharyngeal carcinoma cells; LACTB does not influence the proliferation of nasopharyngeal carcinoma cells. | LACTB activates Erb-B2 receptor tyrosine kinase 3/Epidermal growth factor receptor-extracellular signal-regulated kinase signaling, promoting the metastasis of nasopharyngeal carcinoma; Suppressing LACTB reduced nasopharyngeal carcinoma cellular motility by enhancing histone H3 stability and its acetylation. | [8] |
| Pancreatic adenocarcinoma | The mRNA and protein expressions of LACTB are high in pancreatic adenocarcinoma; Elevated LACTB mRNA expression in pancreatic adenocarcinoma patients is associated with a poor survival rate. | High LACTB expression is correlated with cell cycle-related genes and multiple immune marker sets. | [7] |
2.1. LACTB Drives a Quiescence-like Tumor Suppressive State
2.2. LACTB Mediates Neoplastic Proliferation and Apoptosis
2.3. LACTB Mediates Epithelial–Mesenchymal Transition and Metastasis
2.4. LACTB Predicts Chemotherapeutic Responses
2.5. LACTB Is Potentially Involved in Immunoregulation in Cancer
3. Structural Basis for the Enzymatic Properties of LACTB
3.1. Mammalian Mitochondrial LACTB Shares Conserved Active-Site Motifs with Bacterial Class B Low-Molecular-Weight PBP-βLs
3.2. Tertiary Structure of LACTB Underlies Its Enzymic Properties
3.3. Pathologic Mutations of LACTB Dump Its Catalytic Activity
4. Regulatory Mechanisms of LACTB
4.1. Transcriptional Events
4.2. Post-Transcriptional Regulators
4.3. Post-Translational Modifications
5. Summary
Author Contributions
Funding
Conflicts of Interest
References
- Gray, M.W. Mitochondrial evolution. Cold Spring Harb. Perspect Biol. 2012, 4, a011403. [Google Scholar] [CrossRef] [PubMed]
- Peitsaro, N.; Polianskyte, Z.; Tuimala, J.; Porn-Ares, I.; Liobikas, J.; Speer, O.; Lindholm, D.; Thompson, J.; Eriksson, O. Evolution of a family of metazoan active-site-serine enzymes from penicillin-binding proteins: A novel facet of the bacterial legacy. BMC Evol. Biol. 2008, 8, 26. [Google Scholar] [CrossRef]
- Liobikas, J.; Polianskyte, Z.; Speer, O.; Thompson, J.; Alakoskela, J.; Peitsaro, N.; Franck, M.; Whitehead, M.; Kinnunen, P.; Eriksson, O. Expression and purification of the mitochondrial serine protease LACTB as an N-terminal GST fusion protein in Escherichia coli. Protein Expr. Purif. 2006, 45, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Keckesova, Z.; Donaher, J.; De Cock, J.; Freinkman, E.; Lingrell, S.; Bachovchin, D.; Bierie, B.; Tischler, V.; Noske, A.; Okondo, M.; et al. LACTB is a tumour suppressor that modulates lipid metabolism and cell state. Nature 2017, 543, 681–686. [Google Scholar] [CrossRef]
- Butler, L.; Perone, Y.; Dehairs, J.; Lupien, L.; de Laat, V.; Talebi, A.; Loda, M.; Kinlaw, W.; Swinnen, J. Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv. Drug Deliv. Rev. 2020, 159, 245–293. [Google Scholar] [CrossRef] [PubMed]
- AlphaFold Protein Structure Database. Available online: https://alphafold.ebi.ac.uk/ (accessed on 3 August 2022).
- Xie, J.; Peng, Y.; Chen, X.; Li, Q.; Jian, B.; Wen, Z.; Liu, S. LACTB mRNA expression is increased in pancreatic adenocarcinoma and high expression indicates a poor prognosis. PLoS ONE 2021, 16, e0245908. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Wang, M.; Xie, P.; Yang, J.; Sun, R.; Zheng, L.; Mei, Y.; Meng, D.; Peng, X.; Lang, Y.; et al. LACTB promotes metastasis of nasopharyngeal carcinoma via activation of ERBB3/EGFR-ERK signaling resulting in unfavorable patient survival. Cancer Lett. 2021, 498, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; He, Y.; Yu, Y.; Chen, X.; Cui, G.; Wang, W.; Zhang, X.; Luo, Y.; Li, J.; Ren, F.; et al. Upregulation of miR-374a promotes tumor metastasis and progression by downregulating LACTB and predicts unfavorable prognosis in breast cancer. Cancer Med. 2018, 7, 3351–3362. [Google Scholar] [CrossRef]
- Zeng, K.; Chen, X.; Hu, X.; Liu, X.; Xu, T.; Sun, H.; Pan, Y.; He, B.; Wang, S. LACTB, a novel epigenetic silenced tumor suppressor, inhibits colorectal cancer progression by attenuating MDM2-mediated p53 ubiquitination and degradation. Oncogene 2018, 37, 5534–5551. [Google Scholar] [CrossRef]
- Xu, W.; Yu, M.; Qin, J.; Luo, Y.; Zhong, M. LACTB Regulates PIK3R3 to Promote Autophagy and Inhibit EMT and Proliferation Through the PI3K/AKT/mTOR Signaling Pathway in Colorectal Cancer. Cancer Manag. Res. 2020, 12, 5181–5200. [Google Scholar] [CrossRef]
- Zhou, S.; Miao, L.; Li, T.; Liu, P.; Zhou, H. Clinical Significance of β-Lactamase Expression in Colorectal Cancer. Cancer Biother. Radiopharm. 2020. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Yang, Y.; Feng, H.; Zhou, Q.; Liu, Y. Pinocembrin Inhibits the Proliferation, Migration, Invasiveness, and Epithelial-Mesenchymal Transition of Colorectal Cancer Cells by Regulating LACTB. Cancer Biother. Radiopharm. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Shi, Z.; Hong, Z.; Pan, J.; Chen, Z.; Qiu, C.; Zhuang, H.; Zheng, X. MicroRNA-1276 Promotes Colon Cancer Cell Proliferation by Negatively Regulating LACTB. Cancer Manag. Res. 2020, 12, 12185–12195. [Google Scholar] [CrossRef]
- Yang, F.; Yan, Z.; Nie, W.; Cheng, X.; Liu, Z.; Wang, W.; Shao, C.; Fu, G.; Yu, Y. LACTB induced apoptosis of oxaliplatin-resistant gastric cancer through regulating autophagy-mediated mitochondrial apoptosis pathway. Am. J. Transl. Res. 2021, 13, 601–616. [Google Scholar]
- Yang, F.; Yan, Z.; Nie, W.; Liu, Z.; Cheng, X.; Wang, W.; Shao, C.; Fu, G.; Yu, Y. LACTB and LC3 could serve as potential biomarkers of gastric cancer to neoadjuvant chemotherapy with oxaliplatin plus S-1. Oncol. Lett. 2021, 21, 470. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Dong, D.; Liu, Q.; Xu, Y.; Chen, L. Overexpression of LACTB, a Mitochondrial Protein That Inhibits Proliferation and Invasion in Glioma Cells. Oncol. Res. 2019, 27, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; He, Y.; Zhu, W.; Chen, X.; Yu, Y.; Hu, Q.; Chen, J.; Liu, L.; Ren, F.; Ren, Z.; et al. Low expression of LACTB promotes tumor progression and predicts poor prognosis in hepatocellular carcinoma. Am. J. Transl. Res. 2018, 10, 4152–4162. [Google Scholar]
- Liu, Z.; Yang, F.; Nie, W.; Yan, Z.; Shi, Q.; Yuan, B.; Liu, L. Alternative Splicing Analysis of LACTB Gene and Expression Characteristics of Different Transcripts in Leukemia Cell Lines. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2021, 29, 1019–1027. [Google Scholar] [CrossRef]
- Xu, Y.; Shi, H.; Wang, M.; Huang, P.; Xu, M.; Han, S.; Li, H.; Wang, Y. LACTB suppresses carcinogenesis in lung cancer and regulates the EMT pathway. Exp. Ther. Med. 2022, 23, 247. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, L.; He, F.; Yang, J.; Ding, Y.; Ge, S.; Fan, X.; Zhou, Y.; Xu, X.; Jia, R. LACTB suppresses melanoma progression by attenuating PP1A and YAP interaction. Cancer Lett. 2021, 506, 67–82. [Google Scholar] [CrossRef]
- Liu, J.; Yang, L.; Yuan, X.; Xiong, M.; Zhu, J.; Wu, W.; Ren, M.; Long, J.; Xu, X.; Gou, M. Targeted Nanotherapeutics Using LACTB Gene Therapy Against Melanoma. Int. J. Nanomed. 2021, 16, 7697–7709. [Google Scholar] [CrossRef] [PubMed]
- Nishina, A.; Kimura, H.; Sekiguchi, A.; Fukumoto, R.-H.; Nakajima, S.; Furukawa, S. Lysophosphatidylethanolamine in Grifola frondosa as a neurotrophic activator via activation of MAPK. J. Lipid Res. 2006, 47, 1434–1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fotheringham, J.; Xu, F.Y.; Nemer, M.; Kardami, E.; Choy, P.C.; Hatch, G.M. Lysophosphatidylethanolamine acyltransferase activity is elevated during cardiac cell differentiation. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2000, 1485, 1–10. [Google Scholar] [CrossRef]
- Torrano, V.; Carracedo, A. Quiescence-like metabolism to push cancer out of the race. Cell Metab. 2017, 25, 997–999. [Google Scholar] [CrossRef]
- Yu, L.; Wu, M.; Zhu, G.; Xu, Y. Emerging Roles of the Tumor Suppressor p53 in Metabolism. Front. Cell Dev. Biol. 2021, 9, 762742. [Google Scholar] [CrossRef]
- Bednarz-Knoll, N.; Alix-Panabieres, C.; Pantel, K. Plasticity of disseminating cancer cells in patients with epithelial malignancies. Cancer Metastasis Rev. 2012, 31, 673–687. [Google Scholar] [CrossRef]
- Jin, H.; He, Y.; Zhao, P.; Hu, Y.; Tao, J.; Chen, J.; Huang, Y. Targeting lipid metabolism to overcome EMT-associated drug resistance via integrin β3/FAK pathway and tumor-associated macrophage repolarization using legumain-activatable delivery. Theranostics 2019, 9, 265–278. [Google Scholar] [CrossRef]
- Lai, D.; Visser-Grieve, S.; Yang, X. Tumour suppressor genes in chemotherapeutic drug response. Biosci. Rep. 2012, 32, 361–374. [Google Scholar] [CrossRef]
- Greenlee, J.; Lopez-Cavestany, M.; Ortiz-Otero, N.; Liu, K.; Subramanian, T.; Cagir, B.; King, M. Oxaliplatin resistance in colorectal cancer enhances TRAIL sensitivity via death receptor 4 upregulation and lipid raft localization. Elife 2021, 10. [Google Scholar] [CrossRef]
- Sun, S.; Tang, Q.; Sun, L.; Zhang, J.; Zhang, L.; Xu, M.; Chen, J.; Gong, M.; Liang, X. Ultrasound-mediated immune regulation in tumor immunotherapy. Mater. Today Adv. 2022, 14, 100248. [Google Scholar] [CrossRef]
- Duan, Z.; Luo, Y. Targeting macrophages in cancer immunotherapy. Signal Transduct. Target. Ther. 2021, 6, 127. [Google Scholar] [CrossRef] [PubMed]
- Chaurio, R.; Janko, C.; Muñoz, L.; Frey, B.; Herrmann, M.; Gaipl, U. Phospholipids: Key players in apoptosis and immune regulation. Molecules 2009, 14, 4892–4914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, W.; Fa, H.; Xiao, D.; Wang, J. Targeting phosphatidylserine for Cancer therapy: Prospects and challenges. Theranostics 2020, 10, 9214–9229. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Yao, X.; Xiu, J.; Hu, Y. MicroRNA-125b-5p attenuates lipopolysaccharide-induced monocyte chemoattractant protein-1 production by targeting inhibiting LACTB in THP-1 macrophages. Arch. Biochem. Biophys. 2016, 590, 64–71. [Google Scholar] [CrossRef]
- Lin, J.; Kakkar, V.; Lu, X. Impact of MCP-1 in atherosclerosis. Curr. Pharm. Des. 2014, 20, 4580–4588. [Google Scholar] [CrossRef]
- Kim, S.; Sprung, R.; Chen, Y.; Xu, Y.; Ball, H.; Pei, J.; Cheng, T.; Kho, Y.; Xiao, H.; Xiao, L.; et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 2006, 23, 607–618. [Google Scholar] [CrossRef]
- Rome, S.; Clement, K.; Rabasa-Lhoret, R.; Loizon, E.; Poitou, C.; Barsh, G.S.; Riou, J.P.; Laville, M.; Vidal, H. Microarray profiling of human skeletal muscle reveals that insulin regulates approximately 800 genes during a hyperinsulinemic clamp. J. Biol. Chem. 2003, 278, 18063–18068. [Google Scholar] [CrossRef]
- Lee, J.; Xu, Y.; Chen, Y.; Sprung, R.; Kim, S.C.; Xie, S.; Zhao, Y. Mitochondrial phosphoproteome revealed by an improved IMAC method and MS/MS/MS. Mol. Cell. Proteom. 2007, 6, 669–676. [Google Scholar] [CrossRef]
- Wu, D.; Dasgupta, A.; Read, A.D.; Bentley, R.E.; Motamed, M.; Chen, K.-H.; Al-Qazazi, R.; Mewburn, J.D.; Dunham-Snary, K.J.; Alizadeh, E. Oxygen sensing, mitochondrial biology and experimental therapeutics for pulmonary hypertension and cancer. Free. Radic. Biol. Med. 2021, 170, 150–178. [Google Scholar] [CrossRef]
- Cascone, A.; Lalowski, M.; Lindholm, D.; Eriksson, O. Unveiling the Function of the Mitochondrial Filament-Forming Protein LACTB in Lipid Metabolism and Cancer. Cells 2022, 11, 1703. [Google Scholar] [CrossRef]
- Smith, T.S.; Southan, C.; Ellington, K.; Campbell, D.; Tew, D.G.; Debouck, C. Identification, genomic organization, and mRNA expression of LACTB, encoding a serine β-lactamase-like protein with an amino-terminal transmembrane domain. Genomics 2001, 78, 12–14. [Google Scholar] [CrossRef] [PubMed]
- Apter, A.; Schelleman, H.; Walker, A.; Addya, K.; Rebbeck, T. Clinical and genetic risk factors of self-reported penicillin allergy. J. Allergy Clin. Immunol. 2008, 122, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Polianskyte, Z.; Peitsaro, N.; Dapkunas, A.; Liobikas, J.; Soliymani, R.; Lalowski, M.; Speer, O.; Seitsonen, J.; Butcher, S.; Cereghetti, G.M. LACTB is a filament-forming protein localized in mitochondria. Proc. Natl. Acad. Sci. USA 2009, 106, 18960–18965. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, J.; Lum, P.Y.; Yang, X.; Pinto, S.; MacNeil, D.J.; Zhang, C.; Lamb, J.; Edwards, S.; Sieberts, S.K. Variations in DNA elucidate molecular networks that cause disease. Nature 2008, 452, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Deignan, J.L.; Qi, H.; Zhu, J.; Qian, S.; Zhong, J.; Torosyan, G.; Majid, S.; Falkard, B.; Kleinhanz, R.R. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat. Genet. 2009, 41, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Franquesa, A.; Stocks, B.; Chubanava, S.; Hattel, H.B.; Moreno-Justicia, R.; Peijs, L.; Treebak, J.T.; Zierath, J.R.; Deshmukh, A.S. Mass-spectrometry-based proteomics reveals mitochondrial supercomplexome plasticity. Cell Rep. 2021, 35, 109180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, L.; Guo, R.; Xiao, C.; Yin, J.; Zhang, S.; Yang, M. Structural basis for the catalytic activity of filamentous human serine beta-lactamase-like protein LACTB. Structure 2022, 30, 685–696.e685. [Google Scholar] [CrossRef]
- Cho, H.; Cho, Y.; Shim, M.; Lee, J.; Lee, H.; Kang, H. Mitochondria-targeted drug delivery in cancers. Biochim. Et Biophys. Acta Mol. Basis Dis. 2020, 1866, 165808. [Google Scholar] [CrossRef]
- Shakhova, O.; Zingg, D.; Schaefer, S.M.; Hari, L.; Civenni, G.; Blunschi, J.; Claudinot, S.; Okoniewski, M.; Beermann, F.; Mihic-Probst, D. Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nat. Cell Biol. 2012, 14, 882–890. [Google Scholar] [CrossRef]
- Du, J.; Zhang, P.; Zhao, X.; He, J.; Xu, Y.; Zou, Q.; Luo, J.; Shen, L.; Gu, H.; Tang, Q.; et al. MicroRNA-351-5p mediates skeletal myogenesis by directly targeting lactamase-β and is regulated by lnc-mg. FASEB J. 2019, 33, 1911–1926. [Google Scholar] [CrossRef]
- Héberlé, É.; Bardet, A.F. Sensitivity of transcription factors to DNA methylation. Essays Biochem. 2019, 63, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Bonnal, S.; López-Oreja, I.; Valcárcel, J. Roles and mechanisms of alternative splicing in cancer-implications for care. Nat. Rev. Clin. Oncol. 2020, 17, 457–474. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Zaidi, S.; Fang, Y.; Uversky, V.; Radivojac, P.; Oldfield, C.; Cortese, M.; Sickmeier, M.; LeGall, T.; Obradovic, Z.; et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl. Acad. Sci. USA 2006, 103, 8390–8395. [Google Scholar] [CrossRef] [PubMed]
- Otmani, K.; Lewalle, P. Tumor Suppressor miRNA in Cancer Cells and the Tumor Microenvironment: Mechanism of Deregulation and Clinical Implications. Front. Oncol. 2021, 11, 708765. [Google Scholar] [CrossRef]
- Sharma, B.; Prabhakaran, V.; Desai, A.; Bajpai, J.; Verma, R.; Swain, P. Post-translational modifications (PTMs), from a cancer perspective: An overview. Oncogen 2019, 2, 12. [Google Scholar] [CrossRef]
- Schwer, B.; Bunkenborg, J.; Verdin, R.O.; Andersen, J.S.; Verdin, E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. USA 2006, 103, 10224–10229. [Google Scholar] [CrossRef]
- Bhatt, H.; Kiran Rompicharla, S.; Ghosh, B.; Torchilin, V.; Biswas, S. Transferrin/α-tocopherol modified poly(amidoamine) dendrimers for improved tumor targeting and anticancer activity of paclitaxel. Nanomedicine 2019, 14, 3159–3176. [Google Scholar] [CrossRef]
- Deshpande, P.; Jhaveri, A.; Pattni, B.; Biswas, S.; Torchilin, V. Transferrin and octaarginine modified dual-functional liposomes with improved cancer cell targeting and enhanced intracellular delivery for the treatment of ovarian cancer. Drug Deliv. 2018, 25, 517–532. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Ren, Z.; Huang, X.; Yu, T. LACTB, a Metabolic Therapeutic Target in Clinical Cancer Application. Cells 2022, 11, 2749. https://doi.org/10.3390/cells11172749
Li X, Ren Z, Huang X, Yu T. LACTB, a Metabolic Therapeutic Target in Clinical Cancer Application. Cells. 2022; 11(17):2749. https://doi.org/10.3390/cells11172749
Chicago/Turabian StyleLi, Xiaohua, Zhongkai Ren, Xiaohong Huang, and Tengbo Yu. 2022. "LACTB, a Metabolic Therapeutic Target in Clinical Cancer Application" Cells 11, no. 17: 2749. https://doi.org/10.3390/cells11172749
APA StyleLi, X., Ren, Z., Huang, X., & Yu, T. (2022). LACTB, a Metabolic Therapeutic Target in Clinical Cancer Application. Cells, 11(17), 2749. https://doi.org/10.3390/cells11172749







