Overexpression of Terpenoid Biosynthesis Genes Modifies Root Growth and Nodulation in Soybean (Glycine max)
Abstract
:1. Introduction
2. Materials and Methods
2.1. In-Silico Differntial Gene Expression and Phylogenetic Analysis
2.2. Cloning of Full-Length Terpenoid Synthase cDNAs
2.3. Differential Gene Expression by qRT-PCR
2.4. Quantitative GC-MS of Terpenoids
2.5. Statistical Analyses
3. Results
3.1. Identification of Terpenoid Biosynthesis Genes from Soybean and Sage Plants
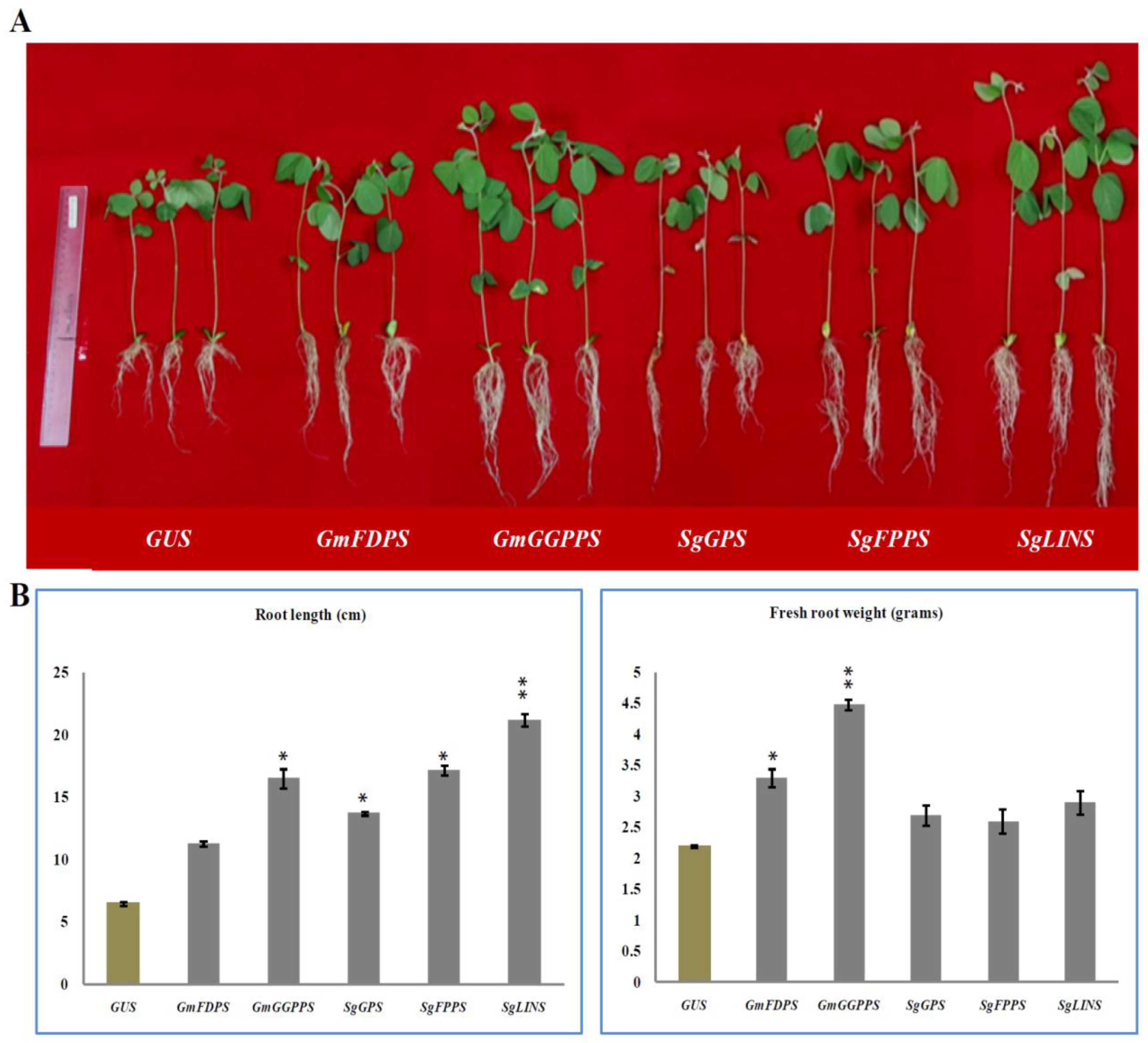
3.2. Overexpression of Terpenoid Genes Changed Soybean Root Growth
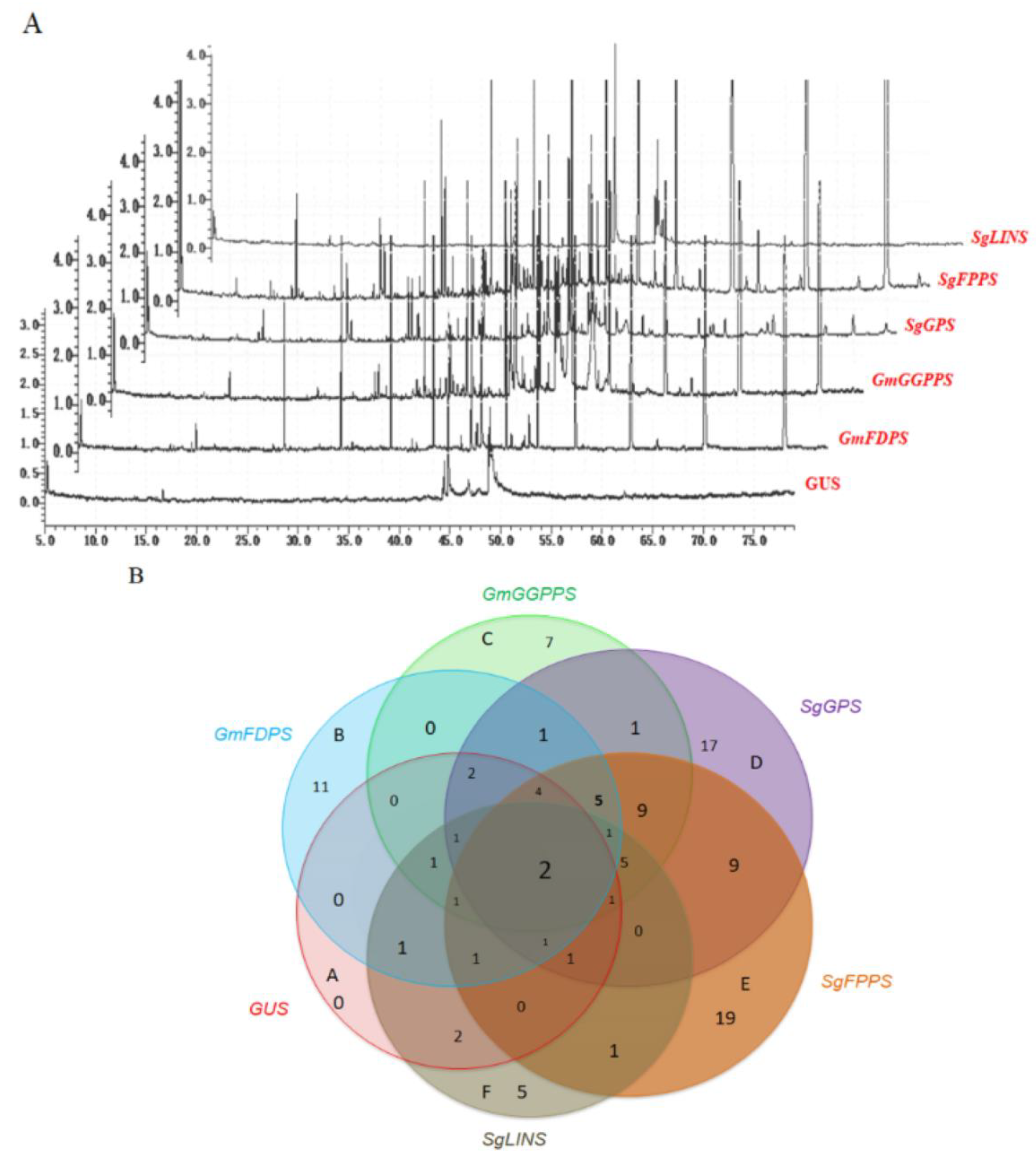
3.3. Overexpression of Terpene Synthase Genes Changed the Terpene Profiles in Transgenic Soybean Hairy Roots
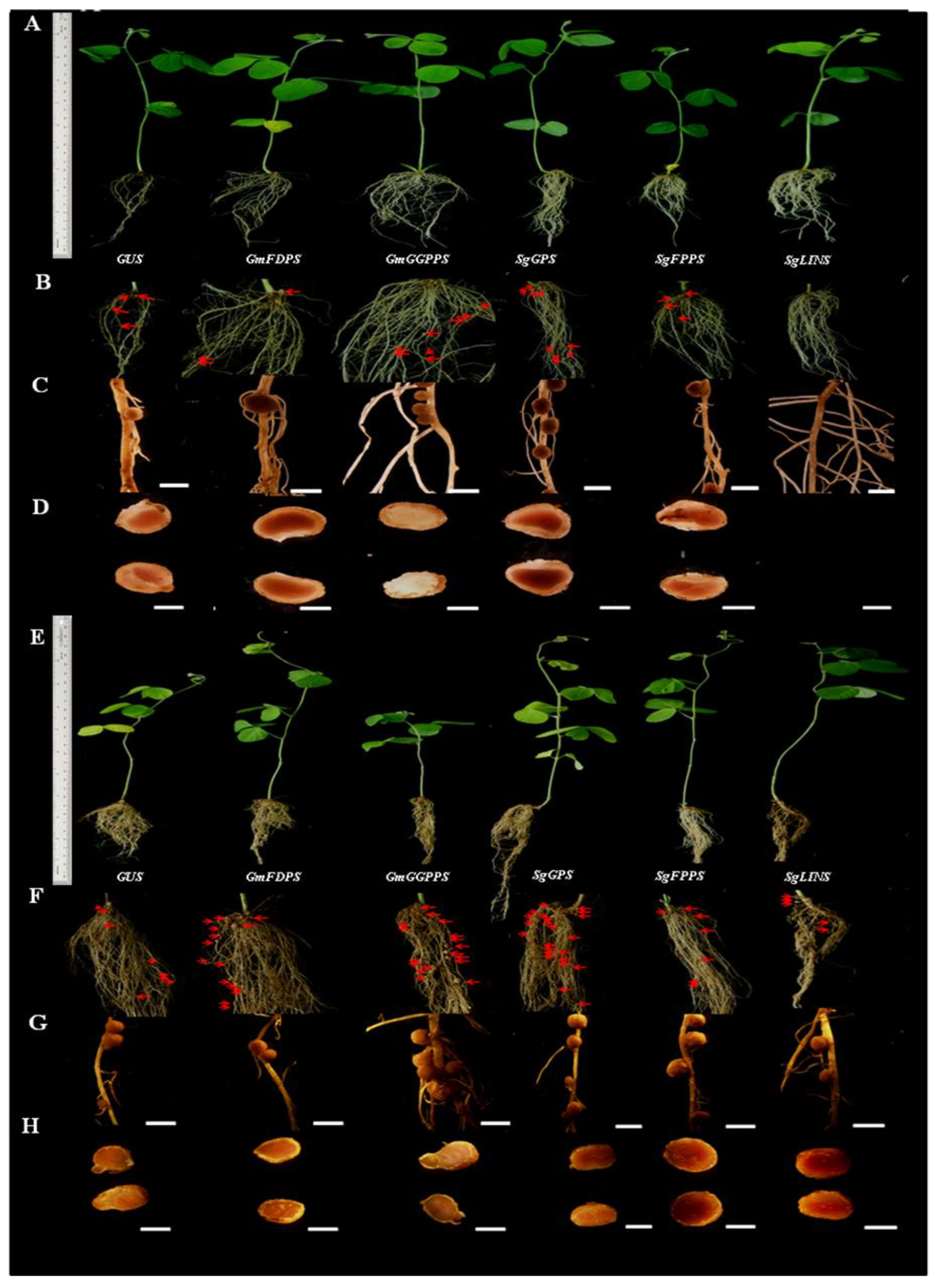
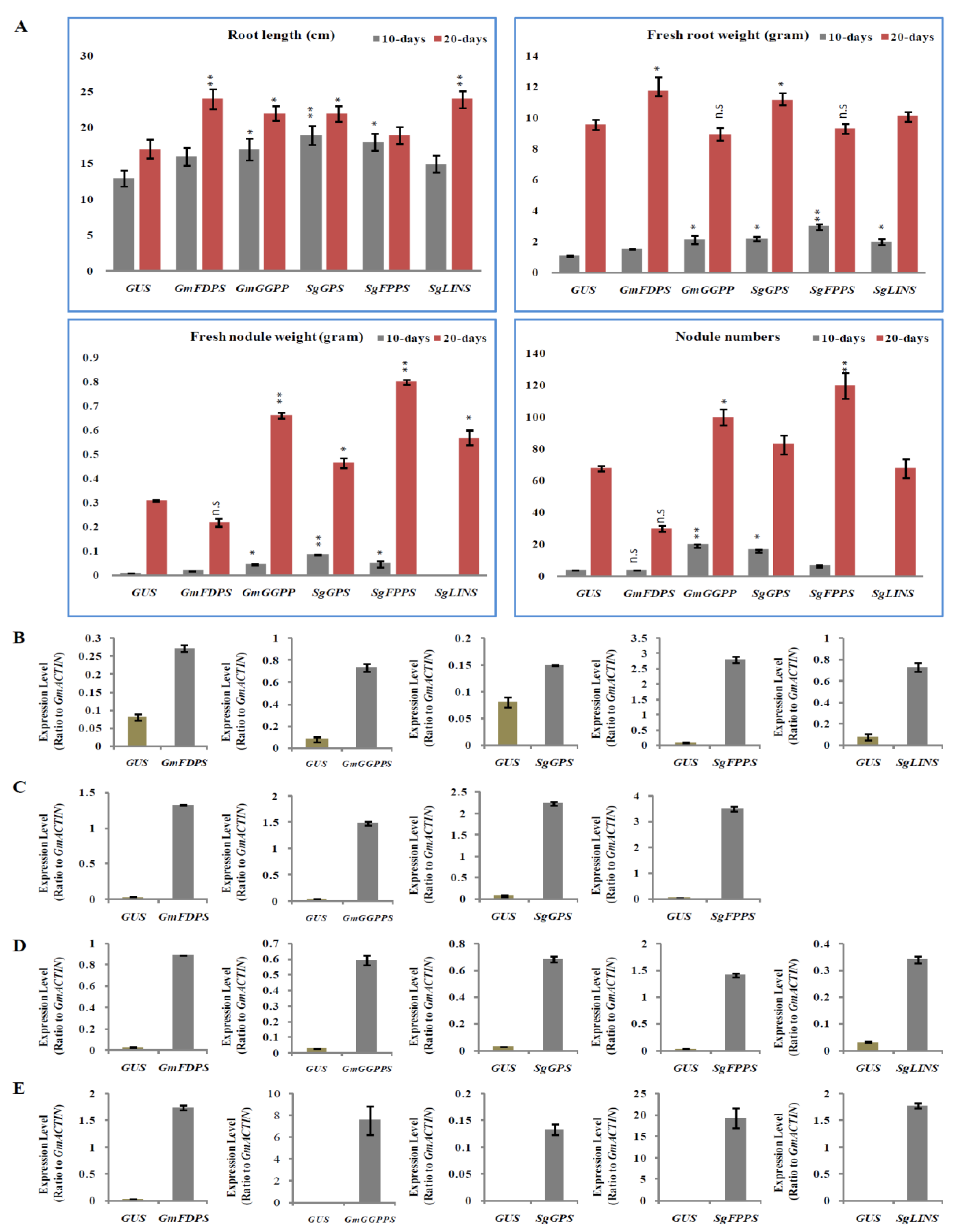
3.4. Overexpression of Terpenoid Biosynthesis Genes after Soybean Hairy Root Nodulation
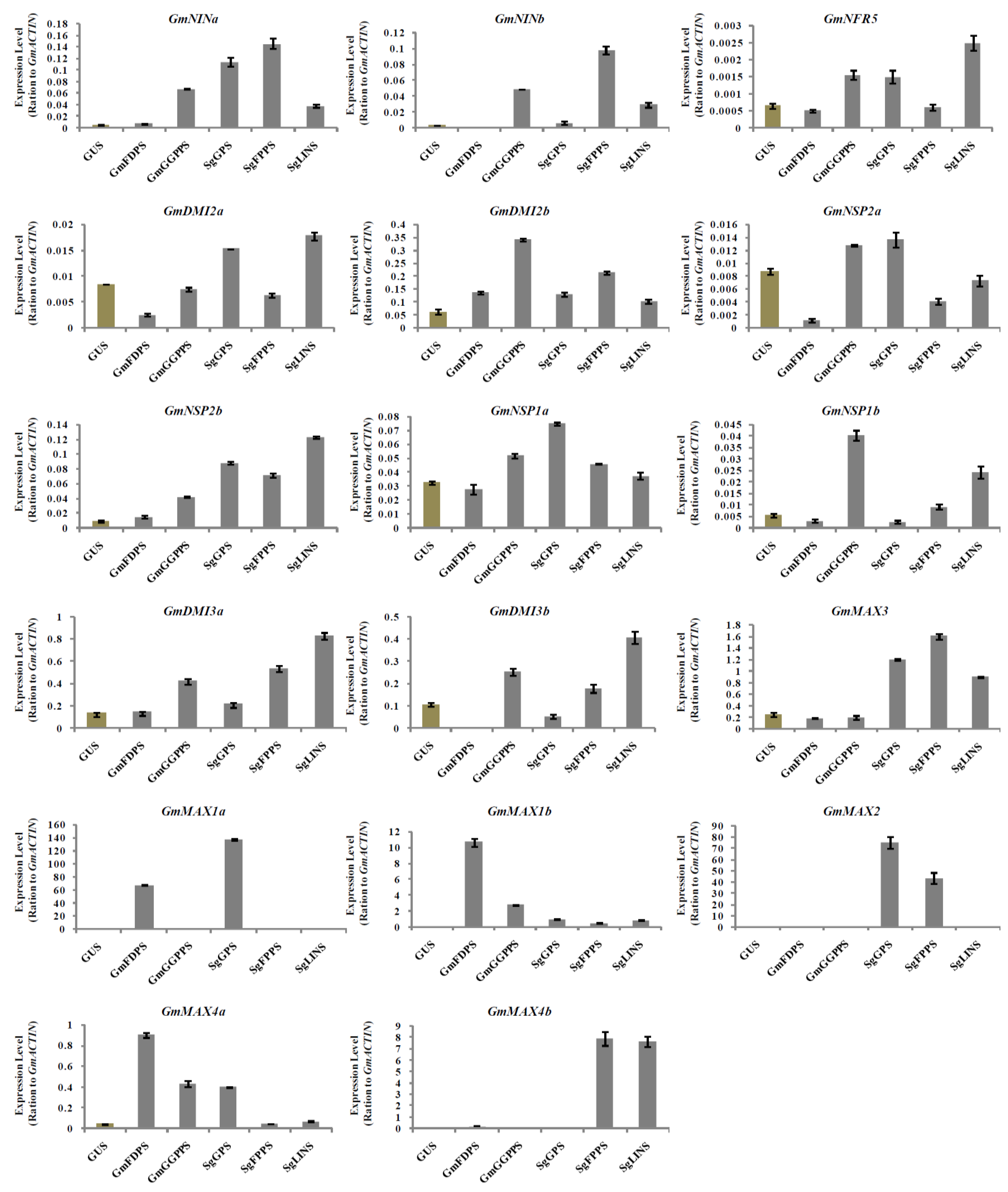
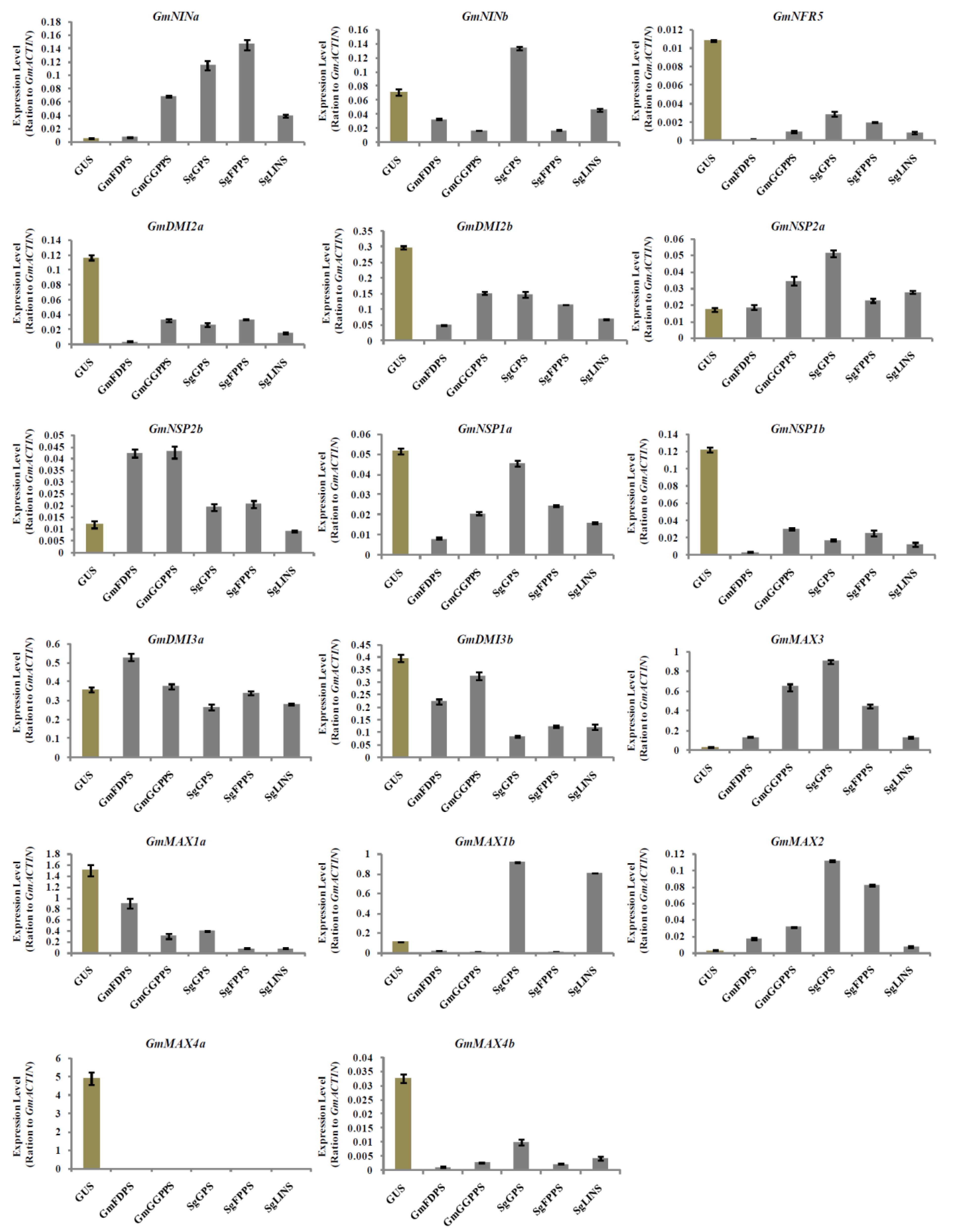
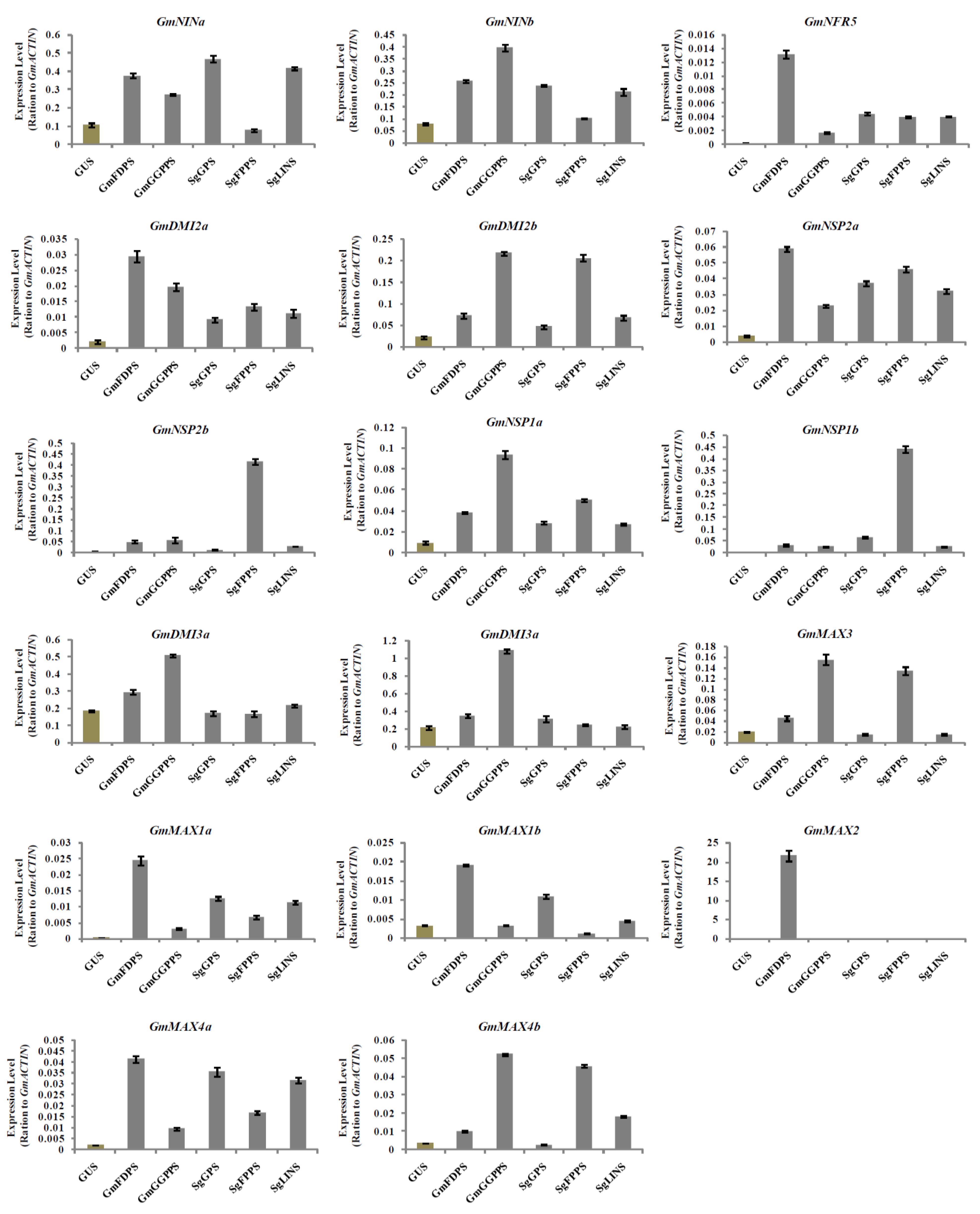
3.5. Relative Expression Analysis of Nodulation and Strigolactone Biosynthesis Genes in Transgenic Soybean Hairy Roots at 10 DAI by B. japonicum
3.6. Relative Expression Analysis of Nodulation and Strigolactone Biosynthesis Genes in Transgenic Soybean Hairy Roots at 20 DAI by B. japonicum
4. Discussion
4.1. Characterization, Putative Expression Patterns, and Subcellular Localization of Terpenoid Genes from Soybean and Anise-Scented Sage Plants
4.2. Terpenoid Genes Overexpression Enhances Terpene Accumulation in Transgenic Soybean Hairy Roots
4.3. Effect of Terpenoid Genes Overexpressing in Hairy Roots Growth and Soybean Nodulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, J.; Huang, F.; Wang, X.; Zhang, M.; Zheng, R.; Wang, J.; Yu, D. Genome-wide analysis of terpene synthases in soybean: Functional characterization of GmTPS3. Gene 2014, 544, 83–92. [Google Scholar] [CrossRef]
- Ahmad, M.Z.; Rehman, N.U.; Yu, S.; Zhou, Y.; Haq, B.U.; Wang, J.; Li, P.; Zeng, Z.; Zhao, J. GmMAX2-D14 and -KAI interaction-mediated SL and KAR signaling play essential roles in soybean root nodulation. Plant J. 2020, 101, 334–351. [Google Scholar] [CrossRef]
- Ferguson, B.J.; Ross, J.J.; Reid, J.B. Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol. 2005, 138, 2396–2405. [Google Scholar] [CrossRef]
- Takanashi, K.; Takahashi, H.; Sakurai, N.; Sugiyama, A.; Suzuki, H.; Shibata, D.; Nakazono, M.; Yazaki, K. Tissue-specific transcriptome analysis in nodules of Lotus japonicus. Mol. Plant Microbe Interact. 2012, 25, 869–876. [Google Scholar] [CrossRef]
- Kevei, Z.; Lougnon, G.; Mergaert, P.; Horváth, G.V.; Kereszt, A.; Jayaraman, D.; Zaman, N.; Marcel, F.; Regulski, K.; Kiss, G.B.; et al. Ané JM. 3-hydroxy-3-methylglutaryl coenzyme a reductase 1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell 2007, 19, 3974–3989. [Google Scholar] [CrossRef]
- Ali, M.; Miao, L.; Hou, Q.; Darwish, D.B.; Alrdahe, S.S.; Ali, A.; Benedito, V.A.; Tadege, M.; Wang, X.B.; Zhao, J. Overexpression of Terpenoid Biosynthesis Genes From Garden Sage (Salvia officinalis) Modulates Rhizobia Interaction and Nodulation in Soybean. Front. Plant Sci. 2021, 12, 783269. [Google Scholar] [CrossRef]
- Srividya, N.; Davis, E.M.; Croteau, R.B.; Lange, B.M. Functional analysis of (4 S)-limonene synthase mutants reveals determinants of catalytic outcome in a model monoterpene synthase. Proc. Natl. Acad. Sci. USA 2015, 112, 3332–3337. [Google Scholar] [CrossRef]
- Ali, M.; Li, P.; She, G.; Chen, D.; Wan, X.; Zhao, J. Transcriptome and metabolite analyses reveal the complex metabolic genes involved in volatile terpenoid biosynthesis in garden sage (Salvia officinalis). Sci. Rep. 2017, 7, 16074. [Google Scholar] [CrossRef]
- Ali, M.; Hussain, R.M.; Rehman, N.U.; She, G.; Li, P.; Wan, X.; Guo, L.; Zhao, J. De novo transcriptome sequencing and metabolite profiling analyses reveal the complex metabolic genes involved in the terpenoid biosynthesis in Blue Anise Sage (Salvia guaranitica L.). DNA Res. 2018, 25, 597–617. [Google Scholar] [CrossRef]
- Ali, M.; Nishawy, E.; Ramadan, W.A.; Ewas, M.; Rizk, M.S.; Sief-Eldein, A.G.M.; El-Zayat, M.A.S.; Hassan, A.H.M.; Guo, M.; Hu, G.-W.; et al. Molecular characterization of a Novel NAD+-dependent farnesol dehydrogenase SoFLDH gene involved in sesquiterpenoid synthases from Salvia officinalis. PLoS ONE 2022, 17, e0269045. [Google Scholar] [CrossRef]
- Funaki, A.; Waki, T.; Noguchi, A.; Kawai, Y.; Yamashita, S.; Takahashi, S.; Nakayama, T. Identification of a Highly Specific Isoflavone 7-O-glucosyltransferase in the soybean [Glycine max (L.) Merr.]. Plant Cell Physiol. 2015, 56, 1512–1520. [Google Scholar] [CrossRef]
- Sohn, S.I.; Pandian, S.; Oh, Y.J.; Kang, H.J.; Cho, W.S.; Cho, Y.S. Metabolic Engineering of Isoflavones: An Updated Overview. Front. Plant Sci. 2021, 12, 670103. [Google Scholar] [CrossRef]
- Jung, W.; Yu, O.; Lau, S.-M.C.; O’Keefe, D.; Odell, J.; Fader, G.; Mcgonigle, B. Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat. Biotechnol. 2000, 18, 208–212. [Google Scholar] [CrossRef]
- Zhao, J.; Davis, L.C.; Verpoorte, R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 2005, 23, 283–333. [Google Scholar] [CrossRef]
- Weston, L.A.; Mathesius, U. Flavonoids: Their Structure, Biosynthesis and Role in the Rhizosphere, Including Allelopathy. J. Chem. Ecol. 2013, 39, 283–297. [Google Scholar] [CrossRef]
- Zabala, G.; Zou, J.; Tuteja, J.; Gonzalez, D.O.; Clough, S.J.; Vodkin, L.O. Transcriptome changes in the phenylpropanoid pathway of Glycine max in response to Pseudomonas syringae infection. BMC Plant Biol. 2006, 6, 26. [Google Scholar] [CrossRef]
- Whitham, S.A.; Qi, M.; Innes, R.W.; Ma, W.; Lopes-Caitar, V.; Hewezi, T. Molecular Soybean-Pathogen Interactions. Annu. Rev. Phytopathol. 2016, 54, 443–468. [Google Scholar] [CrossRef]
- Akashi, T.; Sasaki, K.; Aoki, T.; Ayabe, S.; Yazaki, K. Molecular cloning and characterization of a cDNA for pterocarpan 4-dimethylallyltransferase catalyzing the key prenylation step in the biosynthesis of glyceollin, a soybean phytoalexin. Plant Physiol. 2009, 149, 683–693. [Google Scholar] [CrossRef]
- Sukumaran, A.; McDowell, T.; Chen, L.; Renaud, J.; Dhaubhadel, S. Isoflavonoid-specific prenyltransferase gene family in soybean: GmPT01, a pterocarpan 2-dimethylallyltransferase involved in glyceollin biosynthesis. Plant J. 2018, 96, 966–981. [Google Scholar] [CrossRef]
- Lin, H.; Wang, R.; Qian, Q.; Yan, M.; Meng, X.; Fu, Z.; Yan, C.; Jiang, B.; Su, Z.; Li, J.; et al. DWARF27, an Iron-Containing Protein Required for the Biosynthesis of Strigolactones, Regulates Rice Tiller Bud Outgrowth. Plant Cell 2009, 21, 1512–1525. [Google Scholar] [CrossRef]
- Waldie, T.; McCulloch, H.; Leyser, O. Strigolactones and the control of plant development: Lessons from shoot branching. Plant J. 2014, 79, 607–622. [Google Scholar] [CrossRef]
- Bennett, T.; Liang, Y.; Seale, M.; Ward, S.; Müller, D.; Leyser, O. Strigolactone regulates shoot development through a core signalling pathway. Biol. Open 2016, 5, 1806–1820. [Google Scholar] [CrossRef]
- Brewer, P.B.; Yoneyama, K.; Filardo, F.; Meyers, E.; Scaffidi, A.; Frickey, T.; Akiyama, K.; Seto, Y.; Dun, E.A.; Cremer, J.E.; et al. Lateral branching oxidoreductase acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 6301–6306. [Google Scholar] [CrossRef]
- Foo, E.; Yoneyama, K.; Hugill, C.J.; Quittenden, L.J.; Reid, J.B. Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol. Plant 2013, 6, 76–87. [Google Scholar] [CrossRef]
- Foo, E.; Davies, N.W. Strigolactones promote nodulation in pea. Planta 2011, 234, 1073–1081. [Google Scholar] [CrossRef]
- Aliche, E.B.; Screpanti, C.; De Mesmaeker, A.; Munnik, T.; Bouwmeester, H.J. Science and application of strigolactones. New Phytol. 2020, 227, 1001–1011. [Google Scholar] [CrossRef]
- Rehman, N.U.; Ali, M.; Ahmad, M.Z.; Liang, G.; Zhao, J. Strigolactones promote rhizobia interaction and increase nodulation in soybean (Glycine max). Microb. Pathog. 2018, 114, 420–430. [Google Scholar] [CrossRef]
- Kereszt, A.; Li, D.; Indrasumunar, A.; Nguyen, C.D.; Nontachaiyapoom, S.; Kinkema, M.; Gresshoff, P.M. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007, 2, 948–952. [Google Scholar] [CrossRef]
- Ahmad, M.Z.; Li, P.; Wang, J.; Rehman, N.U.; Zhao, J. Isoflavone Malonyltransferases GmIMaT1 and GmIMaT3 Differently Modify Isoflavone Glucosides in Soybean (Glycine max) under Various Stresses. Front. Plant Sci. 2017, 8, 735. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Venkateshwaran, M.; Jayaraman, D.; Chabaud, M.; Genre, A.; Balloon, A.J.; Maeda, J.; Forshey, K.; Os, D.D.; Kwiecien, N.W.; Coon, J.J.; et al. A role for the mevalonate pathway in early plant symbiotic signaling. Proc. Natl. Acad. Sci. USA 2015, 112, 9781–9786. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Sharma, R.A. Plant terpenes: Defense responses, phylogenetic analysis, regulation and clinical applications. 3 Biotech 2015, 5, 129–151. [Google Scholar] [CrossRef] [PubMed]
- Pazouki, L.; Niinemets, Ü. Multi-Substrate Terpene Synthases: Their Occurrence and Physiological Significance. Front. Plant Sci. 2016, 7, 1019. [Google Scholar] [CrossRef] [PubMed]
- Samudin, S.; Kuswantoro, H. Effect of Rhizobium inoculation to nodulation and growth of soybean [Glycine max (L.) Merrill] germplasm. Legume Res. 2018, 41, 303–310. [Google Scholar] [CrossRef]
- Field, B.; Osbourn, A.E. Metabolic diversification--independent assembly of operon-like gene clusters in different plants. Science 2008, 320, 543–547. [Google Scholar] [CrossRef]
- Field, B.; Fiston-Lavier, A.S.; Kemen, A.; Geisler, K.; Quesneville, H.; Osbourn, A.E. Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc. Natl. Acad. Sci. USA 2011, 108, 16116–16121. [Google Scholar] [CrossRef]
- Kampranis, S.C.; Ioannidis, D.; Purvis, A.; Mahrez, W.; Ninga, E.; Katerelos, N.A.; Anssour, S.; Dunwell, J.M.; Degenhardt, J.; Makris, A.M.; et al. Rational conversion of substrate and product specificity in a Salvia monoterpene synthase: Structural insights into the evolution of terpene synthase function. Plant Cell 2007, 19, 1994–2005. [Google Scholar] [CrossRef]
- Wang, Q.; Jia, M.; Huh, J.H.; Muchlinski, A.; Peters, R.J.; Tholl, D. Identification of a Dolabellane Type Diterpene Synthase and other Root-Expressed Diterpene Synthases in Arabidopsis. Front. Plant Sci. 2016, 7, 1761. [Google Scholar] [CrossRef]
- Ro, D.-K.; Ehlting, J.; Keeling, C.I.; Lin, R.; Mattheus, N.; Bohlmann, J. Microarray expression profiling and functional characterization of AtTPS genes: Duplicated Arabidopsis thaliana sesquiterpene synthase genes At4g13280 and At4g13300 encode root-specific and wound-inducible (Z)-γ-bisabolene synthases. Arch. Biochem. Biophys. 2006, 448, 104–116. [Google Scholar] [CrossRef]
- Vaughan, M.M.; Wang, Q.; Webster, F.X.; Kiemle, D.; Hong, Y.J.; Tantillo, D.J.; Coates, R.M.; Wray, A.T.; Askew, W.; O’Donnell, C.; et al. Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. Plant Cell 2013, 25, 1108–1125. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Ro, D.K.; Petri, J.; Gershenzon, J.; Bohlmann, J.; Pichersky, E.; Tholl, D. Characterization of a root-specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1, 8-cineole. Plant Physiol. 2004, 135, 1956–1966. [Google Scholar] [CrossRef] [PubMed]
- Tajima, R.; Lee, O.N.; Abe, J.; Lux, A.; Morita, S. Nitrogen-Fixing Activity of Root Nodules in Relation to Their Size in Peanut (Arachis hypogaea L.). Plant Prod. Sci. 2007, 10, 423–429. [Google Scholar] [CrossRef]
- Ferguson, B.J.; Mathesius, U. Phytohormone regulation of legume-rhizobia interactions. J. Chem. Ecol. 2014, 40, 770–790. [Google Scholar] [CrossRef]
- Rastogi, S.; Meena, S.; Bhattacharya, A.; Ghosh, S.; Shukla, R.K.; Sangwan, N.S.; Lal, R.K.; Gupta, M.M.; Lavania, U.C.; Gupta, V.; et al. De novo sequencing and comparative analysis of holy and sweet basil transcriptomes. BMC Genom. 2014, 15, 588. [Google Scholar] [CrossRef]
- Yang, D.; Du, X.; Liang, X.; Han, R.; Liang, Z.; Liu, Y.; Liu, F.; Zhao, J. Different roles of the mevalonate and methylerythritol phosphate pathways in cell growth and tanshinone production of Salvia miltiorrhiza hairy roots. PLoS ONE 2012, 7, e46797. [Google Scholar] [CrossRef]
- Kai, G.; Liao, P.; Wang, J.; Zhou, C.; Zhou, W.; Qi, Y.; Xiao, J.; Wang, Y.; Zhang, L. Molecular mechanism of elicitor-induced tanshinone accumulation in Salvia miltiorrhiza hairy root cultures. Acta Physiol. Plant 2012, 34, 1421–1433. [Google Scholar] [CrossRef]
- Shi, C.Y.; Yang, H.; Wei, C.L.; Yu, O.; Zhang, Z.Z.; Jiang, C.J.; Sun, J.; Li, Y.Y.; Chen, Q.; Xia, T.; et al. Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genom. 2011, 28, 12–131. [Google Scholar] [CrossRef]
- Abeed, A.H.A.; Ali, M.; Ali, E.F.; Majrashi, A.; Eissa, M.A. Induction of Catharanthus roseus Secondary Metabolites When Calotropis procera Was Used as Bio-Stimulant. Plants 2021, 10, 1623. [Google Scholar] [CrossRef]
- Kildegaard, K.R.; Arnesen, J.A.; Adiego-Pérez, B.; Rago, D.; Kristensen, M.; Klitgaard, A.K.; Hansen, E.H.; Hansen, J.; Borodina, I. Tailored biosynthesis of gibberellin plant hormones in yeast. Metab. Eng. 2021, 66, 1–11. [Google Scholar] [CrossRef]
- Ryu, H.; Cho, H.; Choi, D.; Hwang, I. Plant hormonal regulation of nitrogen-fixing nodule organogenesis. Mol. Cells 2012, 34, 117–126. [Google Scholar] [CrossRef]
- Breakspear, A.; Liu, C.; Roy, S.; Stacey, N.; Rogers, C.; Trick, M.; Morieri, G.; Mysore, K.S.; Wen, J.; Oldroyd, G.E.; et al. The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell 2014, 26, 4680–4701. [Google Scholar] [CrossRef]
- van Zeijl, A.; Liu, W.; Xiao, T.T.; Kohlen, W.; Yang, W.C.; Bisseling, T.; Geurts, R. The strigolactone biosynthesis gene DWARF27 is co-opted in rhizobium symbiosis. BMC Plant Biol. 2015, 15, 260. [Google Scholar] [CrossRef]
- McAdam, E.L.; Hugill, C.; Fort, S.; Samain, E.; Cottaz, S.; Davies, N.W.; Reid, J.B.; Foo, E. Determining the Site of Action of Strigolactones during Nodulation. Plant Physiol. 2017, 175, 529–542. [Google Scholar] [CrossRef]
- Haq, B.U.; Ahmad, M.Z.; Ur Rehman, N.; Wang, J.; Li, P.; Li, D.; Zhao, J. Functional characterization of soybean strigolactone biosynthesis and signaling genes in Arabidopsis MAX mutants and GmMAX3 in soybean nodulation. BMC Plant Biol. 2017, 17, 259. [Google Scholar] [CrossRef]
- Wei, M.; Yokoyama, T.; Minamisawa, K.; Mitsui, H.; Itakura, M.; Kaneko, T.; Tabata, S.; Saeki, K.; Omori, H.; Tajima, S.; et al. Soybean seed extracts preferentially express genomic loci of Bradyrhizobium japonicum in the initial interaction with soybean, Glycine max (L.) Merr. DNA Res. 2008, 15, 201–214. [Google Scholar] [CrossRef]
- Subramanian, S.; Stacey, G.; Yu, O. Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J. 2006, 48, 261–273. [Google Scholar] [CrossRef]
- Limpens, E.; Franken, C.; Smit, P.; Willemse, J.; Bisseling, T.; Geurts, R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 2003, 302, 630–633. [Google Scholar] [CrossRef]
- Madsen, E.B.; Madsen, L.H.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 2003, 425, 637–640. [Google Scholar] [CrossRef]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Felle, H.H.; Umehara, Y.; Grønlund, M.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N.; et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003, 425, 585–592. [Google Scholar] [CrossRef]
- Arrighi, J.-F.; Barre, A.; Ben Amor, B.; Bersoult, A.; Soriano, L.C.; Mirabella, R.; de Carvalho-Niebel, F.; Journet, E.-P.; Ghérardi, M.; Huguet, T.; et al. The Medicago truncatula Lysine Motif-Receptor-Like Kinase Gene Family Includes NFP and New Nodule-Expressed Genes. Plant Physiol. 2006, 142, 265–279. [Google Scholar] [CrossRef]
- Smit, P.; Limpens, E.; Geurts, R.; Fedorova, E.; Dolgikh, E.; Gough, C.; Bisseling, T. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007, 145, 183–191. [Google Scholar] [CrossRef]
- Xu, S.; Song, S.; Dong, X.; Wang, X.; Wu, J.; Ren, Z.; Wu, X.; Lu, J.; Yuan, H.; Wu, X.; et al. GmbZIP1 negatively regulates ABA-induced inhibition of nodulation by targeting GmENOD40-1 in soybean. BMC Plant Biol. 2021, 9, 1–12. [Google Scholar] [CrossRef]
- Ahmad, M.Z.; Zhang, Y.; Zeng, X.; Li, P.; Wang, X.; Benedito, V.A.; Zhao, J. Isoflavone malonyl-CoA acyltransferase GmMaT2 is involved in nodulation of soybean by modifying synthesis and secretion of isoflavones. J. Exp. Bot. 2020, 72, 1349–1369. [Google Scholar] [CrossRef]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Jurkiewicz, A.; Fukai, E.; Quistgaard, E.M.; Albrektsen, A.S.; James, E.K.; Thirup, S.; Stougaard, J. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007, 26, 3923–3935. [Google Scholar] [CrossRef]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.D.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.A.; Dickstein, R.; Udvardi, M.K. Celebrating 20 Years of Genetic Discoveries in Legume Nodulation and Symbiotic Nitrogen Fixation. Plant Cell 2019, 32, 15–41. [Google Scholar] [CrossRef]
- Murakami, E.; Cheng, J.; Gysel, K.; Bozsoki, Z.; Kawaharada, Y.; Hjuler, C.T.; Sørensen, K.K.; Tao, K.; Kelly, S.; Venice, F.; et al. Epidermal LysM receptor ensures robust symbiotic signalling in Lotus japonicus. Elife 2018, 7, e33506. [Google Scholar] [CrossRef]
- Schauser, L.; Roussis, A.; Stiller, J.; Stougaard, J. A plant regulator controlling development of symbiotic root nodules. Nature 1999, 402, 191–195. [Google Scholar] [CrossRef]
- Chaulagain, D.; Frugoli, J. The Regulation of Nodule Number in Legumes Is a Balance of Three Signal Transduction Pathways. Int. J. Mol. Sci. 2021, 22, 1117. [Google Scholar] [CrossRef]
- Stougaard, J. Regulators and regulation of legume root nodule development. Plant Physiol. 2000, 124, 531–540. [Google Scholar] [CrossRef]
- Dessein, R.; Gironella, M.; Vignal, C.; Peyrin-Biroulet, L.; Sokol, H.; Secher, T.; Lacas-Gervais, S.; Gratadoux, J.J.; Lafont, F.; Dagorn, J.C.; et al. Toll-like receptor 2 is critical for induction of Reg3 beta expression and intestinal clearance of Yersinia pseudotuberculosis. Gut 2009, 58, 771–776. [Google Scholar] [CrossRef]
- Cerri, M.R.; Frances, L.; Laloum, T.; Auriac, M.C.; Niebel, A.; Oldroyd, G.E.; Barker, D.G.; Fournier, J.; de Carvalho-Niebel, F. Medicago truncatula ERN transcription factors: Regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol. 2012, 160, 2155–2172. [Google Scholar] [CrossRef]
- Hirsch, S.; Kim, J.; Muñoz, A.; Heckmann, A.B.; Downie, J.A.; Oldroyd, G.E. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 2009, 21, 545–557. [Google Scholar] [CrossRef]
- Liu, F.; Rice, J.H.; Lopes, V.; Grewal, P.; Lebeis, S.L.; Hewezi, T.; Staton, M.E. Overexpression of Strigolactone-Associated Genes Exerts Fine-Tuning Selection on Soybean Rhizosphere Bacterial and Fungal Microbiome. Phytobiomes J. 2020, 4, 239–251. [Google Scholar] [CrossRef]
- Ferguson, B.J.; Foo, E.; Ross, J.J.; Reid, J.B. Relationship between gibberellin, ethylene and nodulation in Pisum sativum. New Phytol. 2011, 189, 829–842. [Google Scholar] [CrossRef]
- Wang, G.; Weng, L.; Li, M.; Xiao, H. Response of Gene Expression and Alternative Splicing to Distinct Growth Environments in Tomato. Int. J. Mol. Sci. 2017, 18, 475. [Google Scholar] [CrossRef]
- Buitrago-Bitar, M.; Cortés, A.; López-Hernández, F.; Londoño-Caicedo, J.; Muñoz-Florez, J.; Muñoz, L.; Blair, M. Allelic Diversity at Abiotic Stress Responsive Genes in Relationship to Ecological Drought Indices for Cultivated Tepary Bean, Phaseolus acutifolius A. Gray, and Its Wild Relatives. Genes 2021, 12, 556. [Google Scholar] [CrossRef]
- Cortés, A.; López-Hernández, F. Harnessing Crop Wild Diversity for Climate Change Adaptation. Genes 2021, 12, 783. [Google Scholar] [CrossRef]
- Burbano-Erazo, E.; León-Pacheco, R.I.; Cordero-Cordero, C.C.; López-Hernández, F.; Cortés, A.J.; Tofiño-Rivera, A.P. Multi-Environment Yield Components in Advanced Common Bean (Phaseolus vulgaris L.) × Tepary Bean (P. acutifolius A. Gray) Interspecific Lines for Heat and Drought Tolerance. Agronomy 2021, 11, 1978. [Google Scholar] [CrossRef]
- Cortés, A.J.; López-Hernández, F.; Osorio-Rodriguez, D. Predicting Thermal Adaptation by Looking into Populations’ Genomic Past. Front. Genet. 2020, 11, 564515. [Google Scholar] [CrossRef]


| N | Compound Name | R.T. | Terpene Type | GUS | GmFDPS | GmGGPPS | SgGPS | SgFPPS | SgLINS |
|---|---|---|---|---|---|---|---|---|---|
| 1 | (8)Annulene | 5.238 | 0.3 | 0.94 | 1.07 | 3.83 | 1.59 | 4.07 | |
| 2 | (−)-Isopulegol | 7.376 | Mono- | 0.02 | |||||
| 3 | Artificial Almond Oil | 8.382 | Organic | 0.03 | |||||
| 4 | Dihomo-γ-linolenic acid | 9.448 | 0.25 | 0.03 | |||||
| 5 | cis-Verbenol | 10.786 | Mono- | 0.09 | 0.37 | 0.13 | |||
| 6 | Dihydrocarveol | 14.176 | Mono- | 0.33 | 0.12 | ||||
| 7 | Limonene dioxide | 16.262 | Mono- | 0.52 | 0.1 | ||||
| 8 | Isomenthol | 16.686 | Mono- | 0.06 | 0.54 | 0.37 | 1.79 | 0.75 | 1.44 |
| 9 | L-trans-Pinocarveol | 17.335 | Mono- | 0.26 | |||||
| 10 | Pinocarveol | 17.503 | Mono- | ||||||
| 11 | 3-Acetylpyridine | 18.997 | 0.24 | 0.04 | |||||
| 12 | α-Terpineol | 19.305 | Mono- | ||||||
| 13 | Naphthalene | 20.446 | 0.25 | 0.1 | |||||
| 14 | Farnesan | 24.283 | Sesqui | ||||||
| 15 | Indole | 24.961 | 4.57 | 0.59 | |||||
| 16 | Dodecamethylcyclohexasiloxane | 25.403 | 3.09 | 0.15 | 1.12 | ||||
| 17 | Farnesan | 27.982 | Sesqui | 0.08 | 0.03 | ||||
| 18 | n-Cetane | 28.864 | 0.07 | 0.15 | |||||
| 19 | Caryophyllene oxide | 31.027 | Sesqui | 11.79 | 3.72 | 1.3 | |||
| 20 | 4-Caranol | 31.391 | Mono- | 0.47 | 3.59 | ||||
| 21 | trans-β-Terpineol | 32.127 | Mono | 0.17 | 0.29 | ||||
| 22 | Isopulegol | 35.93 | Mono | 9.91 | 3.59 | 1.29 | 1.65 | ||
| 23 | β-Eudesmol | 36.513 | sesqui | 0.12 | |||||
| 24 | dihydrophytol | 38.002 | Diter | ||||||
| 25 | β-Carotene; β-Carotene, all-trans | 39.66 | 1.35 | ||||||
| 26 | delta.-Cadinol | 42.248 | Sesqui | 0.13 | 0.76 | 0.22 | |||
| 27 | Stearic acid; n-Octadecanoic acid; | 42.285 | |||||||
| 28 | Linoleyl alcohol | 42.974 | 0.17 | ||||||
| 29 | Erucic acid | 44.418 | 7.83 | ||||||
| 30 | Palmitic acid | 44.858 | 3.95 | 8.01 | 11.85 | 60.56 | |||
| 31 | trans-Phytol | 45.597 | Diter | 0.49 | 0.74 | ||||
| 32 | Ethyl palmitate | 45.617 | |||||||
| 33 | cis,cis-Linoleic acid | 46.84 | |||||||
| 34 | δ13-cis-Docosenoic acid | 46.865 | 0.55 | 10.44 | |||||
| 35 | Arachic acid | 47.235 | |||||||
| 36 | Phytol | 47.305 | Diter | 9.62 | 11.6 | 1.11 | 12.26 | ||
| 37 | Adamantane, 1,3-dimethyl | 48.167 | 0.65 | ||||||
| 38 | 9-Octadecyne | 48.778 | 0.95 | ||||||
| 39 | Oleic Acid | 49.096 | 16.88 | ||||||
| 40 | Stearic acid | 49.545 | 0.66 | 1.47 | 2.9 | ||||
| 41 | trans-Linalool oxide | 49.893 | Mono- | 0.86 | 0.04 | ||||
| 42 | γ-Sitosterol | 50.424 | 11.24 | 14.66 | 15.55 | ||||
| 43 | dl-Isopulegol | 52.064 | Mono- | 0.27 | |||||
| 44 | Mandelic acid | 52.514 | 0.72 | ||||||
| 45 | Isopropyl linoleate | 53.073 | 0.17 | ||||||
| 46 | Octadeamethyl-cyclononasiloxane | 54.167 | 8.79 | 1.19 | 12.73 | ||||
| 47 | Oleyl amide | 54.838 | 0.09 | 0.09 | 0.36 | 0.09 | 0.49 | ||
| 48 | n-Heneicosane | 56.512 | 0.17 | 1.02 | 0.16 | ||||
| 49 | Methoprene | 59.701 | 6.56 | 8.04 | 1.12 | 10.79 | |||
| 50 | Pulegol | 61.087 | Mono- | 0.09 | 0.7 | 0.1 | |||
| 51 | Phthalic acid dioctyl ester | 62.27 | 0.22 | 0.97 | 0.46 | ||||
| 52 | β,β-Carotene | 63.348 | 0.04 | ||||||
| 53 | Octadeamethyl-cyclononasiloxane | 67.034 | 6.31 | 7.47 | 1.06 | 10.68 | |||
| 54 | Farnesane | 72.172 | Sesqui | 0.09 | |||||
| 55 | Levomenthol | 78.19 | Mono- | 0.07 | |||||
| Total percentage (%) of monoterpenes | 0.6 | 10.54 | 4.29 | 6.12 | 3.47 | 1.44 | |||
| Total percentage (%) of sesquiterpenes | 11.87 | 0.25 | 4.83 | 1.72 | |||||
| Total percentage (%) of diterpenes | 9.62 | 12.09 | 1.11 | 13.0 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, M.; Miao, L.; Soudy, F.A.; Darwish, D.B.E.; Alrdahe, S.S.; Alshehri, D.; Benedito, V.A.; Tadege, M.; Wang, X.; Zhao, J. Overexpression of Terpenoid Biosynthesis Genes Modifies Root Growth and Nodulation in Soybean (Glycine max). Cells 2022, 11, 2622. https://doi.org/10.3390/cells11172622
Ali M, Miao L, Soudy FA, Darwish DBE, Alrdahe SS, Alshehri D, Benedito VA, Tadege M, Wang X, Zhao J. Overexpression of Terpenoid Biosynthesis Genes Modifies Root Growth and Nodulation in Soybean (Glycine max). Cells. 2022; 11(17):2622. https://doi.org/10.3390/cells11172622
Chicago/Turabian StyleAli, Mohammed, Long Miao, Fathia A. Soudy, Doaa Bahaa Eldin Darwish, Salma Saleh Alrdahe, Dikhnah Alshehri, Vagner A. Benedito, Million Tadege, Xiaobo Wang, and Jian Zhao. 2022. "Overexpression of Terpenoid Biosynthesis Genes Modifies Root Growth and Nodulation in Soybean (Glycine max)" Cells 11, no. 17: 2622. https://doi.org/10.3390/cells11172622
APA StyleAli, M., Miao, L., Soudy, F. A., Darwish, D. B. E., Alrdahe, S. S., Alshehri, D., Benedito, V. A., Tadege, M., Wang, X., & Zhao, J. (2022). Overexpression of Terpenoid Biosynthesis Genes Modifies Root Growth and Nodulation in Soybean (Glycine max). Cells, 11(17), 2622. https://doi.org/10.3390/cells11172622







