Abstract
Autophagy is an evolutionally conserved degradation mechanism for maintaining cell homeostasis whereby cytoplasmic components are wrapped in autophagosomes and subsequently delivered to lysosomes for degradation. This process requires the concerted actions of multiple autophagy-related proteins and accessory regulators. In neurons, autophagy is dynamically regulated in different compartments including soma, axons, and dendrites. It determines the turnover of selected materials in a spatiotemporal control manner, which facilitates the formation of specialized neuronal functions. It is not surprising, therefore, that dysfunctional autophagy occurs in epilepsy, mainly caused by an imbalance between excitation and inhibition in the brain. In recent years, much attention has been focused on how autophagy may cause the development of epilepsy. In this article, we overview the historical landmarks and distinct types of autophagy, recent progress in the core machinery and regulation of autophagy, and biological roles of autophagy in homeostatic maintenance of neuronal structures and functions, with a particular focus on synaptic plasticity. We also discuss the relevance of autophagy mechanisms to the pathophysiology of epileptogenesis.
1. Introduction
Autophagy is a highly conserved catabolic process that ensures cellular homeostasis through the digestion and recycling of misfolded proteins, damaged organelles, and invasive pathogens [1]. Among three mechanistically distinct types, macroautophagy is the major and multistep process that involves the formation of double-membrane autophagosome, which is different from microautophagy and chaperone-mediated autophagy (CMA). In macroautophagy, selected cytoplasmic components are sequestrated in autophagosomes via a series of dynamic membrane events, such as appearances, expansion, and closure of phagophore, and maturation, trafficking, and fusions of the autophagosome, as well as release and reuse of degradation products [2]. This process virtually occurs in almost all mammalian cells, including rodents and primates, implicating in cell quality control, development and differentiation, activity-dependent function, survival, aging, and other cell activities [3].
Unlike other cell types, neurons are post-mitotic cells and have an unusual, compartmentalized morphology, such as axons, dendrites, and synaptic connections, which play a key role in neurotransmission, neural circuits, synaptic plasticity, and other neuronal physiology [4]. These unique characteristics determine that neurons require high demands on autophagy to maintain homeostasis and ensure survival over many decades [5]. Although neuronal autophagy shares basal machinery with non-neuronal cells, some neuron-specific proteins including endophilin A and synaptojanin1 are involved in the autophagy mechanisms for maintaining synaptic morphology and functions, particularly at axon terminals [6]. A growing body of evidence suggests that activity-dependent synaptic plasticity, the critical basis of brain adaptation, relies on neuronal autophagy [7,8]. Along with this prominent role in neuronal homeostasis, autophagy is also essential for the occurrence and progression of neurological and neurodegenerative diseases, such as epilepsy, which is characterized by spontaneously synchronized abnormal neuronal firing activity [9]. Recent studies have shown a close link between epilepsy and autophagy [10]. Pathological changes in epilepsy manifestations such as synaptic structural and functional changes, neuronal excitatory and inhibitory imbalances, and abnormal connections in neural circuits are also regulated by autophagy [10,11]. However, the potential role of autophagy in epilepsy has been relatively unexplored.
In this review, we describe the historical landmarks, distinct types, and core machinery of autophagy in the cell and the interconnection between neuronal subcellular compartments and autophagy. We will then discuss the physiology regulations of autophagy in synapse plasticity and explore the recent progress in neuronal autophagy in epilepsy.
2. The Historical Landmarks of Autophagy
Soon after the observation of the existence of cytoplasmic organelles within membrane-limited vacuoles by Clark in 1957, Ashford and Poter first found that after glucagon stimulation, the components from other organelles such as mitochondria were detected in some lysosomes that moved to the center of the hepatic cell, and appeared to be progressive decomposition [12,13]. Unfortunately, they mistakenly believed that the above phenomenon was the process of lysosome formation and that lysosomes were not organelles located in the cytoplasm like mitochondria. In 1963, Hruban et al. reported the ultrastructure of focal cytoplasmic degradation including three consecutive steps from cytoplasmic fusion to lysosome formation. They proposed that this process is not only induced by the injury stage but also in the physiological stage of organelle disposal, cytoplasmic component reuse, and cell differentiation [14]. Based on Hurban’s discoveries, de Duve’s laboratory named this phenomenon “autophagy” and appreciated that lysosomes play a vital role by fusion with prelysosomes containing sequestered cytoplasm in the cellular autophagy triggered in the rat liver by glucagon [15,16,17]. Remarkably, Clarke proposed a new death type for dying cells, autophagic cell death (ACD), distinct from apoptosis or caspase-independent cell death [18]. Although the importance of autophagy and lysosomes for maintaining cell homeostasis had been noted by researchers at the time, it was mainly regarded as a system for non-specific cleaning up and scavenging debris.
Degradation of cytoplasmic components was considered to be one of the key functions of the vacuoles of yeast cells, but it remained largely unresolved how cellular components are delivered to the vacuole. Many decades later, Ohsumi and colleagues identified the extensive autophagic degradation of cytosolic components in the vacuoles of yeast cells and named spherical bodies “autophagic bodies”, which contained cytoplasmic ribosomes, rough endoplasmic reticulum (ER), mitochondria, lipid granules, and glycogen granules [19]. Accumulation of autophagic bodies in the vacuoles of yeast cells was induced not only by nutrient-deficient conditions but also by disruption of the PRB1 gene (PRB1 gene encodes proteinase B associated with protein degradation with the vacuoles) or deficiency of multiple proteinases. Simultaneously, they isolated a mutagenized proteinase-deficient strain, apg1, which is defective in the accumulation of the autophagic bodies in the vacuoles, and confirmed that at least 15 APG genes are involved in autophagy, which is indispensable for protein degradation via autophagic bodies in the vacuoles of yeast cells under starvation conditions [20]. This result was arguably the most important historical event in the field of autophagy (Figure 1).
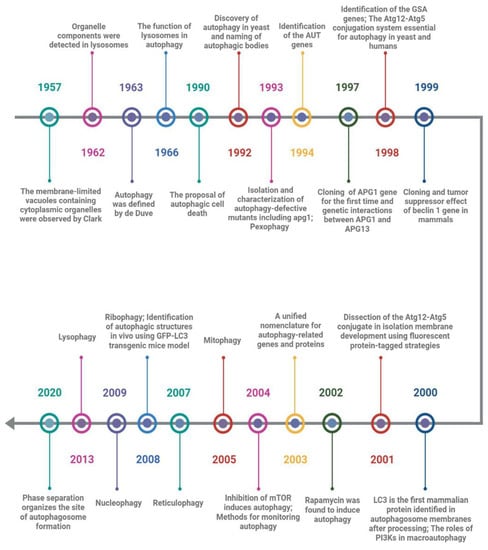
Figure 1.
Timeline of historical events in autophagy. This timeline depicts the key discoveries of autophagy. Unfortunately, it is not possible to include all discoveries due to limited space.
Following the introduction of genetics, autophagy-related genes had also been identified by other researchers independently. For example, Thumm et al. identified AUT genes (found to be identical to APG genes later) from autophagocytosis mutants of Saccharomyces cerevisiae [21]; Sakai et al. identified GSA genes during a search for underlying molecular involved in peroxisomal degradation [22]. Due to confusion about the names of the field, these autophagy-related genes and proteins had been unified as “ATG” and “Atg” in 2003, respectively, which stand for “autophagy-related” [23]. Strikingly, the Ohsumi laboratory cloned the ATG1 gene for the first time and explored the genetic interaction between ATG1 and ATG13 in yeast [24], and reported a well-conserved Atg12-Atg5 conjugation system that uses a ubiquitination-like conjugation model and is essential for autophagy in yeast and humans [25,26]. Meanwhile, beclin 1, a mammalian homolog of yeast Atg6/Vps30, was cloned by Liang et al. in 1999, and decreased expression of which may contribute to the development of human breast carcinoma and other malignancies [27]; Microtubule-associated protein 1 light chain 3 (LC3), a mammalian homolog of yeast Atg8p, was found to be specifically associated with autophagosome membranes after processing and has since been used as a marker of autophagy [28].
To determine how the isolation membranes are formed in autophagy, the Yoshimori laboratory showed that the Atg12-Atg5 conjugation system plays a critical role in isolation membrane development using green fluorescent protein-tagged Atg5-deficient mouse embryonic stem cells [29]. This is the first study on the molecular mechanism of autophagosome formation in mammals. Simultaneously, the signaling pathways in which distinct classes of phosphatidylinositol 3’-kinases (PI3K) control macroautophagy of HT-29 cells in opposite directions were discovered by Petiot et al. [30]; Rapamycin, a specific inhibitor of the mammalian target of rapamycin (mTOR), was found to induce autophagy and attenuate polyglutamine protein accumulation in cell models of Huntington’s disease [31,32]. This method of rapamycin-inducing autophagy has been widely used in the field of autophagy. Furthermore, the development of tools for monitoring autophagy by GFP-LC3 transgenic mice and useful marker proteins has accelerated the expansion of autophagy research across many fields such as neuropsychiatric disease, structural biology, developmental biology, aging biology, cell metabolism, inheritance and evolution, cancer, drug addiction, immunity, infection, and inflammation, to name a few [33,34,35,36,37,38,39]. In addition, the findings of organelle-specific autophagy reflect a corresponding multiplicity of intracellular events, such as pexophagy, mitophagy, reticulophagy, nucleophagy, lysophagy, and ribophagy [16,40,41,42,43,44,45,46,47,48].
Recent work has demonstrated that the pre-autophagosomal structure (PAS) in yeast is a liquid-like condensate of Atg protein complex and that inhibiting phase separation impairs PAS formation in vivo, indicating that phase separation plays an active, key role in autophagy, whereby it organizes the autophagy machinery at the PAS formation [49]. However, accumulating data has suggested that the ATG genes have functions in pathways other than autophagy and that some ATG elements may not be absolutely necessary for autophagy [50,51]. Even autophagy regulation shows obvious differences among different creatures [52]. Therefore, elucidating the meticulous structure and function of each component of distinct stages of autophagy will help us fully comprehend the autophagic system’s diversity.
3. The Distinct Types of Autophagy
Since its initial description, the autophagy process has mainly been referred to induction, sequestration, transportation, and degradation of unwarranted components of the cell [53]. Although the exact membrane resources (such as mitochondria, ER, Golgi, and plasma membranes) for autophagosomes remain a contentious question in the field, lysosomal digestion of the substrates has been widely recognized to date [2,54,55,56]. After the initiation of the process, different pathways for delivery of cargo to the lysosomes distinguish the three categories within autophagy in cells: CMA, microautophagy, and macroautophagy [57,58,59,60,61].
CMA was the first discovered process by which intracellular proteins are selectively degraded [62]. It involves the discrimination of degradation targets, the assembly of chaperone complexes, and other sophisticated mechanisms, which unfold and translocate the selected substrate across the lysosomal membrane, to then be digested by lysosomal hydrolases [61]. Specifically, target components for CMA were determined by a canonical KFERQ-like motif, which contains one of the negatively charged D or E residues, one or two of the positive residues K and R, one or two of the hydrophobic residues F, I, L, or V, and generally flanked by glutamine on one of the sides. On binding to the KFERQ-like motif, targeted proteins were docked to a LAMP2A-containing lysosomal membrane complex for degradation via the cytosolic chaperone HSC70 and cochaperones, such as HSP70-HSP90 organizing protein (HOP), the carboxyl terminus of HSC70-interacting protein (CHIP), and heat shock protein 40 (HSP40) [63]. By degrading specific target substrates promptly, CMA is involved in a variety of cellular activities.
Microautophagy is a constitutive process by which cytosolic components are directly engulfed by the invagination of endosomal, lysosomes, or vacuole membrane (in yeast) [64]. Due to the inability to detect an invagination-like process and no conserved function of yeast-microautophagy genes in mammals, the study of microautophagy has been least well elucidated.
Macroautophagy is an intricate and sequential process in which intracellular proteins and organelles are sequestered within the autophagosome, and then finally delivered to the lysosome for degradation [65,66]. During this process, phagophore, a double-membrane sequestering structure from several plausible sources of the cellular membrane, emerges first and becomes specialized as a PAS [67]. In the cooperation of two conserved ubiquitin-like conjugation systems, a portion of cytoplasm, including invasive pathogens, superfluous and damaged organelles, and cytosolic protein aggregates, is enveloped by elongation of PAS, ultimately forming a new vesicle, the autophagosome. Following delivery and fusion with the lysosome, the cargo including contents and its inner membrane is degraded by lysosomal hydrolases, and the breakdown products such as amino acids are released back to the cytoplasm for recycling the macromolecular constituents to reuse as building blocks or a source of energy under unfavorable conditions (Figure 2) [68]. In addition, ATG proteins are involved in each of these steps and mechanistically regulate the autophagic flux along autophagosome and autophagolysosome biogenesis [25,26,49]. Due to the highest efficient degradative capacity, macroautophagy is a predominant and prevalent degradative mechanism in various aspects of cellular activities.

Figure 2.
Schematic for macroautophagy. Macroautophagy is a multistep process that involves the induction/initiation, nucleation/expansion, maturation, trafficking of autophagosome, formation of autophagolysosome, and release and reuse of degradation products.
Moreover, some researchers divided autophagy into non-selective and selective autophagy according to distinct nutritional statuses. For example, selective autophagy included mitochondrial autophagy (Mitophagy), endoplasmic reticulum autophagy, peroxisome autophagy under nutrient-rich conditions, etc. [69]. Notably, autophagy in this review is macroautophagy.
4. The Molecular Machinery of Autophagy
Autophagy induction is generally determined by the cellular state. In nutrient-deficient conditions, AMP-activated protein kinase (AMPK) is activated by high AMP levels, thereby increasing the functions of the tuberous sclerosis complex (TSC1) and TSC2, which in turn inhibits the activity of the GTP-binding protein Rheb and phosphorylation of the mammalian target of rapamycin (mTOR) kinase [70,71]. Once mTOR is inactivated, it promotes the unc-51-like autophagy activating kinase 1 (ULK1) autophosphorylation, initiating PAS membrane assembly for phagophores biogenesis [72]. Regardless of the indirect mechanism, rapamycin, low amino acid levels, and energy stores also suppress mTOR phosphorylation. Furthermore, TSC1 and TSC2 are alternatively activated by the class I PI3K/Akt/protein kinase B (PKB) signal cascade, which is suppressed by the upstream growth factors or insulin, subsequently facilitating ULK1 phosphorylation by the Rheb/ mTOR pathway (Figure 3) [30]. In this situation, these modifications finally bring about the accumulation of phosphorylated ATG protein ULK1.

Figure 3.
Overview of upstream signaling pathways that regulate macroautophagy. Autophagy induction is initiated when mTOR signaling is suppressed by a range of stress modalities and an upstream signaling cascade. Abbreviations: PKB, protein kinase B; AMPK, AMP-activated protein kinase; TSC1, tuberous sclerosis complex 2; TSC2, tuberous sclerosis complex 2; mTOR, mammalian target of rapamycin.
ULK1, a protein with serine/threonine kinase activity, is the sole kinase of the core autophagy component in mammals [73]. Active ULK1 phosphorylates ATG13 and FIP200 (or RB1CC1), and recruits Atg101 (or C12orf44), a stabilizer of ATG13 and FIP200, forming the stable ULK1-ATG13-FIP200-Atg101 complex in a hierarchical manner (Figure 3) [74,75]. ATG13 is the regulatory subunit in this complex and identifies highly curved lipid membranes. In addition, the interaction between ULK1 and ATG13 is stabilized by the scaffold protein FIP200 [76]. In entire ULK1 complex-activated conditions, it initiates the assembly of phagophore membranes and facilitates the transitions into the PAS structures. Although the ULK1 complex is believed to be the core molecular machine of autophagy in the induction phase, the innate character of the PAS about its membrane origins and protein composition is not known.
Vesicle nucleation of autophagy is mainly regulated by the ATG9 cycling system and BECN1-ATG14L-VPS15-VPS34 complex (Figure 4) [66,77,78]. ATG9, which is a transmembrane protein consisting of six transmembrane domains, is phosphorylated by active ULK1 in PAS, subsequently forming a cycling system with ATG2 and WIPI1/2 (WIPI: WD-repeat protein interacting with phosphoinositides) [79]. Activated ATG9 shuttles between new growing PAS and membrane donors, such as the Golgi complex, endoplasmic reticulum, mitochondria, and other resources, and binds to the lipid-binding protein ATG13 in the ULK1 complex, thus directing membranes to the nascent PAS [80]. As for ATG2, it not only brings lipids to the growing PAS membrane as a possible lipid transporter but also interacts with ATG9 and WIPI1/2, in which WIPI1 and WIPI2 may oligomerize and interact with membranes for autophagy flux [81].

Figure 4.
Schematic diagram of vesicle nucleation in macroautophagy. Vesicle expansion is orchestrated by Atg proteins and complexes, such as the class III PI3K complex. Abbreviations: 3-MA, 3-methyl adenine.
Simultaneously, the WIPI also links VPS34 (or PIK3C3) for phagophores expansion [82]. VPS34, the type III phosphatidylinositol-3 kinase (PI3K) that can be inhibited by 3-methyl adenine (3-MA), forms the class III phosphoinositide 3-kinase complexes with VPS15 (or PIK3R4), ATG14L and BECN1 (or VPS30) [83]. The activation and recruitment of the class III PI3K complex are dependent on the phosphorylation of BECN1 via active ULK1 [84]. It has been suggested that the class III PI3K complex binds to the C-terminus of the Atg101 HORMA domain in the PAS membrane, and generates the phosphatidylinositol-3 phosphate (PI3P), an essential component of the vesicle nucleation of phagophore during biogenesis [85]. The generation of PI3P facilitates the recruitment of PI3P effectors and PI3P-binding proteins, thus involving in “isolation” membrane formation, curvature shaping, and phagophores expansion [86]. In addition, ATG14 is a critical component for the proper recruitment and localization of the class III PI3K nucleation complex as well as the creation of an “isolation” membrane (Figure 4) [87].
Remarkably, BECN1, namely Beclin1, includes three key functional domains, the central coiled-coil domain (CCD), the Bcl2-homology3 (BH3) binding site located at the N-terminus of the protein framework, and the evolutionarily conserved and C-terminus domain, playing a core role in coordinating the formation of the class PI3K complex and the process of membrane transport [85,88]. More research has revealed that BECN1 is specifically required for autophagy vesicle nucleation, inflammation response, cell apoptosis, longevity and other cellular activities, and is considered as the central link of these signal pathways [89,90]. For instance, using BECN1-induced autophagy, the oncolytic viruses significantly increased BCR/ABL protein expression, and significantly improved the pathology of chronic myeloid leukemia, suggesting that BECN1 may offer a new target for clinical implications [91]. Meanwhile, the BECN1-related complex contains at least three different complexes, with the catalytic core region BECN1-VPS34 (Figure 4) [92]. BECN1-ATG14L-VPS34 complex, the classical autophagy pathway in vesicle nucleation, interacts with the phagocytic membrane of the endoplasmic reticulum, and is stimulated and inhibited by the binding of AMBRA1 and Bcl-2/Bcl-xL, respectively [66,93]; BECN1-VPS15-Vps34-UVRAG complex is the endocytic pathway of the autophagosome, and constitutively displaces AMBRA1 and ATG14 components with UVRAG and its regulatory element SH3GLB1, leading to the direct phagocytic effect and the accelerated maturation of autophagosomes [94]; The third complex is an inhibitory complex, in which the regulator SH3GLB1 is switched to KIAA0226/Rubicon. Rubicon can inhibit the UVRAG and VPS34 activities, finally playing a negative role in PAS membrane expansion and autophagosome maturation [95,96]. Additionally, Rubicon and ATG14L competitively interact with the central coiled-coil domain of BECN1, thereby maintaining the neutral state between endocytosis and autophagy [97].
Elongation of the phagophores membrane is main mediated by two ubiquitin-like (Ubl) conjugation systems, namely the microtubule-associated protein 1A/1B-light chain (LC3) Ubl conjugation and the ATG12 Ubl conjugation (Figure 5) [98]. Initially, under the effects of the E1-like ubiquitin ligase ATG7 and E2-like ubiquitin ligase ATG10, the carboxy-terminal Ubl structure of Ubl protein ATG12 conjugates to the lysine of the ATG5, forming the ATG12-ATG5 conjugate [99]. This conjugate increases the affinity for ATG16L1, which safeguards the ATG12-ATG5 complex against ubiquitin-like modification and protease degradation by binding to free ATG5, promoting the complicated assembly of the ATG12-ATG5-ATG16L1 system [100]. In this system, ATG16L1 plays the role of dimerization protein through binding to two ATG12-ATG5 conjugates and constitutes ATG12-ATG5-ATG16L1 homodimers [101]. ATG12 is one of two Ubl proteins in autophagy and it directs the ATG12 Ubl conjugation system to the outer membrane of the phagophores by an auxiliary connection [98]. Moreover, two hydrophobic residues in the ATG12 ubiquitin-fold region are necessary for the ATG12 Ubl conjugation system. The mutation at Y149 reduces the catalytic capability of ATG10 and severely impedes the conjugate formation of the ATG12-ATG5 complex, which is not affected by the F154 mutation [102]. In response to the mutation at F154, the LC3 II-phosphatidylethanolamine (PE) conjugation is significantly stopped, failing vesicle elongation [103].
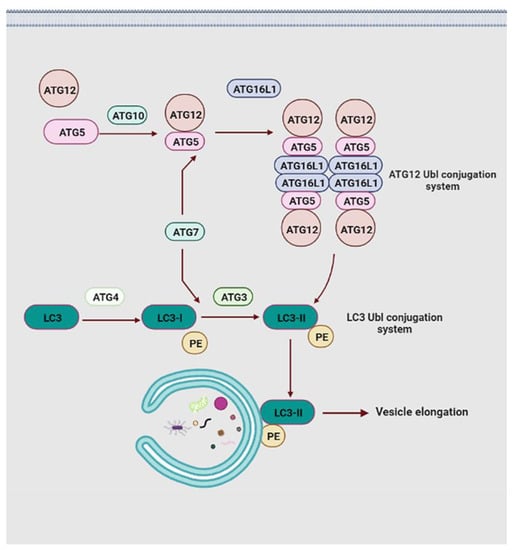
Figure 5.
Schematic representations of vesicle elongation events. The elongation of the isolation membrane is mainly directed by two ubiquitin-like protein conjugation pathways. Abbreviations: PE, phosphatidylethanolamine.
In the LC3 Ubl conjugation system, the carboxy-terminal of LC3 is cleaved by ATG4 protease to form LC3-I, and then it is transferred into LC3-II formation via the action of E1-like ligase ATG7 and E2-like ligase ATG3 [104]. Subsequently, the ATG12 Ubl conjugation system interacts with LC3-II in a manner similar to the E3-like enzyme and facilitates the generation of LC3 II-PE conjugation through the amine headgroup binding of LC3-II and PE. LC3 II-PE conjugation relies on the ATG12 component to accomplish membrane positioning with the expanded phagophores membrane [105]. Lipidated LC3 not only discriminates against specific cargo through the LC3 interaction region domain but also participates in the elongation and closure of the PAS membranes and further autophagosome maturation (Figure 5) [104]. Moreover, LC3 is one of two subfamilies of yeast ATG8 homologs in mammals, the other being GABARAP [106]. Previous studies have shown that the LC3 proteins act at the stage of vesicle elongation, whereas the GABARAP functions at a later stage of autophagosome maturation [107]. Notably, LC3 or GABARAP proteins interact with a series of adaptor proteins, such as p62/sequestosome 1 (SQSTM1) (hereafter p62) and calcium-binding and coiled-coil domain-containing protein 2 (also called NDP52), facilitating autophagosome formation around specific cargoes [108,109,110]. p62 is one of the most well-characterized of these adaptors and is a multidomain protein that modulates distinct signaling pathways through different binding partners. Prominent p62 domains encompass the N-terminus Phox/Bem1p (PB1), the LC3-interacting region (LIR), the tumor necrosis factor receptor-associated factor 6 (TRAF6)-binding sequence (TBS), the ZZ-type zinc finger (ZZ) domain, the Kelch-like ECH-associated protein 1 (Keap1)-interacting region (KIR), and a C-terminal ubiquitin associated (UBA) domain. p62 interacts directly with LC3 and GABARAP protein family members through its LIR domain, and binds directly to selected cargoes through its C-terminal UBA domain [111]. Moreover, the PB1 domain-mediated homopolymerization of p62 results in its co-aggregation with the selected cargo, which promotes the close interaction between the p62-binging cargo and lipidated LC3 proteins (or GABARAP proteins) at the phagophore [112]. Not only are these interactions indispensable for delivery of selected substrates to autophagosomes, but they also potentially play a key role in the formation of autophagosomes themselves. Additionally, p62 degradation also relies on autophagy, that is, the level of p62 accumulates in response to the inhibition of autophagy, and the level of p62 decreases in response to the induction of autophagy [108,109]. Apart from this, accessory ATG proteins are related to the ultimate steps of autophagosome biogenesis, but their concrete roles are not explicitly known yet.
After the elongation, the cup-shaped PAS membrane encloses a portion of the cytoplasm or damaged organelles through the endosomal sorting complex required for transport (ESCRT) mechanism, finally forming the autophagosome [113]. Nascent autophagosomes are randomly dispersed throughout the cytoplasm, and recruit the molecular switch Rab GTPases to the autophagosome membrane, promoting the interaction between the autophagosome and dynein (Figure 6) [114]. Generally, Rab GTPases are critical regulators of eukaryotic membrane trafficking, such as Rab2A, Rab5, Rab7, and Drosophila ADP-ribosylation factor (Arf)-like 8 (Arl8) (key players in autophagy), and can hydrolyze GTP to GDP as well as bind to target membranes on GTP binding, thereby serving as the molecular identity of the target membrane (lysosomes, autophagosomes, late endosomes, and Golgi-derived vesicles) or molecular switches [115]. For instance, autophagosomes recruit Rab7 during maturation in a Monensin sensitivity protein 1-Caffeine, calcium, and zinc 1 (Mon1-Ccz1) dependent manner, and also play a key role during endolysosome formation or endosome maturation [113,116,117]. These proteins bind mainly to effectors required to drive the fusion of autophagosome-lysosome in mammals, such as cytoskeleton-associated motor proteins, tethering factors, and adaptors [118]. As is known to all, autophagosome and lysosome transport depends on the microtubular system and related motor proteins. The minus-end directed dynein-dynactin motor complex drives the autophagosome to the perinuclear region, whereas plus-end directed kinesins move late endosomes/lysosomes to the cell periphery, promoting each other’s successful encounters [119,120]. Rab GTPases including Rab7 bind to several motor adaptor proteins, such as FYVE and coiled-coil domain-containing 1 (FYCO1, mediate outward transport), Rab-interacting lysosomal protein (RILP, mediate inward transport), and oxysterol-binding protein-related protein 1L (ORP1L, mediate inward transport), pleckstrin homology and RUN domain containing M1 (PLEKHM1), and BORC complex [113,121,122,123]. These effectors can link lysosomes or autophagosomes to cytoskeleton-associated motor proteins (such as the light chain of kinesin 1 (KLC2), Kinesin-3, and kinesin-13), so they can mediate both plus-end and/or minus-end transport of these vesicles or autophagosome [124,125]. Of note, motor adaptor proteins have been shown to directly bind LC3 or GABARAP proteins to promote dynein-mediated autophagosome transport [126].

Figure 6.
Schematic for the trafficking of autophagosome, formation of autophagolysosome, and release and reuse of degradation products in macroautophagy. Abbreviations: SNAREs, the soluble N-ethylmaleimide sensitive fusion protein attachment protein receptor.
Through dynein-dependent mechanisms, autophagosomes and lysosomes are transported along the intracellular microtubules toward the perinuclear region where the autophagolysosome formation is mediated via the soluble N-ethylmaleimide sensitive fusion protein attachment protein receptor (SNAREs), the membrane-tethering proteins and other fusion-related protein families [113,127]. Specifically, autophagosome-lysosome (vacuole in yeast) fusion requires tethering the lysosome and autophagosome together, which is usually mediated by multisubunit membrane-tethering complexes or large coiled-coil proteins [128]. These proteins usually bind to GTP-loaded Rab GTPases and/or vesicle membranes of autophagosomes and late endosomes/lysosomes, and collaborate with Sec1/Munc-18 (SM) family proteins to promote SNAREs complex assembly and zippering [129]. The most well-studied tethering protein in autophagosome-lysosome fusion is the heterohexameric homotypic fusion and vacuole protein sorting (HOPS) complex, which comprises six subunits: the two Rab7 (Ypt7 in yeast) binding subunits (Vps41 and Vps39) and the 4-subunit class C core [130]. Accumulated pieces of evidence have indicated that HOPS is critical for nearly all lysosome-related fusion events, such as autophagosome-lysosome fusion, late endosome-lysosome fusion, and secretory granule-lysosome fusion during crinophagy and lysosome-related organelle biogenesis [127,131,132,133]. HOPS can directly (or indirectly) bind to Rab7 (through adaptors Plekhm1 and RILP), Rab2 (or RAB2A), and Arl8, and be recruited to lysosomes through Arl8 and the BORC complex and to autophagosomes through Rab2 or STX17. In addition, HOPS also acts as an SM protein via subunit Vps33A [134,135,136,137]. Besides HOPS, the tethering of autophagosomes with lysosomes is also meditated by additional proteins, such as BRUCE and RUN and FYVE domain-containing protein 4 (RUFY4), ectopic P-granules 5 (EPG-5), ATG14, Golgi reassembly stacking protein 55 (GRASP55), and tectonin beta-propeller repeat containing 1 (TECPR1) [138,139,140,141]. Remarkably, some tether proteins have also been found to bind to the autophagosome PI3P and LC3/GABARAP family proteins [142].
Tethered proteins cross-link the lysosomal or late endosomal membranes to autophagosome membranes, whereby SNARE complexes carry out autophagosome-lysosome fusion [143]. Approximately forty soluble SNARE proteins are essential for intracellular membrane fusion and they are classified into vesicular/v-SNAREs (R-SNAREs) and target/t-SNAREs (Q-SNARE-s). Q-SNAREs are usually grouped into Qa-, Qb-, and Qc-SNARE-s, and some are Qbc-SNARE-s [144,145]. Autophagosome-lysosome fusion is driven by a trans-SNARE complex, which consists of three different Q-SNAREs (Qa, Qb, and Qc-SNAREs) anchored in a vesicle membrane and an R-SNARE anchored to the membrane of another vesicle [144,146]. Interestingly, these SNARE proteins are assembled into multiple SNARE complexes, which are not conserved from yeast to mammalian cells [143]. For instance, the autophagosomal R-SNARE Ykt6, the vacuolar Qa-SNARE vacuole morphology3 (Vam3), and the Qc-SNARE Vam7 and the Qb-SNARE Vps10 Interacting1 (Vti1) form a trans-SNARE complex that promotes the membrane fusion of autophagosomes with the vacuole in yeast [147,148,149,150]. Conversely, autophagosome-lysosome fusion in mammals is mainly mediated by two different SNARE complexes: the autophagosomal Qa-SNARE Syntaxin17 (STX17), the lysosomal/late endosomal R SNARE Vamp7 or Vamp8, and two Qbc-SNARE SNAP-29; and the autophagosomal R SNARE YKT6, the lysosomal/late endosomal Qa SNARE STX7, and two Qbc-SNARE SNAP-29 [151,152,153,154,155]. Following the fusion with lysosomes or late endosomes, autophagosome components are digested by activating lysosomal hydrolase, and the degradation of macromolecules is transported back into the cytoplasm for recycling [156]. The ultimate destination of autophagolysosomes is to become residual bodies (Figure 6) [157].
Notably, the protein LC3-II and p62 levels have been widely applied to the assessment of autophagy flux in vivo and in vitro [158]. Autophagic flux is an entire intracellular process including acquisition, packaging, trafficking, degradation, and release of unwanted or damaged substrates in the lysosomal system. Monitoring autophagic flux is critically important for dissecting the biology of autophagy, renewing the understanding of how autophagy regulates physiological and pathological conditions, and screening for autophagy-modulating drugs or other molecular compounds [159]. To date, autophagic flux is usually measured by the comparison of the LC3-II level or the autophagosome number between samples or individuals with and without lysosomal inhibitors (chloroquine, bafilomycin A1, or other protease inhibitors) using fluorescence visualization strategies (such as red fluorescent protein, or GFP or mCherry) [160]. In parallel with monitoring LC3-II, the localization and level of p62 are used to reflect autophagic activity. In addition, several detection methods are currently available, such as flow cytometry, western blots, pulse-chase measurement of half-life, immunofluorescence (subcellular localization pattern) and electron microscopy, and live cell imaging with fluorescence protein tags (pH-sensitive or not) [161]. However, accurate measurement of autophagic flux is still challenging.
5. The Regulation of Autophagy in Neurons
Neurons are a class of post-mitotic, polarized, hypermetabolic, long-lived, and highly specialized cells that were initially elucidated by Cajal in the early 19th century. Unlike somatic cells, neurons have a unique architecture with three subcellular compartments: the soma, dendrites, and axons, which are critical to their highly specialized functions (such as signaling transmission and synapse plasticity) (Figure 7). Axons usually grow from the axon hillock and extend multiple branches and elaborate terminal arbors from growth cones, which connect widely divergent synaptic targets in brain regions, finally forming complex neural circuits and topographic maps [162]. In this network, synapses are the fundamental units of communication between two neurons and are established by the dendrites of one neuron and axons of the ascending projection neuron.

Figure 7.
Neurons have unique architectures and characteristics.
Although the neuronal architectures and characteristics (such as hypermetabolic, long-lived, and highly specialized) rely on cell determination, autophagy is required for the maintenance of these properties and functions. Firstly, the neuron is a long-lived and post-mitotic cell, whereby it has no ability to dilute the accumulated effects of damaged proteins, organelles, and themselves through mitosis. Autophagy can dominate the turnover of RNA, lipids, proteins, and organelles, which maintain neuronal survival throughout the lifetime of most organisms [163,164]. Secondly, the neuron is a polarized cell and has highly polarized structures, such as axons and dendrites. Autophagy plays a key role in these aspects (such as complex neural circuits and topographic maps). For instance, during early growth and active developmental refinement of axons and dendrites (such as orphan neurites are pruned), autophagy regulates new protein synthesis, protein degradation, and neurite elimination (axon or dendrite pruning), which is important for the formation of specific neuronal structures [163,165,166]. Thirdly, the elimination and establishment of synapses (synapse plasticity, and neuronal activity) are highly dynamically regulated in response to activity-dependent plasticity or Hebb’s synaptic plasticity that involves cytoskeletal dynamic mechanisms and multiple intracellular signaling cascades [167]. This process needs a lot of molecules and/or proteins (such as neurotransmitters, RNA, and lipids) that are synthesized, delivered, and degraded for their dynamic functions in synaptic growth and synaptic activity. These are also regulated by autophagy (see below) [168]. Thus, we can say that autophagy is critical to maintaining the specialized functions of the neuron.
In addition, neurons must function continuously for as long as possible in a starved state, which is exceedingly different from other cell types. Brain neurons perform the functions of receiving, transmitting, integrating, processing, and outputting information, actively managing the general and cognitive activities of the organism. These processes place high energy and metabolic demand on neurons, especially in axons and dendrites located far away from the soma. Previous studies have suggested that neurons can use several different energy sources optimally, receive additional nutrients and neurotrophin support from glial cells, and benefit from the hypothalamic regulation of peripheral nutrient supplies [169]. Autophagy can also regulate nutrient supply under extreme starvation or diabetes.
Notably, the activity-dependent refinement of synapses (including the neuronal circuit change) occurs during the development and learning stages, and shares features with diseases such as epilepsy. For instance, the structural remodeling of mossy fiber synapses and the formation of aberrant synaptic contacts (such as synapse plasticity) in the dentate gyrus are common features in experimental models of epilepsy and human TLE [170,171,172,173]. In addition, amino acid metabolism (such as GABA) may be critical for epilepsy due to its role as the neurotransmitter (synapse transmission) and energy source [11]. Epileptic seizures increase the energy requirements of metabolically already compromised neurons, establishing a vicious cycle that results in worsening energy failure and neuronal death [174]. Changes in action potentials and ion channels are also related to the epileptogenesis of both genetic and acquired epilepsies [175]. Thus, changes in these physiological states may lead to the occurrence and development of epilepsy pathology (see below).
5.1. Autophagy in Axons
Axons have a powerful capability to regulate growth, which controls the axonal extension to the accurate sites of descending projection neurons and the timely termination of unwanted growth, ensuring the successful establishment of functional neural networks [162]. Simultaneously, synaptic vesicles, fusing with the presynaptic membrane of the axon terminals, trigger the release of neurotransmitters into the synaptic cleft (or Postsynaptic density, PSD), and evoke the activity of postsynaptic neurons by binding to the postsynaptic membrane receptors. Then, axon integrity (including membranes recycling, protein composition, synaptic connection, etc.) and synaptic stability are also maintained over time [176]. Moreover, experience-dependent synaptic plasticity, such as long-term depression (LTD) and long-term potentiation (LTP), results in long-lasting structural changes via synaptic pruning, a process of depleting highly intracellular ATP [177]. All these processes in axons require autophagic recycling of damaged or no longer needed contents to repurpose materials for structural alteration and growth. Thus, autophagy is a critical mechanism for maintaining synaptic functions, particularly in presynaptic compartments of the axon.
Autophagy in axons is initially identified in cultured peripheral neurons, in which large autophagosomes are trafficked retrogradely toward the soma and fused with lysosomes, suggesting that the formation of autophagy appears constitutively in distal axons [178]. Subsequently, autophagosome biogenesis at the axonal tip was corroborated in the cultured dorsal root ganglion (DRG) neurons as well as the embryonic cortical and hippocampal neurons [179]. These findings have consistently occurred in multiple model organisms, such as Caenorhabditis elegans, yeast, worms, Drosophila, Zebrafish, rats, mouse, humans, and other mammalians, demonstrating that the biogenesis and trafficking of axonal autophagosome in vivo are evolutionarily conserved [180,181,182,183]. Conditional ablation of autophagy proteins ULK/UNC-51, ATG5, ATG7, ATG9A, and FIP200 indicated that impairing autophagy resulted in abnormal axon growth and morphology, selective accumulation of tubular ER, and increased neurotransmitter release, finally inducing axon degeneration and behavioral deficits in vivo [184,185,186,187]. In cultured motor neurons, Pleckstrin homology containing family member 5 (Plekhg5, the guanine exchange factor for Rab26 GTPase) gene inactivation decreased axon outgrowth and increased accumulation of synaptic vesicles at the axon tip, whereas constitutively active Rab26 GTPase mitigates functional deficits in axon growth and autophagy [188].
A recent study has shown that Rab26 and ATG16L regulate neurotransmitter release through autophagy. Rab26 localizes on synaptic vesicles and is preferentially oligomerized to its GDP-bound form, which binds to ATG16L1 (Figure 8) [189]. This complex links synaptic vesicle clusters to the autophagy pathway and may assist in the migration of synaptic vesicles to nearby active sites, involving the regulation of presynaptic autophagy and neuronal activity [190,191]. Collectively, accumulated evidence implicated that axonal autophagy shares core machinery with non-neuronal cells.

Figure 8.
The cross-regulation between neuronal autophagy and synaptic vesicles at the pre-synaptic and post-synaptic terminals. Neuronal activity induces autophagosome formation at the presynaptic terminal by regulating several presynaptically enriched adaptors (such as Synaptojanin1, Endophilin A, and Bassoon) and regulatory proteins (a). At the postsynaptic terminal, neuronal activity also upregulates neuronal autophagy, resulting in endocytic removal of neurotransmitter receptors (such as GABAAR, NMDAR, and AMPAR) from the plasma membrane, presumably by modulating receptor adaptor proteins (b).
Apart from the general autophagy mechanisms, several neuron-specific proteins are involved in autophagy regulation at axon terminals. Endophilin A, a synapse-enriched adaptor that can be phosphorylated by the kinase LRRK2 at serine 75 of the BAR domain, generates zones of highly curved membranes [192]. These membranes serve as a docking platform to recruit E2-like ligase ATG3, facilitating autophagosome biogenesis (Figure 8a) [193]. The functional disturbance of phosphorylated Endophilin A accelerates the accumulation of clathrin-coated vesicles, and impedes synaptic transmission, thus resulting in severe activity-dependent neurodegenerations and seizures [194]. Furthermore, Endophilin A also interacts with GTPase dynamin and synaptojanin 1 by its SH3 domain for autophagy [195]. Dynamin is a core component of the endocytic machinery and interacts directly or indirectly with multiple accessory proteins to coordinate vesicle membrane fission, promoting the constriction of coated pits (Figure 8) [196]. Synaptojanin1 is a lipid phosphoinositide phosphatase that is localized to axon terminals and contains two different phosphatase domains, SAC1 and 5-phosphatase, playing a critical role in synaptic vesicle trafficking [197]. The SAC1 hydrolyses phosphoinositide phosphate PI(3)P, PI(4)P, and PI(3,5)P2, while the 5-phosphatase mainly targets the PI(4,5)P2 as an enzyme substrate [191,192]. The genetic disruption of synaptojanin1 enzymatic domains has confirmed that the 5-phosphatase domain reduces the membrane-binding affinity of the clathrin adaptors and uncoat nascent vesicles, rather than the SAC1 domain [198]. Interestingly, recent work has suggested that the SAC1 domain can demolish the WIPI2/Atg18a, which recognizes and binds the protein PI(3)P/PI(3,5)P2, and is essential for autophagosome biogenesis at presynaptic terminals, not the 5-phosphatase domain [191,199]. The mutations of synaptojanin1 R258Q caused the accumulation of Atg18a-positive structures at newly formed synaptic autophagosomes, and impaired the autophagosome maturation, finally inducing the loss of dopaminergic neurons in flies and humans [199,200]. In addition, the E3-like ubiquitin ligase FBXO32/atrogin-1 tabulates vesicle membranes and localizes to clathrin-coated regions for autophagosome formation in a manner similar to endophilinA [201].
Several studies have shown that autophagy deficiency induced by impairing positive regulators (such as EPG5, AP4, TECPR2, Alfy, VAMP7, PTPRσ, and other accessory proteins) exhibits different results in axon growth and neurological disease, accompanied by confirmations of gain or loss of function [202,203,204,205,206,207]. Some cause neuroaxonal dystrophy and hypoplasia, while others promote axonal growth. For instance, AP-4 is a member of the heterotetrameric adaptor protein (AP) family involved in protein sorting in the endomembrane system of eukaryotic cells (including neurons) [208]. δ2 glutamate receptor (δ2R) and amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-type glutamate receptors (AMPAR) are also sorted by AP-4 as cargos [209,210]. The AP-4 ε (the ε subunit of AP-4) KO mice display a thin corpus callosum and axonal swellings in various brain regions and spinal cord, such as hippocampal and Purkinje neurons, with a defect in autophagosome maturation. In addition, AP-4 ε KO mice also display impaired performance on the rotarod, and other motor and behavioral abnormalities, such as reduced grip strength, hindlimb clasping, increased ambulation, and enhanced acoustic startle response [210,211]. The disruption of the gene encoding the β subunit of AP-4 can increase the accumulation of axonal autophagosomes that contain AMPAR and transmembrane AMPA receptor regulatory proteins (TARPs) in axons of cerebellar Purkinje cells and hippocampal neurons both in vivo and in vitro. In fact, AP-4 is associated with TARPs that tightly bind to all subunits of AMPA receptors GluR1-GluR4, whereby the mutation of AP-4 may regulate the proper somatodendritic-specific distribution of AMPA receptor, TARP, and other cargo in different neurons (such as the hippocampus, spinal cord, and other brain regions) [204,211]. Moreover, knockouts of Atg5 and Atg7 in Purkinje neurons and rhodopsin neurons of the retina, and Atg7 in agouti-related peptide (AgRP) neurons of the hypothalamus have also been well-studied [212,213,214,215,216]. For instance, the disruption of the genes Atg5 and Atg7 caused Purkinje axonal swellings, followed by progressive dystrophy and degeneration in the axon [212,213]. Thus, the above studies suggest that autophagy deficiency induced by impairing positive regulators Ap4 (or ATG5, or ATG7) contributes to neuroaxonal dystrophy, axonal degeneration, and abnormal neuron functions, such as AMPAR-mediated synapse efficiency, particularly in Purkinje and hippocampal neurons.
However, Ban et al. reported that the inhibition of autophagy by atg7 small interfering RNA (siRNA) resulted in the elongation of axons, while activation of autophagy by rapamycin suppressed axon growth in cortical neurons [217]. Consistent with this, the mutation of Mir505-3p, which is an autophagic regulator for axonal elongation and branching, consistently enhanced the autophagy signaling cascade for axon growth and autophagosome biogenesis [218]. So, why does knocking out a positive regulator of autophagy has the opposite effect? We note that the mammalian central nervous system needs autophagy to maintain its normal functions and homeostasis, particularly in the axons [163,165,168,219]. Different brain regions have distinct types of neurons, synapse connections, and biological functions, which depend on dissimilar autophagy needs in different neurons [165,168]. The deletion of positive regulators (such as AP-4, ATG5, and ATG7) displays axonal degeneration in different neurons of brain regions, particularly in hippocampal neurons, whereas the deletion of positive regulators (such as ATG7) in cortical neurons displays axon growth. Thus, this apparent paradox may suggest that different types of neurons (such as hippocampal, Purkinje, hypothalamic, and rhodopsin neurons) have different structures for specific neuron functions, and different neuron types have dissimilar autophagy needs during axon regulation, which results in different effects for deletion of positive regulators.
In contrast, Bassoon and Piccolo negatively regulate autophagy by inhibiting autophagosome biogenesis. Specifically, Bassoon is a scaffold protein in the presynaptic active zone and interacts, sequesters, and prevents ATG5, an E3-like ligase that is essential for the binding of LC3 to PAS membranes, actively interfering in the formation of autophagosomes [220]. Functional loss of Bassoon in primary embryonic hippocampal neurons leads to the accumulation of LC3-positive complexes and increased autophagy activities at axon terminals, and impaired synaptic integrity. Yet, it is accompanied by reduced numbers of synaptic vesicles, implicating that presynaptic autophagy still actively hydrolyses synaptic vesicles as well as their components in the absence of Bassoon [221]. Moreover, the ubiquitin ligase RPM-1 restricts UNC-51/ULK kinase activity, and inhibits the formation of autophagosomes for axon termination, negatively regulating the initiation of autophagy, synapse maintenance, and behavioral habituation (Figure 8a) [222].
Additionally, the brain-derived neurotrophic factor (BDNF) is not only an important mediator of activity-dependent modifications of synaptic strength in synapses but also a key regulator of autophagy (Figure 8a). BDNF increases the accumulation of synaptic vesicles at active zones and the quantal neurotransmitter release within the presynaptic terminal, and specifically suppresses axonal autophagy by inhibiting autophagosome biogenesis [223]. The conditional deletion of BDNF exhibits an overabundance of autophagosomes in the axon terminals, as confirmed by electron microscopy in the hippocampal neurons [224]. Taken together, these shreds of evidence indicate that autophagy directly regulates axon growth and presynaptic neurotransmitter release, via the mechanisms that remain to be clearly elucidated.
5.2. Autophagy in Dendrites
Dendrites are the foremost sites for information input and integration from presynaptic neurotransmitters. Function impairments of dendrites lead to abnormal neuronal activities and neuronal circuits. Under native conditions, autophagy functions as a scavenger to clear cytotoxic materials and recycle the degraded components, especially in dendrite spine pruning [225]. Autophagic dysfunction has been linked to dendritic atrophy as well as neuronal cell death. However, little is known about the underlying mechanisms of dendrite autophagy in neurons.
In Drosophila, basal ATG genes direct dendritic terminal branching and arborization by positive regulation of the homeodomain transcription factor Cut. The genetic disruption of Atg genes boosts type-specific neuronal dendritic arborization, whereas the function gain of these genes dramatically reduces the dendritic complexity in multidendritic sensory neurons, such as deficient dendritic branching and decreased arbor growth [226]. In addition, the specific deletion of ATG7 in dopaminergic neurons results in delayed dendritic neurodegeneration, accompanied by large dendritic ubiquitinated inclusions and dendritic dystrophy [227]. Whole-brain knock-out of Atg7 significantly impairs dendritic spine pruning, promoting social behavioral disorders in mice [228]. These results show that constitutive autophagy is essential for dendrite arborization, branching, and spine fine-pruning.
Moreover, dendritic autophagy also mediates the intracellular trafficking of postsynaptic membrane receptors. Previous studies have suggested that the lapidated GABARAP subfamily of mATG8s facilitates intracellular trafficking of Gamma-aminobutyric acid A (GABAA) receptor, and increases the GABAA receptor surface expression in primary hippocampal neurons, PC12 cells, and Xenopus oocytes (Figure 8b) [183,229]. Dendritic autophagy degrades the GABAA receptor at postsynaptic terminals in the absence of presynaptic innervation, with the diffuse distribution of the GABAA receptor on the plasma membrane of the muscle. Genetic disruption of autophagy rescues neurotransmission defects in non-innervated muscles. Interestingly, in C. elegans, dendritic autophagy reduces the surface expression of the GABAA receptor in non-innervated muscles [230]. The deletion of autophagy does not increase acetylcholine (ACh) currents, accompanied by non-colocalization with autophagosome markers [231]. These results indicate that autophagy selectively may modulate neuronal excitation and inhibition.
Autophagy also coordinates synaptic activity in the dendrites [191]. In cultured hippocampal neurons, KCl depolarization and brief low-dose N-methyl-D-aspartate (NMDA) transiently increases the number of LC3-II puncta and autophagosomes in spines and accelerates the degradation of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors, which are partially recovered by the NMDA receptor inhibitor APV. The short hairpin RNA-induced knockdown of ATG7 and the autophagy inhibitor wortmannin inhibits chemical-LTD-mediated autophagy through the PI3K-Akt-mTOR pathway [232]. Additionally, BDNF functions as a chemical inducer of LTP and modulates dendritic autophagy in hippocampal neurons. BDNF deficiency leads to LTP deficits and memory impairment in mice, which are alleviated by the suppression of autophagy [224]. Several scaffolding proteins including SHANK3, PSD-95, and PICK1 in dendrites are also colocalized in autophagosomes, and their degradation may regulate synaptic plasticity (Figure 8) [233].
5.3. Autophagy in Soma
Little is known about neuronal autophagy in the soma. Previous evidence indicates that autophagosome biogenesis events are enriched in the distal axon, and are infrequently observed in the soma of the hippocampal cultures and the primary dorsal root ganglion via live-cell imaging [183]. The soma in the hippocampal neurons contains the bulk of autophagosomes, which are defined by differences in dynamics as well as maturation state and are generated from different neuronal compartments and are enriched by populations of proteolytically active lysosomes enabling cargo degradation [234]. The autophagosomes derived from the neuronal soma tend to cluster and are less mobile.
The most common form of autophagy in neuronal soma is mitophagy. Mitochondria play a key role in neuronal function and survival, and mitophagy is essential for eliminating damaged and aged mitochondria. The Parkin-mediated and mitochondrial Δψm-induced mitophagy in cortical neurons is the unique process where the Parkin-labeled mitochondria are accumulated at a somatodendritic site and subsequently degraded by the autophagy-lysosomal pathway in soma [235]. In addition, the functional deletion of genes in the PINK1/Parkin pathway results in diminished mitochondrial membrane potential and compromised somatic mitochondrial integrity, but does not increase the mitochondrial density or length in axons, suggesting that mitophagy-dependent mitochondrial turnover events may be restricted to the neuronal soma in vivo [236]. Overall, several studies have shown that autophagy in soma directly regulates mitochondria quality control and involves neuronal senescence and multiple neurological disorders, by mechanisms that remain to be further elucidated.
5.4. Neuronal Autophagy and Synaptic Plasticities
Synaptic plasticity, the activity-dependent change in the efficacy of synaptic connection, has long been thought to represent a fundamental property of learning and memory and an important component of brain adaptation [237]. Synaptic plasticity processes rely heavily on the quality control of protein-protein interactions and organelles, and autophagy is crucial to the homeostatic maintenance of synaptic plasticity, spine pruning, and axon guidance [238]. Recent studies indicated that autophagy plays a key role in information encoding, memory processing, and cognitive functions. When dysregulated, autophagy is associated with disrupted synaptic plasticity, neurological disorders, such as autism spectrum disorders and stroke, and neurodegenerative diseases, such as Alzheimer’s disease, Parkinson’s disease, multiple sclerosis, and others [239]. However, little is known about how autophagy regulates synaptic plasticity.
Loss of Atg7 in mouse neurons and microglial cells damages synaptosome degradation, results in larger axonal profiles, increases dendritic spines, and alters the strength of synaptic connection, similar to the results of cultured Atg7-deficient hippocampal neurons [228,240]. In animal models, autophagy-dependent changes in synaptic plasticity elicit a series of behavioral phenotypes, including learning and memory deficits, anxiety-like behaviors, social-behavioral defects, and cognitive deficits [240,241,242,243]. For instance, conditional knockdown of autophagy-related genes including BCN1, Atg12, and FIP200, or pharmacological inhibition of autophagy-related activities in mouse hippocampal neurons impairs the behavioral performance in contextual fear conditioning and novel object recognition tasks, implying the role of autophagy in learning and memory [241].
There is growing evidence for autophagy in LTP and LTD, two major forms of synaptic plasticity. Autophagy activator rapamycin can block late LTP induced by theta burst stimulation in the mouse cortex, leading to impaired synaptic plasticity [244]. In addition, some evidence suggests that the pharmacological inhibition of neuronal autophagy with Spautin-1 also administers the induction of LTP in CA1, whereas autophagy inhibition with BDNF is found to permit LTP in hippocampal regions [224,241]. These contradictory results may be caused by different animal backgrounds, surgical procedures, animal age, side effects of molecular drugs, or other unknown mechanisms.
Hyperactivation of mTORC1 by delivery of shRNA into the CA1 results in decreased autophagy and increased dendritic spine density with an aberrant morphology, and exaggerated LTD in mouse models of fragile X syndrome [243]. The induction of NMDAR-dependent LTD triggers a modified reorganization of the postsynaptic scaffolding protein PSD-95. This process is driven by the autophagic machinery that removes the T19 phosphorylated form of PSD-95 in the synaptic cleft, increasing the surface mobility of the glutamate receptor AMPAR, thereby attenuating synaptic efficacy and increasing short-term plasticity [233]. Notably, several key kinases regulating autophagy activity are also associated with synaptic plasticity, such as AMPK, Akt, and mTOR [245]. In brief, autophagy alters the abundance of selective synaptic proteins in a spatiotemporal-dependent manner, ultimately enabling fine-tuning of synaptic structure, function, and plasticity.
6. Epilepsy and Autophagy
6.1. Epilepsy
Epilepsy is a serious chronic neurological disease characterized clinically by recurrent, unprovoked epileptic seizures, which lead to cognitive decline, temporary neurological dysfunction, brain damage, accidental injury, and other consequences [246]. A survey of international studies showed that the prevalence globally is 6.38 per 1000 persons for active epilepsy, and 7.60 per 1000 persons for lifetime prevalence [247]. The annual incidence rate of epilepsy is 61.44 per 100,000 persons, while the long-term recurrence is 83.6% at 10 years [247,248]. Similarly, epilepsy is associated with patient stigma, neurological and psychiatric comorbidities, and high medical costs, seriously affecting the patient’s health and life quality, particularly in low-income countries.
Seizures are considered as a transient occurrence of clinical symptoms or signs due to synchronous, high-frequency, or abnormal excessive neuronal firing activity in the central nervous system, and actually are caused by multiple syndromes of different etiologies, which are related to congenital or acquired brain malformations, structural injuries or lesions, cerebral cysticercosis, and other brain diseases or disorders [249,250]. During epileptogenesis, the patient’s brain manifests an enduring and pathologic tendency to have spontaneous, recurrent seizures, facilitating neuronal hyperexcitability and abnormal synaptic plasticity [251]. The process is capable of disrupting normal neuronal information processing, the imbalance between excitatory and inhibitory neuronal networks and other networks. For example, epileptogenic networks in generalized epilepsies involve bilateral thalamocortical structures and are widely distributed in the brain, while focal epilepsies implicate the neural circuits in one cerebral hemisphere, commonly the neocortical or limbic cortex [249]. Despite breakthroughs having been made in the genetic basis of most generalized epilepsies and structural cerebral abnormalities in focal epilepsies, the underlying pathophysiological mechanisms remain incompletely understood [252,253].
6.2. Neuronal Autophagy and Epilepsy
The mesial temporal sclerosis in epilepsy patients is the best-ascertained lesion, which is characterized by neuron loss in specific hippocampal subfields, collateral axonal sprouting, synaptic reorganization of neural circuits, and functional and structural alterations in the glial [254]. Autophagy is thought to be a potential mechanism for these processes or targets for disease-modifying therapies. Most studies suggest that neuronal mTOR hyperactivation, which is mediated by loss-of-function gene mutations of mTOR inhibitor proteins including phosphatase and tensin homolog (PTEN), TSC1, TSC2, and STE20-related kinase alpha (STRADα), links to the severity of epilepsy in an animal model of epileptogenic cortical malformations, TSC, and systemic lupus erythematosus, the most common genetic etiology of epilepsy and neurobehavioral disabilities [255,256,257]. Some evidence also exhibits that the mTOR signal participates in temporal lobe epilepsy (TLE), chemoconvulsive compounds-mediated experimental epilepsy, and other epileptogenesis forms of genetic or acquired epilepsy, such as traumatic brain injury and Lafora disease (LD) [258] (Table 1). Inhibition of mTOR activity is capable of preventing epilepsy-induced neuronal alterations and the development of epilepsy [256]. Recent evidence supports the solid correction between mTOR-dependent autophagy and multiple seizure models and epilepsies [258]. However, although the close relationship between mTOR signal and autophagy activity has been automatically validated by many studies, mTOR plays a variety of roles in neurogenesis, neuronal development, synaptic plasticity, neuronal excitability, protein expression of different neuronal molecules, and other metabolic activities [259]. These may induce atypical neuronal circuits eliciting epileptogenesis independently from autophagy impairments. Thus, the overemphasized effects of mTOR for autophagy activity may mislead the interpretation of mTOR-related experimental results, and ignore the autophagy-unrelated contribution of mTOR to epilepsy.
In fact, reliable conclusions may only be drawn from studies that directly assess the autophagy condition associated with epileptogenesis and epilepsy-induced brain alterations. A recent study shows that autophagy markers p62 and LC3, which usually represent the status of autophagy activation, exhibit an obvious increase of autophagy flux in autosomal dominant lateral temporal epilepsy, which can be incompletely rehabilitated by small-molecule correctors of autophagy-related proteins Reelin [260]. Nevertheless, autophagy markers are also found to increase on the blockade of autophagic flux and impairment of autophagy progression. The paradoxical increase of autophagy markers in the context of autophagy disruption is the case for mTOR-independent autophagy inhibition, which should also be taken into account when monitoring the autophagic status of epilepsy [261]. In static encephalopathy of childhood with neurodegeneration in adulthood (SENDA), de novo mutations of the autophagy-related gene WIPI4 (also known as WDR45) severely impair its protein expression, and they lower the autophagic activity with the aberrant accumulation of LC3-positive structures at an early stage of autophagosome formation, which directly contributes to the occurrence of encephalopathic seizures [262]. In addition, the conditional ablation of Atg7 in mouse forebrain neurons induces the development of spontaneous seizures [263]. These two studies demonstrate that the blockage of the autophagy flux and the accumulation of aberrant autophagic structures occur along with lower autophagy activity in epilepsy, but with normal mTOR signaling.
Consistently, research on epilepsy animal models also shows the increased ratio of LC3 II to LC3 I and decreased p62 protein levels after epileptic seizure onset, reflecting an abnormal increase in autophagic activity in kainic acid (KA)-induced rats and mice [264,265]. This may trigger chronic hyperactivation of glutamate receptors, slow degradation of GABAA receptors, and formation of protein aggregates, and in turn, further exacerbate neuronal damage in epilepsy animals, creating a vicious cycle between autophagy and epilepsy [266,267].
Some genetic sequencing studies for epilepsy patients and animal models suggest that the mutations in TBCK (encoding the TBC1-domain-containing kinase and inhibiting mTORC1 signaling), VSP15 (perturbing endosomal-lysosomal trafficking and autophagy flux), EPG5 (encoding a tethering factor that regulates the specific fusion of APs with late lysosomes/endosomes), SNX14 (encoding the sorting nexin family protein that mediating lysosome-AP fusion), ATP6V1A (encoding for the “A” subunit of the v1 sub-complex of V-ATPase, affecting lysosomal homeostasis and autophagy), ATP6AP2 (encoding a key regulator of v-ATPase, and its loss leads to lysosomal and autophagic defects), DMXL2 (encoding a member of the WD40 protein family that regulates v-ATPase trafficking and activity), or FAM134B (encoding the ER autophagy receptor) may contribute to the occurrence of epilepsy in space- and time-dependent manners [268,269] (Table 1). Notably, recent studies have demonstrated that microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) involve the post-transcriptional regulation of autophagy-related proteins and the development of epilepsy pathophysiologies, such as miR-101, miR-181b, miR-134, miR-142, miR-421, miR-223, and Zinc finger antisense 1 (ZFAS1), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), respectively [270,271,272,273,274,275]. Moreover, glia including astrocytes and microglia also plays a multi-faceted role in autophagy-mediated mechanisms that determine seizures and epileptogenesis [276,277].
Collectively, current research suggests that abnormal autophagy occurs in epilepsy, and alters the physiological state of certain neurons [258,263,265,266,278,279,280,281] (Table 1). These state anomalies may encompass the changes at the molecular and cellular levels as well as at the circuit level [11,258,263,265,279,281,282]. For instance, Changes in cell signaling (such as mTOR and Atg7) alter the neuron’s overall ability to receive, integrate, and process inputs from different types of molecular cues, and the synapse morphology and function that decrease or increase the neuron’s ability to stimulate its synaptic partners [11,258,263,268,279,281,282]. Aberrant autophagy in epilepsy also alters the expression or function of ion channels (such as the GABAA receptor or glutamate receptor), resulting in decreased or increased neuronal excitability, and therefore decreased or increased ability to conduct action potentials or other electrical signals [11]. Rapid mobilization of neuronal growth processes that establish new synaptic connections or neurite retraction that removes existing connections may be disrupted by abnormal autophagy [279,280,282]. However, the underlying molecular mechanisms by which autophagic activity regulates these alterations to promote epileptogenesis remain unclear.

Table 1.
The autophagy-related molecular changes in epilepsy animal models and patients.
Table 1.
The autophagy-related molecular changes in epilepsy animal models and patients.
| Models/Patients | Molecular Changes | Pathological Changes | References |
|---|---|---|---|
| Lafora disease mice | Increased Rab5, p62 protein level, decreased LC3-II levels | Generalized stimulus-sensitive tonic-clonic seizures | Puri and Suzuki., 2012 [283]; Criado et al., 2012 [284] |
| Pilocarpine-induced model mice | Increased levels of Beclin 1, ATG5, ATG7 and the ratio of LC3II/I | Epilepsy | Ying et al., 2020 [285] |
| N-ethyl-N-nitrosourea (ENU)-induced mice mutants | Vps15 mutation, decreased LC3-II/LC3-I ratio | Cortical atrophy, dysplasia, and epilepsy | Gstrein et al., 2018 [286] |
| TSC1/PTEN KO mice | mTOR hyperactivation, increased Ulk1 phosphorylation | Epileptogenesis | Yasin et al., 2013 [287] |
| Kainic acid treatment mice | Increased LC3-II levels, elevated ratios of phospho-mTOR/mTOR | Repeated seizures | Shacka et al., 2007 [265] |
| Atg7 KO mice | p62 accumulation | Spontaneous seizures | McMahon et al., 2012 [263] |
| Depdc5 KO mice | Increased mTORC1 signaling | Spontaneous seizures | Yuskaitis et al., 2018 [288] |
| PTEN KO mice + mTOR inhibition | Decreased mTOR activity | Decreased the seizure frequency and death rate | Kwon et al., 2003 [289] |
| Pilocarpine-induced model rats | Increased LC3-II/LC3-I ratio and beclin1 level | Status epilepticus | Cao et al., 2009 [290] |
| Kainic acid treatment rats | mTOR activation | Status epilepticus | Macias et al., 2013 [291] |
| Kainic acid treatment rats + rapamycin | Decreased mTOR activity | Reduced epilepsy | Zeng et al., 2009 [292] |
| Pilocarpine-induced model rats | mTOR activation | Status epilepticus | Buckmaster et al., 2009 [279] |
| Pilocarpine-induced model rats + rapamycin | Decreased mTOR activity | Reduced seizure activity | Huang et al., 2010 [293] |
| Infantile spams/West syndrome rats | mTORC1 pathway overactivation | Spasms, epileptic encephalopathies | Raffo et al., 2011 [294] |
| VPS15 mutation in humans | p62 accumulation | Cortical atrophy, late-onset epilepsy | Gstrein et al., 2018 [286] |
| Beta-propeller protein-associated neurodegeneration patients | De novo mutation in WDR45 | Developmental and epileptic encephalopathies | Carvill et al., 2018 [295] |
| Autosomal dominant lateral temporal epilepsy patients | Reelin mutation | Epilepsy | Dazzo and Nobile., 2022 [260] |
| Vici syndrome patients | EPG5 mutation | Severe seizure disorder, progressive neurodegeneration | Byrne et al., 2016 [296] |
| Pediatric-onset ataxias patients | SNX14 mutation | Progressive cerebellar neurodegeneration, developmental delay, intellectual disability, and seizures | Akizu et al., 2015 [297] |
| Ohtahara syndrome patients | DMXL2 mutation | Intractable seizures and profound developmental disability | Esposito et al., 2019 [298] |
| Children with TBCK p.R126X mutations | Increased LC3-II/LC3-I ratio | Focal and generalized seizures | Ortiz-González et al., 2018 [299] |
| Epilepsy patients | ATG5 gene variant, ATP6V1A/ ATP6AP2 mutation, increased Beclin1 expression | Late-onset epilepsy, temporal lobe epilepsy | Zhang et al., 2021 [300]; Van Damme et al., 2020 [301]; Hirose et al., 2019 [302]; Yang et al., 2022 [303] |
| Focal cortical dysplasia in childhood | mTOR activation, p62 accumulation, TSC1/TSC2 mutation | Epilepsy | Yasin et al., 2013 [287] |
| Human TSC patients | Increased in Ulk1 phosphorylation, p62 accumulation | Cognitive dysfunction, early-onset, intractable epilepsy | McMahon et al., 2012 [263] |
| Hippocampal neuronal culture model of acquired epilepsy | Elevated LC3-II/LC3-I ratio | Acquired epilepsy | Xie et al., 2020 [269] |
7. Conclusions
Converging lines of research support a model whereby all cells, including invertebrate animals, insects, yeasts, and cultured cells, have evolved highly specific mechanisms to ensure protein quality control and maintenance of cell homeostasis, namely autophagy. These mechanisms involve the biogenesis and trafficking of the autophagosome, formation, and degradation of the autolysosome, and intricate regulation of autophagy-related molecular proteins in space- and time-dependent manners, particularly in the neuron. Recent findings have driven forward our understanding of how neuronal autophagy regulates the growth and development of axons and dendrites and influences spine pruning, neurotransmitter release, synapse plasticity, and other neuronal functions. Structural and functional integrity in neurons is more sensitive to changes in autophagy than that of non-neuronal cells due to the highly specialized compartments necessary for intercellular communications. Autophagy impairments likely induce accumulation of toxic protein aggregates and worn organelles, as well as interruption of synapse pruning, ultimately affecting the normal physiological function of the neuron. In the past decades, autophagy alterations have begun to be applied to the field of epileptogenesis besides neurodegenerative disorders. Recent studies have suggested that hyperactivation of the mTOR signaling pathway and abnormal autophagy activity occur in different epilepsy animal models and epilepsy-related patients, whereas inhibition of mTOR activity can ameliorate seizures in various epilepsy models. For instance, the special deletion of TSC1 or PTEN genes in mouse neurons results in seizures and compromised autophagy activity, consistent with human TSC patients. Genetic sequencing of multiple types of epilepsy patients and seizure models indicated that gene mutations in autophagy-related pathways may contribute to the occurrence of epilepsy, such as TBCK, EPG5, VSP15, etc. In addition, impaired autophagy is sufficient to induce epilepsy in rodent models, such as genetic inactivation of Atg7 in mice. However, it represents an underexplored research avenue for epilepsy. There are still many important concepts, principles, and issues to be addressed in this field. Promising directions involve working out the details, including seeking: (1) to decipher how synapses signal membrane biogenesis and autophagy initiation, (2) to uncover how neuronal autophagy and synaptic efficacy are cross-regulated in seizures, (3) to uncover the interconnection of autophagy activity with antiepileptic drugs, and (4) to define the diversity and dynamic nature of the autophagic cargo of different neuronal compartments in terms of their physiological states and epilepsy pathologies.
Author Contributions
Writing—original draft preparation, review, editing, and conceptualization, H.Z. and Y.L.; artwork, W.W.; supervision, Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a grant (2022J0703) from the Scientific Research Fund Project of Yunnan Provincial Department of Education, a grant from the Science and Technology Talent Project (Reserve talent) of the First Affiliated Hospital of Dali University, and a grant (2021KBG047) from the Dali Science and Technology Bureau Project.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pohl, C.; Dikic, I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 2019, 366, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Nakatogawa, H. Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 2020, 21, 439–458. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Biological functions of autophagy genes: A disease perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef]
- Luo, L. Architectures of neuronal circuits. Science 2021, 373, eabg7285. [Google Scholar] [CrossRef]
- Li, S.; Sheng, Z.H. Energy matters: Presynaptic metabolism and the maintenance of synaptic transmission. Nat. Rev. Neurosci. 2022, 23, 4–22. [Google Scholar] [CrossRef]
- Decet, M.; Verstreken, P. Presynaptic autophagy and the connection with neurotransmission. Front. Cell Dev. Biol. 2021, 9, 790721. [Google Scholar] [CrossRef]
- Yamamoto, A.; Yue, Z. Autophagy and its normal and pathogenic states in the brain. Annu Rev. Neurosci. 2014, 37, 55–78. [Google Scholar] [CrossRef]
- Fleming, A.; Rubinsztein, D.C. Autophagy in neuronal development and plasticity. Trends Neurosci. 2020, 43, 767–779. [Google Scholar] [CrossRef]
- Leidal, A.M.; Levine, B.; Debnath, J. Autophagy and the cell biology of age-related disease. Nat. Cell Biol. 2018, 20, 1338–1348. [Google Scholar] [CrossRef]
- Lv, M.; Ma, Q. Autophagy and epilepsy. Adv. Exp. Med. Biol 2020, 1207, 163–169. [Google Scholar]
- Bejarano, E.; Rodríguez-Navarro, J.A. Autophagy and amino acid metabolism in the brain: Implications for epilepsy. Amino Acids 2015, 47, 2113–2126. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.L., Jr. Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J. Biophys. Biochem. Cytol. 1957, 3, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Ashford, T.P.; Porter, K.R. Cytoplasmic components in hepatic cell lysosomes. J. Cell Biol. 1962, 12, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Hruban, Z.; Spargo, B.; Swift, H.; Wissler, R.W.; Kleinfeld, R.G. Focal cytoplasmic degradation. Am. J. Pathol. 1963, 42, 657–683. [Google Scholar] [PubMed]
- De Duve, C. The lysosome. Sci. Am. 1963, 208, 64–72. [Google Scholar] [CrossRef] [PubMed]
- De Duve, C.; Wattiaux, R. Functions of lysosomes. Annu. Rev. Physiol. 1966, 28, 435–492. [Google Scholar] [CrossRef]
- Deter, R.L.; Baudhuin, P.; De Duve, C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J. Cell Biol. 1967, 35, C11–C16. [Google Scholar] [CrossRef]
- Clarke, P.G. Developmental cell death: Morphological diversity and multiple mechanisms. Anat. Embryol. 1990, 181, 195–213. [Google Scholar] [CrossRef]
- Takeshige, K.; Baba, M.; Tsuboi, S.; Noda, T.; Ohsumi, Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992, 119, 301–311. [Google Scholar] [CrossRef]
- Sukada, M.; Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993, 333, 169–174. [Google Scholar] [CrossRef]
- Thumm, M.; Egner, R.; Koch, B.; Schlumpberger, M.; Straub, M.; Veenhuis, M.; Wolf, D.H. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994, 349, 275–280. [Google Scholar] [CrossRef]
- Sakai, Y.; Koller, A.; Rangell, L.K.; Keller, G.A.; Subramani, S. Peroxisome degradation by microautophagy in Pichia pastoris: Identification of specific steps and morphological intermediates. J. Cell Biol. 1998, 141, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Cregg, J.M.; Dunn, W.A., Jr.; Emr, S.D.; Sakai, Y.; Sandoval, I.V.; Sibirny, A.; Subramani, S.; Thumm, M.; Veenhuis, M.; et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell 2003, 5, 539–545. [Google Scholar] [CrossRef]
- Funakoshi, T.; Matsuura, A.; Noda, T.; Ohsumi, Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 1997, 192, 207–213. [Google Scholar] [CrossRef]
- Mizushima, N.; Noda, T.; Yoshimori, T.; Tanaka, Y.; Ishii, T.; George, M.D.; Klionsky, D.J.; Ohsumi, M.; Ohsumi, Y. A protein conjugation system essential for autophagy. Nature 1998, 395, 395–398. [Google Scholar] [CrossRef]
- Mizushima, N.; Sugita, H.; Yoshimori, T.; Ohsumi, Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J. Biol. Chem. 1998, 273, 33889–33892. [Google Scholar] [CrossRef]
- Liang, X.H.; Jackson, S.; Seaman, M.; Brown, K.; Kempkes, B.; Hibshoosh, H.; Levine, B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999, 402, 672–676. [Google Scholar] [CrossRef]
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19, 5720–5728. [Google Scholar] [CrossRef]
- Mizushima, N.; Yamamoto, A.; Hatano, M.; Kobayashi, Y.; Kabeya, Y.; Suzuki, K.; Tokuhisa, T.; Ohsumi, Y.; Yoshimori, T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001, 152, 657–668. [Google Scholar] [CrossRef]
- Petiot, A.; Ogier-Denis, E.; Blommaart, E.F.; Meijer, A.J.; Codogno, P. Distinct classes of phosphatidylinositol 3’-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 2000, 275, 992–998. [Google Scholar] [CrossRef]
- Ravikumar, B.; Duden, R.; Rubinsztein, D.C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002, 11, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, B.; Vacher, C.; Berger, Z.; Davies, J.E.; Luo, S.; Oroz, L.G.; Scaravilli, F.; Easton, D.F.; Duden, R.; O’Kane, C.J.; et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004, 36, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 2004, 36, 2491–2502. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Kuma, A. Autophagosomes in GFP-LC3 transgenic mice. Methods Mol. Biol. 2008, 445, 119–124. [Google Scholar] [PubMed]
- Narendra, D.; Tanaka, A.; Suen, D.F.; Youle, R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008, 183, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef]
- Gutierrez, M.G.; Master, S.S.; Singh, S.B.; Taylor, G.A.; Colombo, M.I.; Deretic, V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004, 119, 753–766. [Google Scholar] [CrossRef]
- Paludan, C.; Schmid, D.; Landthaler, M.; Vockerodt, M.; Kube, D.; Tuschl, T.; Münz, C. Endogenous MHC Class II processing of a viral nuclear antigen after autophagy. Science 2005, 307, 593–596. [Google Scholar] [CrossRef]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef]
- Lemasters, J.J. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005, 8, 3–5. [Google Scholar] [CrossRef]
- Kim, I.; Rodriguez-Enriquez, S.; Lemasters, J.J. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 2007, 462, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Bernales, S.; Schuck, S.; Walter, P. ER-phagy: Selective autophagy of the endoplasmic reticulum. Autophagy 2007, 3, 285–287. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.E.; Hayashi, Y.K.; Bonne, G.; Arimura, T.; Noguchi, S.; Nonaka, I.; Nishino, I. Autophagic degradation of nuclear components in mammalian cells. Autophagy 2009, 5, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, D.L.; Lewin, A.S.; Dunn, W.A., Jr. Selective autophagy of peroxisomes in methylotrophic yeasts. Eur J. Cell Biol. 1993, 60, 283–290. [Google Scholar]
- Tuttle, D.L.; Dunn, W.A., Jr. Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J Cell Sci 1995, 108, 25–35. [Google Scholar] [CrossRef]
- Dunn, W.A., Jr.; Cregg, J.M.; Kiel, J.A.; van der Klei, I.J.; Oku, M.; Sakai, Y.; Sibirny, A.A.; Stasyk, O.V.; Veenhuis, M. Pexophagy: The selective autophagy of peroxisomes. Autophagy 2005, 1, 75–83. [Google Scholar] [CrossRef]
- Maejima, I.; Takahashi, A.; Omori, H.; Kimura, T.; Takabatake, Y.; Saitoh, T.; Yamamoto, A.; Hamasaki, M.; Noda, T.; Isaka, Y.; et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013, 32, 2336–2347. [Google Scholar] [CrossRef]
- Kraft, C.; Deplazes, A.; Sohrmann, M.; Peter, M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol 2008, 10, 602–610. [Google Scholar] [CrossRef]
- Fujioka, Y.; Alam, J.M.; Noshiro, D.; Mouri, K.; Ando, T.; Okada, Y.; May, A.I.; Knorr, R.L.; Suzuki, K.; Ohsumi, Y.; et al. Phase separation organizes the site of autophagosome formation. Nature 2020, 578, 301–305. [Google Scholar] [CrossRef]
- Ponpuak, M.; Mandell, M.A.; Kimura, T.; Chauhan, S.; Cleyrat, C.; Deretic, V. Secretory autophagy. Curr. Opin. Cell Biol. 2015, 35, 106–116. [Google Scholar] [CrossRef]
- Codogno, P.; Mehrpour, M.; Proikas-Cezanne, T. Canonical and non-canonical autophagy: Variations on a common theme of self-eating? Nat. Rev. Mol. Cell Biol 2011, 13, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Noda, N.N.; Mizushima, N. Atg101: Not Just an Accessory Subunit in the Autophagy-initiation Complex. Cell Struct. Funct. 2016, 41, 13–20. [Google Scholar] [CrossRef]
- Ohsumi, Y. Historical landmarks of autophagy research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Melia, T.J.; Lystad, A.H.; Simonsen, A. Autophagosome biogenesis: From membrane growth to closure. J. Cell Biol. 2020, 219, e202002085. [Google Scholar] [CrossRef]
- Nguyen, J.A.; Yates, R.M. Better Together: Current Insights Into Phagosome-Lysosome Fusion. Front. Immunol 2021, 12, 636078. [Google Scholar] [CrossRef]
- Leon, C.; Bouchecareilh, M. The Autophagy Pathway: A Critical Route in the Disposal of Alpha 1-Antitrypsin Aggregates That Holds Many Mysteries. Int J. Mol. Sci. 2021, 22, 1875. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Massey, A.C.; Cuervo, A.M. Lysosome membrane lipid microdomains: Novel regulators of chaperone-mediated autophagy. EMBO J. 2006, 25, 3921–3933. [Google Scholar] [CrossRef]
- Massey, A.C.; Kaushik, S.; Sovak, G.; Kiffin, R.; Cuervo, A.M. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA 2006, 103, 5805–5810. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Mau, T.; O’Brien, M.; Garg, S.; Yung, R. Impaired autophagy activity is linked to elevated ER-stress and inflammation in aging adipose tissue. Aging (Albany NY) 2016, 8, 2525–2537. [Google Scholar] [CrossRef]
- Yu, L.; Chen, Y.; Tooze, S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018, 14, 207–215. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 365–381. [Google Scholar] [CrossRef] [PubMed]
- Stolz, A.; Ernst, A.; Dikic, I. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol 2014, 16, 495–501. [Google Scholar] [CrossRef]
- Chiang, H.L.; Terlecky, S.R.; Plant, C.P.; Dice, J.F. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 1989, 246, 382–385. [Google Scholar] [CrossRef] [PubMed]
- Tekirdag, K.; Cuervo, A.M. Chaperone-mediated autophagy and endosomal microautophagy: Joint by a chaperone. J. Biol. Chem. 2018, 293, 5414–5424. [Google Scholar] [CrossRef]
- Klionsky, D.J. Autophagy: From phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 2007, 8, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Ohsumi, Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001, 2, 211–216. [Google Scholar] [CrossRef]
- Kunz, J.B.; Schwarz, H.; Mayer, A. Determination of four sequential stages during microautophagy in vitro. J. Biol. Chem. 2004, 279, 9987–9996. [Google Scholar] [CrossRef]
- Li, W.; He, P.; Huang, Y.; Li, Y.F.; Lu, J.; Li, M.; Kurihara, H.; Luo, Z.; Meng, T.; Onishi, M.; et al. Selective autophagy of intracellular organelles: Recent research advances. Theranostics 2021, 11, 222–256. [Google Scholar] [CrossRef]
- Egan, D.F.; Shackelford, D.B.; Mihaylova, M.M.; Gelino, S.; Kohnz, R.A.; Mair, W.; Vasquez, D.S.; Joshi, A.; Gwinn, D.M.; Taylor, R.; et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011, 331, 456–461. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, H.; Zhang, D.; Luo, W.; Liu, R.; Xu, D.; Diao, L.; Liao, L.; Liu, Z. Phosphorylation of ULK1 affects autophagosome fusion and links chaperone-mediated autophagy to macroautophagy. Nat. Commun. 2018, 9, 3492. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, N.; Sasaki, T.; Iemura, S.; Natsume, T.; Hara, T.; Mizushima, N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 2009, 5, 973–979. [Google Scholar] [CrossRef]
- Hara, T.; Takamura, A.; Kishi, C.; Iemura, S.; Natsume, T.; Guan, J.L.; Mizushima, N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008, 181, 497–510. [Google Scholar] [CrossRef]
- Zhou, C.; Ma, K.; Gao, R.; Mu, C.; Chen, L.; Liu, Q.; Luo, Q.; Feng, D.; Zhu, Y.; Chen, Q. Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 2017, 27, 184–201. [Google Scholar] [CrossRef]
- Ma, B.; Cao, W.; Li, W.; Gao, C.; Qi, Z.; Zhao, Y.; Du, J.; Xue, H.; Peng, J.; Wen, J.; et al. Dapper1 promotes autophagy by enhancing the Beclin1-Vps34-Atg14L complex formation. Cell Res. 2014, 24, 912–924. [Google Scholar] [CrossRef]
- Sawa-Makarska, J.; Baumann, V.; Coudevylle, N.; von Bülow, S.; Nogellova, V.; Abert, C.; Schuschnig, M.; Graef, M.; Hummer, G.; Martens, S. Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science 2020, 369, eaaz7714. [Google Scholar] [CrossRef]
- Reggiori, F.; Shintani, T.; Nair, U.; Klionsky, D.J. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy 2005, 1, 101–109. [Google Scholar] [CrossRef]
- Valverde, D.P.; Yu, S.; Boggavarapu, V.; Kumar, N.; Lees, J.A.; Walz, T.; Reinisch, K.M.; Melia, T.J. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 2019, 218, 1787–1798. [Google Scholar] [CrossRef] [PubMed]
- Krick, R.; Henke, S.; Tolstrup, J.; Thumm, M. Dissecting the localization and function of Atg18, Atg21 and Ygr223c. Autophagy 2008, 4, 896–910. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.M.; Wrobel, L.; Rubinsztein, D.C. Post-translational modifications of Beclin 1 provide multiple strategies for autophagy regulation. Cell Death Differ. 2019, 26, 617–629. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Dooley, H.C.; Razi, M.; Polson, H.E.; Girardin, S.E.; Wilson, M.I.; Tooze, S.A. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell 2014, 55, 238–252. [Google Scholar] [CrossRef]
- Diao, J.; Liu, R.; Rong, Y.; Zhao, M.; Zhang, J.; Lai, Y.; Zhou, Q.; Wilz, L.M.; Li, J.; Vivona, S.; et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 2015, 520, 563–566. [Google Scholar] [CrossRef]
- Ranaghan, M.J.; Durney, M.A.; Mesleh, M.F.; McCarren, P.R.; Garvie, C.W.; Daniels, D.S.; Carey, K.L.; Skepner, A.P.; Levine, B.; Perez, J.R. The Autophagy-Related Beclin-1 Protein Requires the Coiled-Coil and BARA Domains To Form a Homodimer with Submicromolar Affinity. Biochemistry 2017, 56, 6639–6651. [Google Scholar] [CrossRef]
- Fernández, Á.F.; Sebti, S.; Wei, Y.; Zou, Z.; Shi, M.; McMillan, K.L.; He, C.; Ting, T.; Liu, Y.; Chiang, W.C.; et al. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 2018, 558, 136–140. [Google Scholar] [CrossRef]
- Houtman, J.; Freitag, K.; Gimber, N.; Schmoranzer, J.; Heppner, F.L.; Jendrach, M. Beclin1-driven autophagy modulates the inflammatory response of microglia via NLRP3. EMBO J. 2019, 38, e99430. [Google Scholar] [CrossRef]
- Huang, X.; Li, Y.; Shou, L.; Li, L.; Chen, Z.; Ye, X.; Qian, W. The molecular mechanisms underlying BCR/ABL degradation in chronic myeloid leukemia cells promoted by Beclin1-mediated autophagy. Cancer Manag. Res. 2019, 11, 5197–5208. [Google Scholar] [CrossRef]
- Parzych, K.R.; Klionsky, D.J. An overview of autophagy: Morphology, mechanism, and regulation. Antioxid. Redox Signal. 2014, 20, 460–473. [Google Scholar] [CrossRef]
- Lee, E.F.; Perugini, M.A.; Pettikiriarachchi, A.; Evangelista, M.; Keizer, D.W.; Yao, S.; Fairlie, W.D. The BECN1 N-terminal domain is intrinsically disordered. Autophagy 2016, 12, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Itakura, E.; Kishi, C.; Inoue, K.; Mizushima, N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 2008, 19, 5360–5372. [Google Scholar] [CrossRef]
- Kim, Y.M.; Jung, C.H.; Seo, M.; Kim, E.K.; Park, J.M.; Bae, S.S.; Kim, D.H. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol. Cell 2015, 57, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhang, J.; Fan, W.; Wong, K.N.; Ding, X.; Chen, S.; Zhong, Q. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J. Biol. Chem. 2011, 286, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, K.; Saitoh, T.; Tabata, K.; Omori, H.; Satoh, T.; Kurotori, N.; Maejima, I.; Shirahama-Noda, K.; Ichimura, T.; Isobe, T.; et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009, 11, 385–396. [Google Scholar] [CrossRef]
- Geng, J.; Klionsky, D.J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: Beyond the usual suspects’ review series. EMBO Rep. 2008, 9, 859–864. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Noda, N.N.; Yamamoto, H.; Shima, T.; Kumeta, H.; Kobashigawa, Y.; Akada, R.; Ohsumi, Y.; Agaki, F. Structural insights into Atg10-mediated formation of the autophagy-essential Atg12-Atg5 conjugate. Structure 2012, 20, 1244–1254. [Google Scholar] [CrossRef]
- Fujita, N.; Itoh, T.; Omori, H.; Fukuda, M.; Noda, T.; Yoshimori, T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 2008, 19, 2092–2100. [Google Scholar] [CrossRef]
- Lystad, A.H.; Carlsson, S.R.; de la Ballina, L.R.; Kauffman, K.J.; Nag, S.; Yoshimori, T.; Melia, T.J.; Simonsen, A. Distinct functions of ATG16L1 isoforms in membrane binding and LC3B lipidation in autophagy-related processes. Nat. Cell Biol. 2019, 21, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Hanada, T.; Ohsumi, Y. Structure-function relationship of Atg12, a ubiquitin-like modifier essential for autophagy. Autophagy 2005, 1, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Kharaziha, P.; Panaretakis, T. Dynamics of Atg5-Atg12-Atg16L1 Aggregation and Deaggregation. Methods Enzymol. 2017, 587, 247–255. [Google Scholar]
- Kabeya, Y.; Mizushima, N.; Yamamoto, A.; Oshitani-Okamoto, S.; Ohsumi, Y.; Yoshimori, T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004, 117, 2805–2812. [Google Scholar] [CrossRef]
- Lystad, A.H.; Carlsson, S.R.; Simonsen, A. Toward the function of mammalian ATG12-ATG5-ATG16L1 complex in autophagy and related processes. Autophagy 2019, 15, 1485–1486. [Google Scholar] [CrossRef] [PubMed]
- Jatana, N.; Ascher, D.B.; Pires, D.E.V.; Gokhale, R.S.; Thukral, L. Human LC3 and GABARAP subfamily members achieve functional specificity via specific structural modulations. Autophagy 2020, 16, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Weidberg, H.; Shvets, E.; Shpilka, T.; Shimron, F.; Shinder, V.; Elazar, Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010, 29, 1792–1802. [Google Scholar] [CrossRef]
- Bjørkøy, G.; Lamark, T.; Brech, A.; Outzen, H.; Perander, M.; Overvatn, A.; Stenmark, H.; Johansen, T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005, 171, 603–614. [Google Scholar] [CrossRef]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.A.; Outzen, H.; Øvervatn, A.; Bjørkøy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef]
- Thurston, T.L.; Ryzhakov, G.; Bloor, S.; von Muhlinen, N.; Randow, F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 2009, 10, 1215–1221. [Google Scholar] [CrossRef]
- Lin, X.; Li, S.; Zhao, Y.; Ma, X.; Zhang, K.; He, X.; Wang, Z. Interaction domains of p62: A bridge between p62 and selective autophagy. DNA Cell Biol 2013, 32, 220–227. [Google Scholar] [CrossRef]
- Lamark, T.; Svenning, S.; Johansen, T. Regulation of selective autophagy: The p62/SQSTM1 paradigm. Essays Biochem. 2017, 61, 609–624. [Google Scholar] [PubMed]
- Zhao, Y.G.; Codogno, P.; Zhang, H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat. Rev. Mol. Cell Biol. 2021, 22, 733–750. [Google Scholar] [CrossRef]
- Jahreiss, L.; Menzies, F.M.; Rubinsztein, D.C. The itinerary of autophagosomes: From peripheral formation to kiss-and-run fusion with lysosomes. Traffic 2008, 9, 574–587. [Google Scholar] [CrossRef] [PubMed]
- Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, M.G.; Munafó, D.B.; Berón, W.; Colombo, M.I. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci 2004, 117, 2687–2697. [Google Scholar] [CrossRef] [PubMed]
- Hegedűs, K.; Takáts, S.; Boda, A.; Jipa, A.; Nagy, P.; Varga, K.; Kovács, A.L.; Juhász, G. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol. Biol. Cell 2016, 27, 3132–3142. [Google Scholar] [CrossRef]
- Takáts, S.; Boda, A.; Csizmadia, T.; Juhász, G. Small GTPases controlling autophagy-related membrane traffic in yeast and metazoans. Small GTPases 2018, 9, 465–471. [Google Scholar] [CrossRef]
- Kimura, S.; Noda, T.; Yoshimori, T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct. Funct. 2008, 33, 109–122. [Google Scholar] [CrossRef]
- Pu, J.; Guardia, C.M.; Keren-Kaplan, T.; Bonifacino, J.S. Mechanisms and functions of lysosome positioning. J. Cell Sci. 2016, 129, 4329–4339. [Google Scholar] [CrossRef]
- Cantalupo, G.; Alifano, P.; Roberti, V.; Bruni, C.B.; Bucci, C. Rab-interacting lysosomal protein (RILP): The Rab7 effector required for transport to lysosomes. EMBO J. 2001, 20, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Rocha, N.; Kuijl, C.; van der Kant, R.; Janssen, L.; Houben, D.; Janssen, H.; Zwart, W.; Neefjes, J. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J. Cell Biol. 2009, 185, 1209–1225. [Google Scholar] [CrossRef] [PubMed]
- Pankiv, S.; Alemu, E.A.; Brech, A.; Bruun, J.A.; Lamark, T.; Overvatn, A.; Bjørkøy, G.; Johansen, T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 2010, 188, 253–269. [Google Scholar] [CrossRef]
- Cardoso, C.M.; Groth-Pedersen, L.; Høyer-Hansen, M.; Kirkegaard, T.; Corcelle, E.; Andersen, J.S.; Jäättelä, M.; Nylandsted, J. Depletion of kinesin 5B affects lysosomal distribution and stability and induces peri-nuclear accumulation of autophagosomes in cancer cells. PLoS ONE 2009, 4, e4424. [Google Scholar] [CrossRef] [PubMed]
- Korolchuk, V.I.; Saiki, S.; Lichtenberg, M.; Siddiqi, F.H.; Roberts, E.A.; Imarisio, S.; Jahreiss, L.; Sarkar, S.; Futter, M.; Menzies, F.M.; et al. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 2011, 13, 453–460. [Google Scholar] [CrossRef]
- Fu, M.M.; Nirschl, J.J.; Holzbaur, E.L.F. LC3 binding to the scaffolding protein JIP1 regulates processive dynein-driven transport of autophagosomes. Dev. Cell 2014, 29, 577–590. [Google Scholar] [CrossRef]
- Jiang, P.; Nishimura, T.; Sakamaki, Y.; Itakura, E.; Hatta, T.; Natsume, T.; Mizushima, N. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell 2014, 25, 1327–1337. [Google Scholar] [CrossRef]
- Reggiori, F.; Ungermann, C. Autophagosome Maturation and Fusion. J. Mol. Biol. 2017, 429, 486–496. [Google Scholar] [CrossRef]
- Wickner, W.; Schekman, R. Membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 658–664. [Google Scholar] [CrossRef]
- Bröcker, C.; Kuhlee, A.; Gatsogiannis, C.; Balderhaar, H.J.; Hönscher, C.; Engelbrecht-Vandré, S.; Ungermann, C.; Raunser, S. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl. Acad. Sci. USA 2012, 109, 1991–1996. [Google Scholar] [CrossRef]
- Balderhaar, H.J.; Ungermann, C. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J. Cell Sci. 2013, 126, 1307–1316. [Google Scholar] [CrossRef] [PubMed]
- Pols, M.S.; ten Brink, C.; Gosavi, P.; Oorschot, V.; Klumperman, J. The HOPS proteins hVps41 and hVps39 are required for homotypic and heterotypic late endosome fusion. Traffic 2013, 14, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Akbar, M.A.; Ray, S.; Krämer, H. The SM protein Car/Vps33A regulates SNARE-mediated trafficking to lysosomes and lysosome-related organelles. Mol. Biol. Cell 2009, 20, 1705–1714. [Google Scholar] [CrossRef]
- Takáts, S.; Pircs, K.; Nagy, P.; Varga, Á.; Kárpáti, M.; Hegedűs, K.; Kramer, H.; Kovács, A.L.; Sass, M.; Juhász, G. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol. Biol. Cell 2014, 25, 1338–1354. [Google Scholar] [CrossRef]
- Fujita, N.; Huang, W.; Lin, T.H.; Groulx, J.F.; Jean, S.; Nguyen, J.; Kuchitsu, Y.; Koyama-Honda, I.; Mizushima, N.; Fukuda, M.; et al. Genetic screen in Drosophila muscle identifies autophagy-mediated T-tubule remodeling and a Rab2 role in autophagy. Elife 2017, 6, e23367. [Google Scholar] [CrossRef] [PubMed]
- Khatter, D.; Raina, V.B.; Dwivedi, D.; Sindhwani, A.; Bahl, S.; Sharma, M. The small GTPase Arl8b regulates assembly of the mammalian HOPS complex on lysosomes. J. Cell Sci. 2015, 128, 1746–1761. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Guardia, C.M.; Pu, J.; Chen, Y.; Bonifacino, J.S. BORC coordinates encounter and fusion of lysosomes with autophagosomes. Autophagy 2017, 13, 1648–1663. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhao, Y.G.; Wang, X.; Xu, L.; Miao, L.; Feng, D.; Chen, Q.; Kovács, A.L.; Fan, D.; Zhang, H. Mice deficient in Epg5 exhibit selective neuronal vulnerability to degeneration. J. Cell Biol. 2013, 200, 731–741. [Google Scholar] [CrossRef]
- Ogawa, M.; Yoshikawa, Y.; Kobayashi, T.; Mimuro, H.; Fukumatsu, M.; Kiga, K.; Piao, Z.; Ashida, H.; Yoshida, M.; Kakuta, S.; et al. A Tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host Microbe 2011, 9, 376–389. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Lak, B.; Li, J.; Jokitalo, E.; Wang, Y. GRASP55 Senses Glucose Deprivation through O-GlcNAcylation to Promote Autophagosome-Lysosome Fusion. Dev. Cell 2018, 45, 245–261.e6. [Google Scholar] [CrossRef]
- Terawaki, S.; Camosseto, V.; Prete, F.; Wenger, T.; Papadopoulos, A.; Rondeau, C.; Combes, A.; Rodriguez Rodrigues, C.; Vu Manh, T.P.; Fallet, M.; et al. RUN and FYVE domain-containing protein 4 enhances autophagy and lysosome tethering in response to Interleukin-4. J. Cell Biol. 2015, 210, 1133–1152. [Google Scholar] [CrossRef] [PubMed]
- Stroupe, C.; Collins, K.M.; Fratti, R.A.; Wickner, W. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 2006, 25, 1579–1589. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Teng, J.; Chen, J. New insights regarding SNARE proteins in autophagosome-lysosome fusion. Autophagy 2021, 17, 2680–2688. [Google Scholar] [CrossRef]
- Südhof, T.C.; Rothman, J.E. Membrane fusion: Grappling with SNARE and SM proteins. Science 2009, 323, 474–477. [Google Scholar] [CrossRef]
- Baker, R.W.; Hughson, F.M. Chaperoning SNARE assembly and disassembly. Nat. Rev. Mol. Cell Biol. 2016, 17, 465–479. [Google Scholar] [CrossRef]
- Lőrincz, P.; Juhász, G. Autophagosome-Lysosome Fusion. J. Mol. Biol. 2020, 432, 2462–2482. [Google Scholar] [CrossRef] [PubMed]
- Dilcher, M.; Köhler, B.; von Mollard, G.F. Genetic interactions with the yeast Q-SNARE VTI1 reveal novel functions for the R-SNARE YKT6. J. Biol. Chem. 2001, 276, 34537–34544. [Google Scholar] [CrossRef] [PubMed]
- Darsow, T.; Rieder, S.E.; Emr, S.D. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 1997, 138, 517–529. [Google Scholar] [CrossRef]
- Fischer von Mollard, G.; Stevens, T.H. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell 1999, 10, 1719–1732. [Google Scholar] [CrossRef]
- Ishihara, N.; Hamasaki, M.; Yokota, S.; Suzuki, K.; Kamada, Y.; Kihara, A.; Yoshimori, T.; Noda, T.; Ohsumi, Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell 2001, 12, 3690–3702. [Google Scholar] [CrossRef]
- Saleeb, R.S.; Kavanagh, D.M.; Dun, A.R.; Dalgarno, P.A.; Duncan, R.R. A VPS33A-binding motif on syntaxin 17 controls autophagy completion in mammalian cells. J. Biol Chem. 2019, 294, 4188–4201. [Google Scholar] [CrossRef] [PubMed]
- Itakura, E.; Kishi-Itakura, C.; Mizushima, N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012, 151, 1256–1269. [Google Scholar] [CrossRef] [PubMed]
- Takáts, S.; Nagy, P.; Varga, Á.; Pircs, K.; Kárpáti, M.; Varga, K.; Kovács, A.L.; Hegedűs, K.; Juhász, G. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 2013, 201, 531–539. [Google Scholar] [CrossRef]
- Takáts, S.; Glatz, G.; Szenci, G.; Boda, A.; Horváth, G.V.; Hegedűs, K.; Kovács, A.L.; Juhász, G. Non-canonical role of the SNARE protein Ykt6 in autophagosome-lysosome fusion. PLoS Genet. 2018, 14, e1007359. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Jiang, P.; Nakano, S.; Sakamaki, Y.; Yamamoto, H.; Mizushima, N. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J. Cell Biol. 2018, 217, 2633–2645. [Google Scholar] [CrossRef]
- Zhao, Y.G.; Zhang, H. Autophagosome maturation: An epic journey from the ER to lysosomes. J. Cell Biol. 2019, 218, 757–770. [Google Scholar] [CrossRef]
- Zhou, Y.; Peskett, T.R.; Landles, C.; Warner, J.B.; Sathasivam, K.; Smith, E.J.; Chen, S.; Wetzel, R.; Lashuel, H.A.; Bates, G.P.; et al. Correlative light and electron microscopy suggests that mutant huntingtin dysregulates the endolysosomal pathway in presymptomatic Huntington’s disease. Acta Neuropathol. Commun. 2021, 9, 70. [Google Scholar] [CrossRef]
- Yoshii, S.R.; Mizushima, N. Monitoring and Measuring Autophagy. Int. J. Mol. Sci. 2017, 18, 1865. [Google Scholar] [CrossRef]
- Zhang, X.J.; Chen, S.; Huang, K.X.; Le, W.D. Why should autophagic flux be assessed? Acta Pharmacol. Sin. 2013, 34, 595–599. [Google Scholar] [CrossRef]
- Ueno, T.; Komatsu, M. Monitoring Autophagy Flux and Activity: Principles and Applications. Bioessays 2020, 42, e2000122. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1. Autophagy 2021, 17, 1–382. [Google Scholar] [PubMed]
- Kalil, K.; Dent, E.W. Branch management: Mechanisms of axon branching in the developing vertebrate CNS. Nat. Rev. Neurosci. 2014, 15, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.A. Neuronal autophagy: A housekeeper or a fighter in neuronal cell survival? Exp. Neurobiol. 2012, 21, 1–8. [Google Scholar] [CrossRef]
- Crawley, O.; Grill, B. Autophagy in axonal and presynaptic development. Curr. Opin. Neurobiol. 2021, 69, 139–148. [Google Scholar] [CrossRef]
- Xilouri, M.; Stefanis, L. Autophagy in the central nervvous system: Implications for neurodegenerative disorders. CNS Neurol. Disord. Drug Targets 2010, 9, 701–719. [Google Scholar] [CrossRef] [PubMed]
- Nixon, R.A.; Cataldo, A.M. Lysosomal system pathways: Genes to neurodegeneration in Alzheimer’s disease. J. Alzheimers Dis. 2006, 9, 277–289. [Google Scholar] [CrossRef]
- Gallo, G. The cytoskeletal and signaling mechanisms of axon collateral branching. Dev. Neurobiol. 2011, 71, 201–220. [Google Scholar] [CrossRef]
- Hernandez, D.; Torres, C.A.; Setlik, W.; Cebrian, C.; Mosharov, E.V.; Tang, G.; Cheng, H.C.; Kholodilov, N.; Yarygina, O.; Burke, R.E.; et al. Regulation of presynaptic neurotransmission by macroautophagy. Neuron 2012, 74, 277–284. [Google Scholar] [CrossRef]
- Boland, B.; Nixon, R.A. Neuronal macroautophagy: From development to degeneration. Mol. Aspects Med. 2006, 27, 503–519. [Google Scholar] [CrossRef]
- Cronin, J.; Dudek, F.E. Chronic seizures and collateral sprouting of dentate mossy fibers after kainic acid treatment in rats. Brain Res. 1988, 474, 181–184. [Google Scholar] [CrossRef]
- Sutula, T.; He, X.X.; Cavazos, J.; Scott, G. Synaptic reorganization in the hippocampus induced by abnormal functional activity. Science 1988, 239, 1147–1150. [Google Scholar] [CrossRef] [PubMed]
- Babb, T.L.; Kupfer, W.R.; Pretorius, J.K.; Crandall, P.H.; Levesque, M.F. Synaptic reorganization by mossy fibers in human epileptic fascia dentata. Neuroscience 1991, 42, 351–363. [Google Scholar] [CrossRef]
- Mello, L.E.; Cavalheiro, E.A.; Tan, A.M.; Kupfer, W.R.; Pretorius, J.K.; Babb, T.L.; Finch, D.M. Circuit mechanisms of seizures in the pilocarpine model of chronic epilepsy: Cell loss and mossy fiber sprouting. Epilepsia 1993, 34, 985–995. [Google Scholar] [CrossRef]
- Desguerre, I.; Hully, M.; Rio, M.; Nabbout, R. Mitochondrial disorders and epilepsy. Rev. Neurol. 2014, 170, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Armijo, J.A.; Shushtarian, M.; Valdizan, E.M.; Cuadrado, A.; de las Cuevas, I.; Adín, J. Ion channels and epilepsy. Curr. Pharm. Des. 2005, 11, 1975–2003. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, N.C. Activity-dependent neurotransmitter respecification. Nat. Rev. Neurosci. 2012, 13, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Faust, T.E.; Gunner, G.; Schafer, D.P. Mechanisms governing activity-dependent synaptic pruning in the developing mammalian CNS. Nat. Rev. Neurosci. 2021, 22, 657–673. [Google Scholar] [CrossRef] [PubMed]
- Hollenbeck, P.J. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J. Cell Biol. 1993, 121, 305–315. [Google Scholar] [CrossRef]
- Mizushima, N.; Yamamoto, A.; Matsui, M.; Yoshimori, T.; Ohsumi, Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 2004, 15, 1101–1111. [Google Scholar] [CrossRef]
- Stavoe, A.K.; Hill, S.E.; Hall, D.H.; Colón-Ramos, D.A. KIF1A/UNC-104 Transports ATG-9 to Regulate Neurodevelopment and Autophagy at Synapses. Dev. Cell 2016, 38, 171–185. [Google Scholar] [CrossRef]
- Neisch, A.L.; Neufeld, T.P.; Hays, T.S. A STRIPAK complex mediates axonal transport of autophagosomes and dense core vesicles through PP2A regulation. J. Cell Biol. 2017, 216, 441–461. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.E.; Kauffman, K.J.; Krout, M.; Richmond, J.E.; Melia, T.J.; Colón-Ramos, D.A. Maturation and Clearance of Autophagosomes in Neurons Depends on a Specific Cysteine Protease Isoform, ATG-4.2. Dev. Cell 2019, 49, 251–266.e8. [Google Scholar] [CrossRef] [PubMed]
- Stavoe, A.K.H.; Holzbaur, E.L.F. Autophagy in Neurons. Annu. Rev. Cell Dev. Biol. 2019, 35, 477–500. [Google Scholar] [CrossRef]
- Joo, J.H.; Wang, B.; Frankel, E.; Ge, L.; Xu, L.; Iyengar, R.; Li-Harms, X.; Wright, C.; Shaw, T.I.; Lindsten, T.; et al. The Noncanonical Role of ULK/ATG1 in ER-to-Golgi Trafficking Is Essential for Cellular Homeostasis. Mol. Cell 2016, 62, 491–506. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Wang, Q.J.; Holstein, G.R.; Friedrich, V.L., Jr.; Iwata, J.; Kominami, E.; Chait, B.T.; Tanaka, K.; Yue, Z. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. USA 2007, 104, 14489–14494. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.C.; Wang, C.; Peng, X.; Gan, B.; Guan, J.L. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J. Biol. Chem. 2010, 285, 3499–3509. [Google Scholar] [CrossRef]
- Kuijpers, M.; Kochlamazashvili, G.; Stumpf, A.; Puchkov, D.; Swaminathan, A.; Lucht, M.T.; Krause, E.; Maritzen, T.; Schmitz, D.; Haucke, V. Neuronal Autophagy Regulates Presynaptic Neurotransmission by Controlling the Axonal Endoplasmic Reticulum. Neuron 2021, 109, 299–313.e9. [Google Scholar] [CrossRef]
- Lüningschrör, P.; Binotti, B.; Dombert, B.; Heimann, P.; Perez-Lara, A.; Slotta, C.; Thau-Habermann, N.R.; von Collenberg, C.; Karl, F.; Damme, M.; et al. Plekhg5-regulated autophagy of synaptic vesicles reveals a pathogenic mechanism in motoneuron disease. Nat. Commun. 2017, 8, 678. [Google Scholar] [CrossRef]
- Binotti, B.; Pavlos, N.J.; Riedel, D.; Wenzel, D.; Vorbrüggen, G.; Schalk, A.M.; Kühnel, K.; Boyken, J.; Erck, C.; Martens, H.; et al. The GTPase Rab26 links synaptic vesicles to the autophagy pathway. Elife 2015, 4, e05597. [Google Scholar] [CrossRef]
- Pimentel-Muiños, F.X.; Boada-Romero, E. Selective autophagy against membranous compartments: Canonical and unconventional purposes and mechanisms. Autophagy 2014, 10, 397–407. [Google Scholar] [CrossRef]
- Nikoletopoulou, V.; Tavernarakis, N. Regulation and Roles of Autophagy at Synapses. Trends Cell Biol. 2018, 28, 646–661. [Google Scholar] [CrossRef] [PubMed]
- Verstreken, P.; Koh, T.W.; Schulze, K.L.; Zhai, R.G.; Hiesinger, P.R.; Zhou, Y.; Mehta, S.Q.; Cao, Y.; Roos, J.; Bellen, H.J. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron 2003, 40, 733–748. [Google Scholar] [CrossRef]
- Soukup, S.F.; Verstreken, P. EndoA/Endophilin-A creates docking stations for autophagic proteins at synapses. Autophagy 2017, 13, 971–972. [Google Scholar] [CrossRef]
- Milosevic, I.; Giovedi, S.; Lou, X.; Raimondi, A.; Collesi, C.; Shen, H.; Paradise, S.; O’Toole, E.; Ferguson, S.; Cremona, O.; et al. Recruitment of endophilin to clathrin-coated pit necks.s is required for efficient vesicle uncoating after fission. Neuron 2011, 72, 587–601. [Google Scholar] [CrossRef]
- Hohendahl, A.; Talledge, N.; Galli, V.; Shen, P.S.; Humbert, F.; De Camilli, P.; Frost, A.; Roux, A. Structural inhibition of dynamin-mediated membrane fission by endophilin. Elife 2017, 6, e26856. [Google Scholar] [CrossRef]
- Ramachandran, R.; Schmid, S.L. The dynamin superfamily. Curr. Biol. 2018, 28, R411–R416. [Google Scholar] [CrossRef] [PubMed]
- Pan, P.Y.; Zhu, J.; Rizvi, A.; Zhu, X.; Tanaka, H.; Dreyfus, C.F. Synaptojanin1 deficiency upregulates basal autophagosome formation in astrocytes. J. Biol. Chem. 2021, 297, 100873. [Google Scholar] [CrossRef] [PubMed]
- George, A.A.; Hayden, S.; Stanton, G.R.; Brockerhoff, S.E. Arf6 and the 5’phosphatase of synaptojanin 1 regulate autophagy in cone photoreceptors. Bioessays 2016, 38 (Suppl. S1), S119–S135. [Google Scholar] [CrossRef]
- Vanhauwaert, R.; Kuenen, S.; Masius, R.; Bademosi, A.; Manetsberger, J.; Schoovaerts, N.; Bounti, L.; Gontcharenko, S.; Swerts, J.; Vilain, S.; et al. The SAC1 domain in synaptojanin is required for autophagosome maturation at presynaptic terminals. EMBO J. 2017, 36, 1392–1411. [Google Scholar] [CrossRef]
- Cao, M.; Park, D.; Wu, Y.; De Camilli, P. Absence of Sac2/INPP5F enhances the phenotype of a Parkinson’s disease mutation of synaptojanin 1. Proc. Natl. Acad. Sci. USA 2020, 117, 12428–12434. [Google Scholar] [CrossRef]
- Murdoch, J.D.; Rostosky, C.M.; Gowrisankaran, S.; Arora, A.S.; Soukup, S.F.; Vidal, R.; Capece, V.; Freytag, S.; Fischer, A.; Verstreken, P.; et al. Endophilin-A Deficiency Induces the Foxo3a-Fbxo32 Network in the Brain and Causes Dysregulation of Autophagy and the Ubiquitin-Proteasome System. Cell Rep. 2016, 17, 1071–1086. [Google Scholar] [CrossRef] [PubMed]
- Cullup, T.; Kho, A.L.; Dionisi-Vici, C.; Brandmeier, B.; Smith, F.; Urry, Z.; Simpson, M.A.; Yau, S.; Bertini, E.; McClelland, V.; et al. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat. Genet. 2013, 45, 83–87. [Google Scholar] [CrossRef]
- Fraiberg, M.; Tamim-Yecheskel, B.C.; Kokabi, K.; Subic, N.; Heimer, G.; Eck, F.; Nalbach, K.; Behrends, C.; Ben-Zeev, B.; Shatz, O.; et al. Lysosomal targeting of autophagosomes by the TECPR domain of TECPR2. Autophagy 2021, 17, 3096–3108. [Google Scholar] [CrossRef] [PubMed]
- Davies, A.K.; Itzhak, D.N.; Edgar, J.R.; Archuleta, T.L.; Hirst, J.; Jackson, L.P.; Robinson, M.S.; Borner, G.H.H. AP-4 vesicles contribute to spatial control of autophagy via RUSC-dependent peripheral delivery of ATG9A. Nat. Commun. 2018, 9, 3958. [Google Scholar] [CrossRef]
- Dragich, J.M.; Kuwajima, T.; Hirose-Ikeda, M.; Yoon, M.S.; Eenjes, E.; Bosco, J.R.; Fox, L.M.; Lystad, A.H.; Oo, T.F.; Yarygina, O.; et al. Autophagy linked FYVE (Alfy/WDFY3) is required for establishing neuronal connectivity in the mammalian brain. Elife 2016, 5, e14810. [Google Scholar] [CrossRef] [PubMed]
- Wojnacki, J.; Nola, S.; Bun, P.; Cholley, B.; Filippini, F.; Pressé, M.T.; Lipecka, J.; Man Lam, S.; N’guyen, J.; Simon, A.; et al. Role of VAMP7-Dependent Secretion of Reticulon 3 in Neurite Growth. Cell Rep. 2020, 33, 108536. [Google Scholar] [CrossRef]
- Sakamoto, K.; Ozaki, T.; Ko, Y.C.; Tsai, C.F.; Gong, Y.; Morozumi, M.; Ishikawa, Y.; Uchimura, K.; Nadanaka, S.; Kitagawa, H.; et al. Glycan sulfation patterns define autophagy flux at axon tip via PTPRσ-cortactin axis. Nat. Chem. Biol. 2019, 15, 699–709. [Google Scholar] [CrossRef]
- Hirst, J.; Irving, C.; Borner, G.H. Adaptor protein complexes AP-4 and AP-5: New players in endosomal trafficking and progressive spastic paraplegia. Traffic 2013, 14, 153–164. [Google Scholar] [CrossRef]
- Yap, C.C.; Murate, M.; Kishigami, S.; Muto, Y.; Kishida, H.; Hashikawa, T.; Yano, R. Adaptor protein complex-4 (AP-4) is expressed in the central nervous system neurons and interacts with glutamate receptor delta2. Mol. Cell Neurosci. 2003, 24, 283–295. [Google Scholar] [CrossRef]
- Matsuda, S.; Miura, E.; Matsuda, K.; Kakegawa, W.; Kohda, K.; Watanabe, M.; Yuzaki, M. Accumulation of AMPA receptors in autophagosomes in neuronal axons lacking adaptor protein AP-4. Neuron 2008, 57, 730–745. [Google Scholar] [CrossRef]
- De Pace, R.; Skirzewski, M.; Damme, M.; Mattera, R.; Mercurio, J.; Foster, A.M.; Cuitino, L.; Jarnik, M.; Hoffmann, V.; Morris, H.D.; et al. Altered distribution of ATG9A and accumulation of axonal aggregates in neurons from a mouse model of AP-4 deficiency syndrome. PLoS Genet. 2018, 14, e100736. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Chiba, T.; Murata, S.; Iwata, J.; Tanida, I.; Ueno, T.; Koike, M.; Uchiyama, Y.; Kominami, E.; et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006, 441, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, J.; Miura, E.; Mizushima, N.; Watanabe, M.; Yuzaki, M. Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy 2007, 3, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Rodriguez-Navarro, J.A.; Arias, E.; Kiffin, R.; Sahu, S.; Schwartz, G.J.; Cuervo, A.M.; Singh, R. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 2011, 14, 173–183. [Google Scholar] [CrossRef]
- Chen, Y.; Sawada, O.; Kohno, H.; Le, Y.Z.; Subauste, C.; Maeda, T.; Maeda, A. Autophagy protects the retina from light-induced degeneration. J. Biol. Chem. 2013, 288, 7506–7518. [Google Scholar] [CrossRef]
- Zhou, Z.; Doggett, T.A.; Sene, A.; Apte, R.S.; Ferguson, T.A. Autophagy supports survival and phototransduction protein levels in rod photoreceptors. Cell Death Differ. 2015, 22, 488–498. [Google Scholar] [CrossRef]
- Ban, B.K.; Jun, M.H.; Ryu, H.H.; Jang, D.J.; Ahmad, S.T.; Lee, J.A. Autophagy negatively regulates early axon growth in cortical neurons. Mol Cell Biol 2013, 33, 3907–3919. [Google Scholar] [CrossRef]
- Yang, K.; Yu, B.; Cheng, C.; Cheng, T.; Yuan, B.; Li, K.; Xiao, J.; Qiu, Z.; Zhou, Y. Mir505-3p regulates axonal development via inhibiting the autophagy pathway by targeting Atg12. Autophagy 2017, 13, 1679–1696. [Google Scholar] [CrossRef]
- Nixon, R.A. Endosome function and dysfunction in Alzheimer’s disease and other neurodegenerative diseases. Neurobiol. Aging 2005, 26, 373–382. [Google Scholar] [CrossRef]
- Waites, C.L.; Leal-Ortiz, S.A.; Okerlund, N.; Dalke, H.; . Fejtova, A.; Altrock, W.D.; Gundelfinger, E.D.; Garner, C.C. Bassoon and Piccolo maintain synapse integrity by regulating protein ubiquitination and degradation. EMBO J. 2013, 32, 954–969. [Google Scholar] [CrossRef]
- Okerlund, N.D.; Schneider, K.; Leal-Ortiz, S.; Montenegro-Venegas, C.; Kim, S.A.; Garner, L.C.; Waites, C.L.; Gundelfinger, E.D.; Reimer, R.J.; Garner, C.C. Bassoon Controls Presynaptic Autophagy through Atg5. Neuron 2017, 93, 897–913.e7. [Google Scholar] [CrossRef] [PubMed]
- Crawley, O.; Opperman, K.J.; Desbois, M.; Adrados, I.; Borgen, M.A.; Giles, A.C.; Duckett, D.R.; Grill, B. Autophagy is inhibited by ubiquitin ligase activity in the nervous system. Nat. Commun. 2019, 10, 5017. [Google Scholar] [CrossRef] [PubMed]
- Tyler, W.J.; Pozzo-Miller, L.D. BDNF enhances quantal neurotransmitter release and increases the number of docked vesicles at the active zones of hippocampal excitatory synapses. J. Neurosci 2001, 21, 4249–4258. [Google Scholar] [CrossRef]
- Nikoletopoulou, V.; Sidiropoulou, K.; Kallergi, E.; Dalezios, Y.; Tavernarakis, N. Modulation of Autophagy by BDNF Underlies Synaptic Plasticity. Cell Metab. 2017, 26, 230–242.e5. [Google Scholar] [CrossRef] [PubMed]
- Tavosanis, G. Dendrite enlightenment. Curr. Opin. Neurobiol. 2021, 69, 222–230. [Google Scholar] [CrossRef]
- Clark, S.G.; Graybeal, L.L.; Bhattacharjee, S.; Thomas, C.; Bhattacharya, S.; Cox, D.N. Basal autophagy is required for promoting dendritic terminal branching in Drosophila sensory neurons. PLoS ONE 2018, 13, e0206743. [Google Scholar] [CrossRef] [PubMed]
- Friedman, L.G.; Lachenmayer, M.L.; Wang, J.; He, L.; Poulose, S.M.; Komatsu, M.; Holstein, G.R.; Yue, Z. Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of α-synuclein and LRRK2 in the brain. J. Neurosci. 2012, 32, 7585–7593. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Gudsnuk, K.; Kuo, S.H.; Cotrina, M.L.; Rosoklija, G.; Sosunov, A.; Sonders, M.S.; Kanter, E.; Castagna, C.; Yamamoto, A.; et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 2014, 83, 1131–1143. [Google Scholar] [CrossRef]
- Chen, Z.W.; Chang, C.S.; Leil, T.A.; Olsen, R.W. C-terminal modification is required for GABARAP-mediated GABA(A) receptor trafficking. J. Neurosci. 2007, 27, 6655–6663. [Google Scholar] [CrossRef]
- Rowland, A.M.; Richmond, J.E.; Olsen, J.G.; Hall, D.H.; Bamber, B.A. Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. J. Neurosci 2006, 26, 1711–1720. [Google Scholar] [CrossRef]
- Lieberman, O.J.; Sulzer, D. The Synaptic Autophagy Cycle. J. Mol. Biol. 2020, 432, 2589–2604. [Google Scholar] [CrossRef] [PubMed]
- Shehata, M.; Matsumura, H.; Okubo-Suzuki, R.; Ohkawa, N.; Inokuchi, K. Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J. Neurosci. 2012, 32, 10413–10422. [Google Scholar] [CrossRef]
- Compans, B.; Camus, C.; Kallergi, E.; Sposini, S.; Martineau, M.; Butler, C.; Kechkar, A.; Klaassen, R.V.; Retailleau, N.; Sejnowski, T.J.; et al. NMDAR-dependent long-term depression is associated with increased short term plasticity through autophagy mediated loss of PSD-95. Nat. Commun. 2021, 12, 2849. [Google Scholar] [CrossRef] [PubMed]
- Maday, S.; Holzbaur, E.L. Compartment-Specific Regulation of Autophagy in Primary Neurons. J. Neurosci. 2016, 36, 5933–5945. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Zakaria, H.M.; Simone, A.; Sheng, Z.H. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr. Biol. 2012, 22, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Devireddy, S.; Liu, A.; Lampe, T.; Hollenbeck, P.J. The Organization of Mitochondrial Quality Control and Life Cycle in the Nervous System In Vivo in the Absence of PINK1. J. Neurosci. 2015, 35, 9391–9401. [Google Scholar] [CrossRef] [PubMed]
- Magee, J.C.; Grienberger, C. Synaptic Plasticity Forms and Functions. Annu. Rev. Neurosci. 2020, 43, 95–117. [Google Scholar] [CrossRef]
- Hegde, A.N. Proteolysis, synaptic plasticity and memory. Neurobiol. Learn. Mem. 2017, 138, 98–110. [Google Scholar] [CrossRef]
- Tomoda, T.; Yang, K.; Sawa, A. Neuronal Autophagy in Synaptic Functions and Psychiatric Disorders. Biol. Psychiatry 2020, 87, 787–796. [Google Scholar] [CrossRef]
- Kim, H.J.; Cho, M.H.; Shim, W.H.; Kim, J.K.; Jeon, E.Y.; Kim, D.H.; Yoon, S.Y. Deficient autophagy in microglia impairs synaptic pruning and causes social behavioral defects. Mol. Psychiatry 2017, 22, 1576–1584. [Google Scholar] [CrossRef]
- Glatigny, M.; Moriceau, S.; Rivagorda, M.; Ramos-Brossier, M.; Nascimbeni, A.C.; Lante, F.; Shanley, M.R.; Boudarene, N.; Rousseaud, A.; Friedman, A.K.; et al. Autophagy Is Required for Memory Formation and Reverses Age-Related Memory Decline. Curr. Biol. 2019, 29, 435–448.e8. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Shang, X.; Zhai, B.; Zhang, H.; Zhang, T. Nicotine alleviates chronic stress-induced anxiety and depressive-like behavior and hippocampal neuropathology via regulating autophagy signaling. Neurochem. Int. 2018, 114, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Porch, M.W.; Court-Vazquez, B.; Bennett, M.V.L.; Zukin, R.S. Activation of autophagy rescues synaptic and cognitive deficits in fragile X mice. Proc. Natl. Acad. Sci. USA 2018, 115, E9707–E9716. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, L.K.; Le Prieult, F.; Felzen, V.; Thal, S.C.; Engelhard, K.; Behl, C.; Mittmann, T. Proteasome and Autophagy-Mediated Impairment of Late Long-Term Potentiation (l-LTP) after Traumatic Brain Injury in the Somatosensory Cortex of Mice. Int. J. Mol. Sci. 2019, 20, 3048. [Google Scholar] [CrossRef]
- Horwood, J.M.; Dufour, F.; Laroche, S.; Davis, S. Signalling mechanisms mediated by the phosphoinositide 3-kinase/Akt cascade in synaptic plasticity and memory in the rat. Eur. J. Neurosci. 2006, 23, 3375–3384. [Google Scholar] [CrossRef]
- Thijs, R.D.; Surges, R.; O’Brien, T.J.; Sander, J.W. Epilepsy in adults. Lancet 2019, 393, 689–701. [Google Scholar] [CrossRef]
- Fiest, K.M.; Sauro, K.M.; Wiebe, S.; Patten, S.B.; Kwon, C.S.; Dykeman, J.; Pringsheim, T.; Lorenzetti, D.L.; Jetté, N. Prevalence and incidence of epilepsy: A systematic review and meta-analysis of international studies. Neurology 2017, 88, 296–303. [Google Scholar] [CrossRef]
- Beretta, S.; Carone, D.; Zanchi, C.; Bianchi, E.; Pirovano, M.; Trentini, C.; Padovano, G.; Colombo, M.; Cereda, D.; Scanziani, S.; et al. PRO-LONG Study Group. Long-term applicabili.ity of the new ILAE definition of epilepsy. Results from the PRO-LONG study. Epilepsia 2017, 58, 1518–1523. [Google Scholar] [CrossRef]
- Fisher, R.S.; Cross, J.H.; French, J.A.; Higurashi, N.; Hirsch, E.; Jansen, F.E.; Lagae, L.; Moshé, S.L.; Peltola, J.; Roulet Perez, E.; et al. Operational classification of seizure types by the International League Against Epilepsy: Position Paper of the ILAE Commission for Classification and Terminology. Epilepsia 2017, 58, 522–530. [Google Scholar] [CrossRef]
- Balestrini, S.; Arzimanoglou, A.; Blümcke, I.; Scheffer, I.E.; Wiebe, S.; Zelano, J.; Walker, M.C. The aetiologies of epilepsy. Epileptic Disord. 2021, 23, 1–16. [Google Scholar] [CrossRef]
- Fisher, R.S.; Acevedo, C.; Arzimanoglou, A.; Bogacz, A.; Cross, J.H.; Elger, C.E.; Engel, J., Jr.; Forsgren, L.; French, J.A.; Glynn, M.; et al. ILAE official report: A practical clinical definition of epilepsy. Epilepsia 2014, 55, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Helbig, I.; Scheffer, I.E.; Mulley, J.C.; Berkovic, S.F. Navigating the channels and beyond: Unravelling the genetics of the epilepsies. Lancet Neurol. 2008, 7, 231–245. [Google Scholar] [CrossRef]
- Berg, A.T.; Mathern, G.W.; Bronen, R.A.; Fulbright, R.K.; DiMario, F.; Testa, F.M.; Levy, S.R. Frequency, prognosis and surgical treatment of structural abnormalities seen with magnetic resonance imaging in childhood epilepsy. Brain 2009, 132, 2785–2797. [Google Scholar] [CrossRef] [PubMed]
- Vivash, L.; Tostevin, A.; Liu, D.S.; Dalic, L.; Dedeurwaerdere, S.; Hicks, R.J.; Williams, D.A.; Myers, D.E.; O’Brien, T.J. Changes in hippocampal GABAA/cBZR density during limbic epileptogenesis: Relationship to cell loss and mossy fibre sprouting. Neurobiol. Dis. 2011, 41, 227–236. [Google Scholar] [CrossRef]
- Crino, P.B. mTOR: A pathogenic signaling pathway in developmental brain malformations. Trends Mol. Med. 2011, 17, 734–742. [Google Scholar] [CrossRef]
- Nguyen, L.H.; Mahadeo, T.; Bordey, A. mTOR Hyperactivity Levels Influence the Severity of Epilepsy and Associated Neuropathology in an Experimental Model of Tuberous Sclerosis Complex and Focal Cortical Dysplasia. J. Neurosci. 2019, 39, 2762–2773. [Google Scholar] [CrossRef]
- Fernandez, D.; Perl, A. mTOR signaling: A central pathway to pathogenesis in systemic lupus erythematosus? Discov. Med. 2010, 9, 173–178. [Google Scholar]
- Limanaqi, F.; Biagioni, F.; Busceti, C.L.; Fabrizi, C.; Frati, A.; Fornai, F. mTOR-Related Cell-Clearing Systems in Epileptic Seizures, an Update. Int. J. Mol. Sci. 2020, 21, 1642. [Google Scholar] [CrossRef]
- Lasarge, C.L.; Danzer, S.C. Mechanisms regulating neuronal excitability and seizure development following mTOR pathway hyperactivation. Front. Mol. Neurosci. 2014, 7, 18. [Google Scholar] [CrossRef]
- Dazzo, E.; Nobile, C. Epilepsy-causing Reelin mutations result in impaired secretion and intracellular degradation of mutant proteins. Hum. Mol. Genet. 2022, 31, 665–673. [Google Scholar] [CrossRef]
- Giorgi, F.S.; Biagioni, F.; Lenzi, P.; Frati, A.; Fornai, F. The role of autophagy in epileptogenesis and in epilepsy-induced neuronal alterations. J. Neural. Transm. 2015, 122, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Saitsu, H.; Nishimura, T.; Muramatsu, K.; Kodera, H.; Kumada, S.; Sugai, K.; Kasai-Yoshida, E.; Sawaura, N.; Nishida, H.; Hoshino, A.; et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat. Genet. 2013, 45, 445–449. [Google Scholar] [CrossRef] [PubMed]
- McMahon, J.; Huang, X.; Yang, J.; Komatsu, M.; Yue, Z.; Qian, J.; Zhu, X.; Huang, Y. Impaired autophagy in neurons after disinhibition of mammalian target of rapamycin and its contribution to epileptogenesis. J. Neurosci. 2012, 32, 15704–15714. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Han, Y.; Du, J.; Jin, H.; Zhang, J.; Niu, M.; Qin, J. Alterations of apoptosis and autophagy in developing brain of rats with epilepsy: Changes in LC3, P62, Beclin-1 and Bcl-2 levels. Neurosci. Res. 2018, 130, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Shacka, J.J.; Lu, J.; Xie, Z.L.; Uchiyama, Y.; Roth, K.A.; Zhang, J. Kainic acid induces early and transient autophagic stress in mouse hippocampus. Neurosci. Lett. 2007, 414, 57–60. [Google Scholar] [CrossRef]
- Fulceri, F.; Ferrucci, M.; Lazzeri, G.; Paparelli, S.; Bartalucci, A.; Tamburini, I.; Paparelli, A.; Fornai, F. Autophagy activation in glutamate-induced motor neuron loss. Arch. Ital. Biol. 2011, 149, 101–111. [Google Scholar]
- Wong, M. Cleaning up epilepsy and neurodegeneration: The role of autophagy in epileptogenesis. Epilepsy Curr. 2013, 13, 177–178. [Google Scholar] [CrossRef]
- Fassio, A.; Falace, A.; Esposito, A.; Aprile, D.; Guerrini, R.; Benfenati, F. Emerging role of the autophagy/lysosomal degradative pathway in neurodevelopmental disorders with epilepsy. Front. Cell Neurosci. 2020, 14, 39. [Google Scholar] [CrossRef]
- Xie, N.; Li, Y.; Wang, C.; Lian, Y.; Zhang, H.; Li, Y.; Meng, X.; Du, L. FAM134B attenuates seizure-induced apoptosis and endoplasmic reticulum stress in hippocampal neurons by promoting autophagy. Cell Mol. Neurobiol. 2020, 40, 1297–1305. [Google Scholar] [CrossRef]
- Wang, L.; Song, L.F.; Chen, X.Y.; Ma, Y.L.; Suo, J.F.; Shi, J.H.; Chen, G.H. MiR-181b inhibits P38/JNK signaling pathway to attenuate autophagy and apoptosis in juvenile rats with kainic acid-induced epilepsy via targeting TLR4. CNS Neurosci. Ther. 2019, 25, 112–122. [Google Scholar] [CrossRef]
- Sun, J.; Gao, X.; Meng, D.; Xu, Y.; Wang, X.; Gu, X.; Guo, M.; Shao, X.; Yan, H.; Jiang, C.; et al. Antagomirs targeting miroRNA-134 attenuates epilepsy in rats through regulation of oxidative stress, mitochondrial functions and autophagy. Front. Pharmacol. 2017, 8, 524. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Lv, J.; Zheng, Z.; Liu, Y.; Zhang, Y.; Luo, C.; Qi, L.; Qin, B.; Liu, C. Mechanisms of microRNA-142 in mitochondrial autophagy and hippocampal damage in a rat model of epilepsy. Int. J. Mol. Med. 2021, 47, 98. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Chen, H.; Zhong, Y.; Yang, Q.; Wang, X.; Chen, R.; Guo, Y. MicroRNA 223 Targeting ATG16L1 affects microglial autophagy in the kainic acid model of temporal lobe epilepsy. Front. Neurol. 2021, 12, 704550. [Google Scholar] [CrossRef]
- Hu, F.; Shao, L.; Zhang, J.; Zhang, H.; Wen, A.; Zhang, G.P. Knockdown of ZFAS1 inhibits hippocampal neurons apoptosis and autophagy by activating the PI3K/AKT pathway via up-regulating miR-421 in epilepsy. Neurochem. Res. 2020, 45, 2433–2441. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yi, X. Down-regulation of long noncoding RNA MALAT1 protects hippocampal neurons against excessive autophagy and apoptosis via the PI3K/Akt signaling pathway in rats with epilepsy. J. Mol. Neurosci. 2018, 65, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.M.; Zhao, X.F.; Liao, Y.; Mathur, R.; McCallum, S.E.; Mazurkiewicz, J.E.; Adamo, M.A.; Feustel, P.; Belin, S.; Poitelon, Y.; et al. Deficiency of microglial autophagy increases the density of oligodendrocytes and susceptibility to severe forms of seizures. eNeuro 2021, 8, ENEURO.0183-20.2021. [Google Scholar] [CrossRef]
- Peng, J.; Wu, S.; Guo, C.; Guo, K.; Zhang, W.; Liu, R.; Li, J.; Hu, Z. Effect of Ibuprofen on Autophagy of Astrocytes during Pentylenetetrazol-Induced Epilepsy and its Significance: An Experimental Study. Neurochem. Res. 2019, 44, 2566–2576. [Google Scholar] [CrossRef]
- Garyali, P.; Segvich, D.M.; DePaoli-Roach, A.A.; Roach, P.J. Protein degradation and quality control in cells from Laforin and Malin knockout mice. J. Biol. Chem. 2014, 289, 20606–20614. [Google Scholar] [CrossRef]
- Buckmaster, P.S.; Ingram, E.A.; Wen, X. Inhibition of the mammalian target of rapamycin signaling pathway suppresses dentate granule cell axon sprouting in a rodent model of temporal lobe epilepsy. J. Neurosci. 2009, 29, 8259–8269. [Google Scholar] [CrossRef]
- Buckmaster, P.S.; Lew, F.H. Rapamycin suppresses mossy fiber sprouting but not seizure frequency in a mouse model of temporal lobe epilepsy. J. Neurosci. 2011, 31, 2337–2347. [Google Scholar] [CrossRef]
- Wong, M. Mammalian target of rapamycin (mTOR) pathways in neurological diseases. Biomed. J. 2013, 36, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Amegandjin, C.A.; Choudhury, M.; Jadhav, V.; Carriço, J.N.; Quintal, A.; Berryer, M.; Snapyan, M.; Chattopadhyaya, B.; Saghatelyan, A.; Di Cristo, G. Sensitive period for rescuing parvalbumin interneurons connectivity and social behavior deficits caused by TSC1 loss. Nat. Commun. 2021, 12, 3653. [Google Scholar] [CrossRef]
- Puri, R.; Suzuki, T.; Yamakawa, K.; Ganesh, S. Dysfunctions in endosomal-lysosomal and autophagy pathways underlie neuropathology in a mouse model for Lafora disease. Hum. Mol. Genet. 2012, 21, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Criado, O.; Aguado, C.; Gayarre, J.; Duran-Trio, L.; Garcia-Cabrero, A.M.; Vernia, S.; San Millán, B.; Heredia, M.; Romá-Mateo, C.; Mouron, S.; et al. Lafora bodies and neurological defects in malin-deficient mice correlate with impaired autophagy. Hum. Mol. Genet. 2012, 21, 1521–1533. [Google Scholar] [CrossRef] [PubMed]
- Ying, C.; Ying, L.; Yanxia, L.; Le, W.; Lili, C. High mobility group box 1 antibody represses autophagy and alleviates hippocampus damage in pilocarpine-induced mouse epilepsy model. Acta Histochem. 2020, 122, 151485. [Google Scholar] [CrossRef] [PubMed]
- Gstrein, T.; Edwards, A.; Přistoupilová, A.; Leca, I.; Breuss, M.; Pilat-Carotta, S.; Hansen, A.H.; Tripathy, R.; Traunbauer, A.K.; Hochstoeger, T.; et al. Mutations in Vps15 perturb neuronal migration in mice and are associated with neurodevelopmental disease in humans. Nat. Neurosci. 2018, 21, 207–217. [Google Scholar] [CrossRef]
- Yasin, S.A.; Ali, A.M.; Tata, M.; Picker, S.R.; Anderson, G.W.; Latimer-Bowman, E.; Nicholson, S.L.; Harkness, W.; Cross, J.H.; Paine, S.M.; et al. mTOR-dependent abnormalities in autophagy characterize human malformations of cortical development: Evidence from focal cortical dysplasia and tuberous sclerosis. Acta Neuropathol. 2013, 126, 207–218. [Google Scholar] [CrossRef]
- Yuskaitis, C.J.; Jones, B.M.; Wolfson, R.L.; Super, C.E.; Dhamne, S.C.; Rotenberg, A.; Sabatini, D.M.; Sahin, M.; Poduri, A. A mouse model of DEPDC5-related epilepsy: Neuronal loss of Depdc5 causes dysplastic and ectopic neurons, increased mTOR signaling, and seizure susceptibility. Neurobiol. Dis. 2018, 111, 91–101. [Google Scholar] [CrossRef]
- Kwon, C.H.; Zhu, X.; Zhang, J.; Baker, S.J. mTor is required for hypertrophy of Pten-deficient neuronal soma in vivo. Proc. Natl. Acad. Sci. USA 2003, 100, 12923–12928. [Google Scholar] [CrossRef]
- Cao, L.; Xu, J.; Lin, Y.; Zhao, X.; Liu, X.; Chi, Z. Autophagy is upregulated in rats with status epilepticus and partly inhibited by Vitamin, E. Biochem. Biophys. Res. Commun. 2009, 379, 949–953. [Google Scholar] [CrossRef]
- Macias, M.; Blazejczyk, M.; Kazmierska, P.; Caban, B.; Skalecka, A.; Tarkowski, B.; Rodo, A.; Konopacki, J.; Jaworski, J. Spatiotemporal characterization of mTOR kinase activity following kainic acid induced status epilepticus and analysis of rat brain response to chronic rapamycin treatment. PLoS ONE 2013, 8, e64455. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.H.; Rensing, N.R.; Wong, M. The mammalian target of rapamycin signaling pathway mediates epileptogenesis in a model of temporal lobe epilepsy. J. Neurosci. 2009, 29, 6964–6972. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhang, H.; Yang, J.; Wu, J.; McMahon, J.; Lin, Y.; Cao, Z.; Gruenthal, M.; Huang, Y. Pharmacological inhibition of the mammalian target of rapamycin pathway suppresses acquired epilepsy. Neurobiol. Dis. 2010, 40, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Raffo, E.; Coppola, A.; Ono, T.; Briggs, S.W.; Galanopoulou, A.S. A pulse rapamycin therapy for infantile spasms and associated cognitive decline. Neurobiol Dis 2011, 43, 322–329. [Google Scholar] [CrossRef]
- Carvill, G.L.; Liu, A.; Mandelstam, S.; Schneider, A.; Lacroix, A.; Zemel, M.; McMahon, J.M.; Bello-Espinosa, L.; Mackay, M.; Wallace, G.; et al. Severe infantile onset developmental and epileptic encephalopathy caused by mutations in autophagy gene WDR45. Epilepsia 2018, 59, e5–e13. [Google Scholar] [CrossRef]
- Byrne, S.; Jansen, L.; U-King-Im, J.M.; Siddiqui, A.; Lidov, H.G.; Bodi, I.; Smith, L.; Mein, R.; Cullup, T.; Dionisi-Vici, C.; et al. EPG5-related Vici syndrome: A paradigm of neurodevelopmental disorders with defective autophagy. Brain 2016, 139, 765–781. [Google Scholar] [CrossRef] [PubMed]
- Akizu, N.; Cantagrel, V.; Zaki, M.S.; Al-Gazali, L.; Wang, X.; Rosti, R.O.; Dikoglu, E.; Gelot, A.B.; Rosti, B.; Vaux, K.K.; et al. Biallelic mutations in SNX14 cause a syndromic form of cerebellar atrophy and lysosome-autophagosome dysfunction. Nat. Genet. 2015, 47, 528–534. [Google Scholar] [CrossRef]
- Esposito, A.; Falace, A.; Wagner, M.; Gal, M.; Mei, D.; Conti, V.; Pisano, T.; Aprile, D.; Cerullo, M.S.; De Fusco, A.; et al. Biallelic DMXL2 mutations impair autophagy and cause Ohtahara syndrome with progressive course. Brain 2019, 142, 3876–3891. [Google Scholar] [CrossRef]
- Ortiz-González, X.R.; Tintos-Hernández, J.A.; Keller, K.; Li, X.; Foley, A.R.; Bharucha-Goebel, D.X.; Kessler, S.K.; Yum, S.W.; Crino, P.B.; He, M.; et al. Homozygous boricua TBCK mutation causes neurodegeneration and aberrant autophagy. Ann. Neurol. 2018, 83, 153–165. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Qiao, S.; Cai, M.T.; Lai, Q.L.; Shen, C.H.; Ding, M.P. Association between autophagy-related protein 5 gene polymorphisms and epilepsy in Chinese patients. Neurosci. Lett. 2021, 753, 135870. [Google Scholar] [CrossRef]
- Van Damme, T.; Gardeitchik, T.; Mohamed, M.; Guerrero-Castillo, S.; Freisinger, P.; Guillemyn, B.; Kariminejad, A.; Dalloyaux, D.; van Kraaij, S.; Lefeber, D.J.; et al. Mutations in ATP6V1E1 or ATP6V1A Cause Autosomal-Recessive Cutis Laxa. Am. J. Hum. Genet. 2020, 107, 374. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Cabrera-Socorro, A.; Chitayat, D.; Lemonnier, T.; Féraud, O.; Cifuentes-Diaz, C.; Gervasi, N.; Mombereau, C.; Ghosh, T.; Stoica, L.; et al. ATP6AP2 variant impairs CNS development and neuronal survival to cause fulminant neurodegeneration. J. Clin. Invest. 2019, 129, 2145–2162. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Lin, P.; Jing, W.; Guo, H.; Chen, H.; Chen, Y.; Guo, Y.; Gu, Y.; He, M.; Wu, J.; et al. Beclin1 Deficiency Suppresses Epileptic Seizures. Front. Mol. Neurosci. 2022, 15, 807671. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).