Abstract
Epistasis of ERAP1 single nucleotide variations (SNVs) and HLA-B27 has been linked to ankylosing spondylitis susceptibility (AS). The current study examined how prevalent ERAP1 allelic variants (SNV haplotypes) in Taiwan affect ERAP1 functions and AS susceptibility in the presence or absence of HLA-B27. Sanger sequencing was used to discover all ERAP1 coding SNVs and common allelic variants in Taiwanese full-length cDNAs from 45 human patients. For the genetic association investigation, TaqMan genotyping assays were utilized to establish the genotypes of ERAP1 SNVs in 863 AS patients and 1438 healthy controls. Ex vivo biological analysis of peripheral blood mononuclear cells from homozygous donors of two common-risk ERAP1 allelic variants was performed. Two common-risk ERAP1 allelic variants were also cloned and functionally studied. In Taiwanese, eleven frequent ERAP1 SNVs and six major ERAP1 allelic variants were discovered. We discovered that in Taiwanese, the most prevalent ERAP1-001 variant with 56E, 127R, 276I, 349M, 528K, 575D, 725R, and 730Q interacting with HLA-B27 significantly contributed to the development of AS. In HLA-B27 negative group, however, the second most prevalent ERAP1-002 variant with 56E, 127P, 276M, 349M, 528R, 575D, 725R, and 730E was substantially related with an increased risk of AS. Ex vivo and in vitro research demonstrated that ERAP1 allelic variants have a significant impact on ERAP1 functions, suggesting that ERAP1 plays a role in the development of AS. In an HLA-B27-dependent manner, common ERAP1 allelic variants are related with AS susceptibility.
1. Introduction
Ankylosing spondylitis (AS) is a subtype of axial spondylopathy (AxSpA) that primarily affects the axial skeleton and is characterized by the formation of syndesmophytes and spinal ankylosis deformities [1]. The pathophysiology of AS, which has long been thought to be a highly familial and heritable disease [2,3], is complicated by a number of variables. Multiple genes may be involved in the development of AS, based on the heterogeneity of the disease’s progression. Several genes and genomic areas have been linked to AS susceptibility and severity in genome-wide association studies (GWAS), demonstrating that both MHC and non-MHC genes have a role in the illness process [4,5,6,7,8]. Despite breakthroughs in recent years, the genetic and pathophysiology processes of AS remain poorly known.
Antigenic peptides on MHC molecules are scanned by the human immune system to discern infected or sick cells. Antigenic peptides degraded in proteasomes are then trimmed to appropriate lengths on the ER luminal side by aminopeptidases for correct presentation on MHC-I molecules [9,10,11,12,13]. Endoplasmic reticulum aminopeptidase 1 (ERAP1), a unique member of the M1 aminopeptidase family, trims antigen peptides to fit the antigen binding site on MHC-I during the final stage of antigen processing. Peptide–MHC-I complexes on the cell surface reflect the existing proteome of respective cells.
The function of ERAP1 has a major impact on the stability and immunological characteristics of MHC-I molecules because of its critical involvement in processing MHC-I ligands. Low or excessive ERAP1 activity could result in antigen peptides that are unsuitable for MHC-I presentation. As a result, effective ERAP1 function is required for optimal antigen presentation and health maintenance. ERAP1 is a non-MHC gene on chromosome 5q15, and genetic studies have shown that epistasis between ERAP1 SNVs and HLA-B27 is highly linked to AS vulnerability in different ethnic groups [6,14]. The biological functions of ERAP1 allelic variants on the other hand, are largely unexplored. The molecular processes underpinning the link between ERAP1 allelic variations and AS are still being investigated [15]. Furthermore, different combinations of ERAP1 SNVs (or allelic variants) may interact with HLA-B in a unique way to contribute to AS vulnerability in a single ethnic population. The breadth of ERAP1 SNVs and allelic variations in Taiwanese was investigated in this study. The ERAP1-001 variation was discovered to be a key risk factor for AS in Taiwanese people. Furthermore, in an HLA-B27-dependent manner, two prevalent ERAP1 allelic variants are strongly related with AS susceptibility.
2. Materials and Methods
2.1. Study Subjects
A total of 863 AS patients (717 males and 146 females; onset ages ranged from 8 to 72 with the mean onset age of 25.38-year-old) who fulfilled the 1984 revised New York diagnostic criteria for AS were recruited at the Chang Gung Memorial Hospital. HLA-B27 carriers accounted for 801 (92.8%) of the 863 AS patients. As normal healthy controls, a total of 1438 adult healthy blood donors (706 males and 732 females; ages varied from 18 to 65, with the mean age of 40.78-year-old) from the same geographical region were recruited. All human research protocols were approved by the ethics committee of Chang Gung Memorial Hospital, and all subjects gave their informed consent.
2.2. HLA-B27 Determination
Flow cytometry and/or PCR tests were used to determine HLA-B27 positive. Whole blood samples were analyzed by flow cytometry for HLA-B27 using the BDTM HLA-B27 kit (BD Biosciences, Franklin Lakes, NJ, USA), which combines fluorescein-conjugated anti-HLA-B27 and phycoerythrin-conjugated anti-CD3 antibodies.
2.3. HLA-B Allele Determination
On ABI 3730XL DNA Analyzers, HLA-B alleles were determined using a commercial sequencing-based typing (SBT) kit (TBG Biotechnology Corp., New Taipei City, Taiwan). The AccuType HLA SBT Analysis Software assigned HLA-B alleles (TBG Biotechnology Corp, New Taipei City, Taiwan).
2.4. Genomic DNA Isolation
As previously disclosed [16], genomic DNA was recovered from EDTA anticoagulated peripheral blood using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN, USA).
2.5. RNA Isolation and cDNA Synthesis
TRIzol total RNA isolation reagent was used to isolate total RNA from human peripheral blood leukocytes (Invitrogen, Carlsbad, CA, USA). For deep sequencing investigation of ERAP1 coding SNVs and creation of ERAP1 expression constructs, the SuperScript Preamplification system (Invitrogen, Carlsbad, CA, USA) was utilized to synthesize peripheral blood leukocyte cDNA.
2.6. Deep DNA Sequencing Analyses of ERAP1 SNVs in Taiwanese
To identify Taiwanese-specific ERAP1 SNVs for the population genetic study, we carried out RT-PCR to amplify the full-length ERAP1 cDNA (2949 bps) using the sense primer (5′-CCC AGA ACC CCA GGT AGG TA-3′) and the antisense primer (5′-GCT ATG CTT CCA TTC CGT TT-3′). The ERAP1 cDNA sequence reactions were performed using the BigDye Terminator Sequencing kit (Applied Biosystem, Foster City, CA, USA) and the following seven sequencing primers: (1) 5′-AAG TCT CCG AAA GAT TGC CAG CAT-3′, (2) 5′-GGT TTC TGT TTA TGC TGT GCC AGA C-3′, (3) 5′-GAT CTT GTT TGG GTA GGG GAT ACG G-3′, (4) 5′-GCA AGA GCA CTA CAT GAA GGG CTC-3′, (5) 5′-TCA TGC CCA CAT TAA ATT TGA TCC A-3′, (6) 5′-AGG AAA AGC TTC AAT GGC TAC TAG A-3′, and (7) 5′-CAA TTG TCT GTT GGA CAC AAC GGA-3′. Sequencing tracer data were analyzed using SeqMan Pro software (DNAstar Inc., Madison, WI, USA) to identify and verify ERAP1 SNVs.
2.7. ERPA1 SNV Genotype Analyses
The common ERPA1 SNVs identified in Taiwanese population were genotyped using made-to-order allelic discrimination assays from Applied Biosystems (ABI, Foster City, CA, USA). SNV genotyping assays were carried out on a vi7 real-time PCR system (ABI). The SNV genotypes were determined using ABI TaqMan Genotyper software (Foster City, CA, USA) according to the vendor’s instructions.
2.8. ERAP1 Expression Constructs
The Xho I/Xba I-flanked Lir-EGFP DNA fragment from pBacCMV-MCS-Lir-EGFP was amplified by PCR using the forward primer 5′-TTCTCGAGTTAAAACAGCCTGTGGGT-3′ (the Xho I site is underlined) and the reverse primer 5′-GGCTCTAGATTACTTGTACAGCTCGTCCA-3′ (the Xba I site is underlined) and the fragment was subsequently cloned into pcDNA3.1 vector (Invitrogen) at Xho I and Xba I restriction enzyme sites for the generation of pcDNA3.1-Lir-EGFP vector. The full-length ERAP1 cDNAs from ERAP1-001 homozygous donors and ERAP1-002 homozygous donors were amplified using the forward primer 5′-ACGGGAATTCCCACCATGGTGTTTCTGCCCCTCAA-3′ (the EcoR I site is underlined) and the reverse primer 5′-AACGGGCTCGAGTCATCCTGTTGCGTCAGCTTCA-3′ (the Xho I site is underlined). The ERAP1 cDNA inserts were cloned into pcDNA3.1-Lir-EGFP vector digested with EcoR I and Xho I restriction enzymes.
2.9. Peripheral Blood Mononuclear Cell (PBMC) Isolation and Culture
Heparin collection tubes were used to collect whole blood from donors. Centrifugation through FicollPaque Premium (GE Healthcare, Chicago, IL, USA) density gradients was used to isolate PBMCs. PBMCs were cultured for 48 h in RPMI-1640 with 10% FBS and 10 ng/mL phorbol 12-myristate 13-acetate (PMA) before being stimulated for 16 h with LPS (1 µg/mL). The supernatants of PBMC cultures were then collected for IL-23 analysis, and cells were extracted for Western blot analysis.
2.10. Cell Culture
CHO (Chinese hamster ovary cells) cells were grown in DMEM with 10% fetal bovine serum (Sigma, St Louis, MO, USA). U937 (human monocytic leukemia) cells were grown in RPMI 1640 medium, which included 2 mM L-glutamine, 4.5 g/L glucose, 10 mM HEPES, 1 mM sodium pyruvate, 100 U/mL penicillin/streptomycin, and 10% fetal bovine serum. All cells were kept at 37 °C in a 5% CO2 environment.
2.11. Transfection of CHO and U937 Cells
TransIT transfection reagent (Mirus, Madison, WI, USA) was used for transfection of ERAP1 expression constructs into cells. On 6-well plates, 2 × 106 cells were seeded in each well. Before transfection, the cells were washed twice with medium devoid of serum. ERAP1 expression construct plasmid DNA (2.5 g) was diluted in 400 µL serum-free DMEM medium and then combined with TransIT transfection reagent (5 µL) to generate DNA-TransIT complexes. The DNA-TransIT complexes were added to cells in 400 µL of serum-free media per well. After 5 h of incubation, the medium was discarded and replaced with 2 mL of new medium containing 10% fetal bovine serum. After 24 h of transfection, the efficacy of transfection was evaluated by fluorescence microscopy.
2.12. Western Blot Analysis
On a micro Protein III system, cell lysates were subjected to SDS–PAGE for protein separation (Bio-Rad, Hercules, CA, USA). Proteins on the gel were electrotransferred onto a polyvinylidene difluoride (PVDF) membrane following SDS–PAGE. Tris-buffered saline (TTBS; 100 mmol/L Tris (pH 7.4), 100 mmol/L NaCl, and 0.1 percent Tween 20) containing 5% (v/v) non-fat milk was used to block PVDF membrane at room temperature for 1 h with moderate shaking. The membrane was then shaken overnight at 4 °C with primary antibody diluted in TBS containing 0.5 percent (v/v) nonfat milk. The membrane was washed three times for ten minutes at room temperature with TBS solution to remove unbound antibodies. The membrane was then treated at room temperature for one hour with secondary antibodies coupled with horseradish peroxidase (HRP). Enhanced Chemiluminescence kit (Pierce, Rockford, IL, USA) and the imaging equipment were used to detect the HRP-labeled proteins on membrane (UVP ChemStudio PLUS Touch, Analytik Jena US LLC, Cambridge, UK). Band intensities were quantified from the digital image by densitometry using ImageJ software (NIH, Bethesda, MD, USA).
2.13. ERAP1 Protein Immunoprecipitation
Cell pellets (25 × 106 cells for lymphoblastoid B cells or 2 × 106 cells for PBMCs) were lysed on ice for 30 min in lysis buffer (M-PER™ Mammalian Protein Extraction Reagent, Thermo Scientific, Waltham, MA, USA). The lysate supernatants were extracted by centrifuging at 14,000 rpm for 10 min at 4 degrees Celsius. Using the BCA (bicinchoninic acid) technique, protein concentrations were determined prior to immunoprecipitation. The lysates were then precleared by incubation with glycine Sepharose beads prior to treatment with anti-ERAP1 4D2-mAb-conjugated Sepharose beads for at least 2 h. The beads were subsequently washed three times in buffer (50 mM Tris, 150 mM NaCl, pH 7.4) and resuspended in buffer (500 µL) prior to use in the activity experiment.
2.14. ERAP1 Enzyme Activity Determination
The effect of ERAP1 allelic variants on ERAP1 enzyme activity was analyzed using a continuous fluorigenic assay as described [17]. Enzymatic reactions were carried out at 37 °C. In all reactions, the substrate concentration exceeded the enzyme concentration by at least 100-fold. Typically, 10 to 1000 nM ERAP1 protein was combined with 1 to 100 µM peptide, and the fluorescence of the reaction was monitored. On a QuantaMaster 4 spectrofluorimeter (Photon Technology International, Birmingham, NJ, USA) and a Tecan SpectraFluor plate reader, fluorescence measurements were conducted. Excitation at 295 nm and emission at 350 nm were used to acquire fluorescence spectra.
2.15. Cytokine Determination
ELISA kits were obtained from BioLegend (San Diego, CA, USA). All assays were conducted according to the manufacturer’s instructions. Absorbencies were measured using MAXlineMicroplate Readers at 450 nm (Molecular devices, San Jose, CA, USA). The data are presented as the mean ± standard deviation of technical replicates.
2.16. Statistical Analysis
Linear regression analysis and Pearson’s coefficient of correlation were used to analyze the correlation between different parameters. The distributions of SNV alleles and genotypes in patients and controls were analyzed using single-locus analysis. These comparisons were made using three chi-square tests (the genotype case-control test, the allele case-control test, and the Cochran-Armitage trend test). Using the SAS/Genetics software package release 8.2 (SAS Institute, Cary, NC, USA), significant relationships of SNVs with characteristics (p < 0.05) were detected, and p-values, odds ratios (ORs), and 95 percent confidence intervals (CIs) were calculated. The reported probability of false-discovery rate (PFDR) was estimated using Q-VALUE software (http://genomics.princeton.edu/storeylab/qvalue accessed on 6 June 2022) to account for multiple simultaneous statistical comparisons. Haploview 4.2 (Broad Institute, Cambridge, MA, USA; http://www.broad.mit.edu/mpg/haploview accessed on 6 June 2022) was used to calculate linkage disequilibrium (LD) between marker loci and to create haplotype blocks. We employed illness status (case vs. control) and HLA-B27 positivity to test for haplotype-trait association for markers within the same haplotype block (or allelic variations). To study genetic correlations, researchers utilized stepwise logistic regression analysis to account for sex. For all analyses, a 5% level of significance was employed for p-values.
3. Results
3.1. Associations of ERAP1 SNVs with AS Susceptibility
By sequencing the full-length ERAP1 cDNAs from 45 Taiwanese subjects randomly selected for SNV discovery purpose, we identified nine common ERAP1 coding SNVs (cSNVs) including eight non-synonymous cSNVs that cause amino acid substitutions (Table S1). We subsequently carried out genotype analyses of nine ERAP1 cSNVs plus two ERAP1 intron SNVs that were previously reported to associate with AS susceptibility [18,19]. Four ERAP1 cSNVs (rs26653G > C [R127P], rs30187A > G [K528R], rs469783G > A [A637A], and rs27044C > G [Q730E]) and two intron SNVs (rs27980A > C and rs27037T > G) had significantly different genotype and allele distributions between 863 AS patients and 1438 healthy controls (Table S2). The cSNV alleles consisting of rs26653G or 127R (Trend test PFDR (TtPFDR)= 0.0024; additive model adjusted for sex PFDR (addsex PFDR) = 0.0689, OR 1.17 [95% CI 1.01–1.35]), rs30187A (528K) (TtPFDR < 0.00001; addsex PFDR = 0.034, OR 1.20 [95% CI 1.04–1.38]), rs469783G (silent SNP) (TtPFDR < 0.00001; addsex PFDR = 0.0029, OR 1.28 [95% CI 1.11–1.48]), and rs27044C (730Q) (TtPFDR < 0.00001; addsex PFDR = 0.01, OR 1.25 [95% CI 1.08–1.44]), intron SNV rs27980A (TtPFDR = 0.0097; addsex PFDR = 0.0446, OR 1.17 [95% CI 1.03–1.34]), and the intron rs27037T (TtPFDR < 0.00001; addsex PFDR = 0.0029, OR 1.30 [95% CI 1.12–1.50]) alleles were similarly substantially related with AS susceptibility (Table S2).
3.2. Association of ERAP1 SNVs with HLA-B27 Positivity among AS Patients
Because HLA-B27 plays a causal role in the development of AS, we investigated the association between ERAP1 SNVs and HLA-B27 status in AS patients. As shown in Table S3, the allele distributions of six ERAP1 SNVs (rs26653, rs26618, rs30187, rs469783, rs27044, and rs27037) differed significantly between HLA-B27 positive patients (HLA-B27+) and the HLA-B27 negative patients (HLA-B27−). Five cSNV alleles are rs26653G (TtPFDR = 0.0083; addsex PFDR = 0.0115, OR 1.73 [95% CI 1.19–2.52]), rs26618A (TtPFDR = 0.0044; addsex PFDR = 0.0021, OR 2.12 [95% CI 1.43–3.14]), rs30187A (TtPFDR = 0.0083; addsex PFDR = 0.0099, OR 1.74 [95% CI 1.21–2.51]), rs469783G (TtPFDR = 0.0099; addsex PFDR = 0.0126, OR 1.69 [95% CI 1.16–2.440]), and rs27044G (TtPFDR = 0.0077; addsex PFDR = 0.0094, OR 1.80 [95% CI 1.25–2.60]). The intron SNV rs27037T allele was also substantially related with HLA-B27 positive in AS patients (TtPFDR = 0.0369; addsex PFDR = 0.0270, OR 1.60 [95% CI 1.10–2.34]).
3.3. Association of ERAP1 Allelic Variants (SNV Haplotypes) with AS Susceptibility
Linkage disequilibrium (LD) block analysis reveals that the majority of 11 ERAP1 SNVs are in strong linkage disequilibrium (Figure S1). Taiwanese haplotype analysis found that eleven ERAP1 SNVs included six main allelic variations (haplotypes) (Table S1). The ERAP1-001 comprised of rs3734016G-rs26653G-rs26618A-rs2287987A-rs30187A-rs10050860G-rs469783G-rs10050860G-rs469783G-s17482078G-rs27044C-rs27980A-rs27037T is the most prevalent allelic variant in Taiwanese healthy population (Table 1). Five cSNV sites (rs26653G > C [R127P], rs26618A > G [I276M], rs30187A > G [K528R], rs469783G > A [A637A], and rs27044C > G [Q730E]) and two intron SNP sites (rs27980 and rs27037) distinguish ERAP1-001 from ERAP1-002 (Table S1). ERAP1-001 was significantly enriched in AS patients (frequency = 0.4685) compared to healthy controls (frequency = 0.3669) (Adjusted p = 1.39E-011, OR = 1.53 [95% CI 1.35–1.73]), whereas the frequencies of the other five ERAP1 variants did not differ significantly between AS patients and healthy controls (Table 1). According to our findings, the ERAP1-001 variant is a significant risk factor for AS in Taiwanese.

Table 1.
Association of ERAP1 allelic variants with AS susceptibility in Taiwanese.
3.4. ERAP1 Allelic Variants Interact with HLA-B27 to Affect AS Susceptibility
HLA-B27 is a well-established AS risk factor. Following this, we studied the association between ERAP1 SNV haplotypes and HLA-B27 status in AS patients stratified by HLA-B27 positivity. The allele frequencies of ERAP1-001, -002, and -004 differed considerably between HLA-B27+ AS patients and H HLA-B27− AS patients, as indicated in Table 2. The ERAP1-001 frequency was significantly greater (Adjusted p = 0.01472, OR = 1.61 [95% CI: 1.10–2.36]) in HLA-B27+ AS patients (frequency = 0.4777) than in HLA-B27− AS patients (frequency = 0.3680) (Table 2). Intriguingly, the frequency of the rare ERAP1-004, which differs from the ERAP1-001 by only two intron SNVs (Table S1), was significantly higher (Adjusted p = 0.0409, OR 2.90 [95% CI 1.04–8.04]) in HLA-B27+ AS patients (frequency = 0.0818) than in HLA-B27− AS patients (frequency = 0.0310) (Table 2). Notably, the ERAP1-002 was substantially higher (Adjusted p = 0.00064, OR 0.50 [95% CI 0.34–0.75]) in HLA-B27− AS patients (frequency = 0.3395) than in HLA-B27+ AS patients (frequency = 0.2089), showing that ERAP1-002 is a risk factor for AS in HLA-B27− persons. The ERAP1-001 allelic frequency tended to be higher in HLA-B27+ AS patients (frequency = 0.4768) than in HLA-B27+ healthy controls (frequency = 0.4021); however, the difference was not statistically significant, most likely due to the limited sample size of HLA-B27+ healthy controls (Table S4). In contrast, the ERAP1-002 frequency was significantly higher (Adjusted p = 0.008, OR 1.69 [95% CI 1.15–2.49]) in HLA-B27− AS patients (frequency = 0.3453) than in HLA-B27− healthy controls (frequency = 0.2365) (Table 3), indicating that ERAP1-002 is a major risk factor for AS in HLA-B27− individuals. ERAP1-001 and -004 are risk factors for AS in HLA-B27+ persons, but ERAP1-002 is a risk factor for AS in HLA-B27− individuals. Our findings support the hypothesis that ERAP1 variants interact with HLA-B to differentially influence Taiwanese susceptibility to AS.

Table 2.
Association of ERAP1 allelic variants with HLA-B27 positivity among Taiwanese AS patients stratified by HLA-B27 status.

Table 3.
Distributions of ERAP1 allelic variants in HLA-B27− AS patients (AS B27−) and HLA-B27− healthy controls (Normal B27−).
3.5. ERAP1 Variant Genotypes Affect Phenotypes
Taiwanese ERAP1 variations have a number of amino acid changes (Table S1). To examine the epistaxis effects if ERAP1 haplotypes influence phenotypes in HLA-B27 positive individuals, peripheral blood mononuclear cells (PBMCs) were extracted from 11 human participants (seven ERAP1-001 homozygous AS patients and four ERAP1-002 AS patients) having a single HLA-B2704 allele. As demonstrated in Figure 1, PBMCs from ERAP1-001 homozygous donors produced more ERAP1 protein, MHC class I free heavy chain (FHCs) and FHC dimers, unfolded protein response (UPR) markers (immunoglobulin heavy-chain binding protein (BiP), CCAAT-enhancer-binding protein homologous protein (CHOP) and X-box-binding protein 1 (XBP1), autophagy markers (Beclin-1, LC3 I and LC3 II), and inflammation markers (Caspase 1 and IL-1β) as compared to those from ERAP1-002 homozygous donors. Notably, PBMCs of seven ERAP1-001 homozygous donors produced significantly higher levels of IL-23 p19 and p40 than PBMCs from four ERAP1-002 homozygous donors.
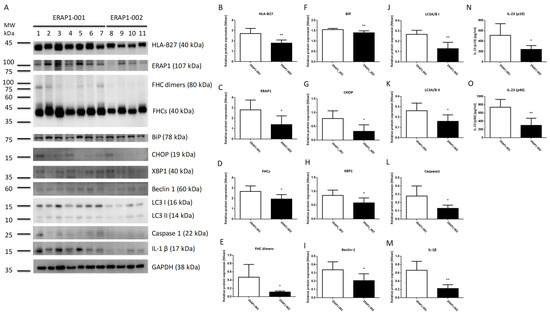
Figure 1.
PBMCs from ERAP1-001 and ERAP1-002 homozygous human subjects produced significantly different levels of ERAP1 and related proteins. Proteins in cell lysates of PBMCs were detected by Western blot analysis and a panel of proteins from seven ERAP1-001 and four ERAP1-002 homozygous donors were analyzed (A). Band intensities were quantified from the digital image by densitometry using Image and were normalized to the loading control. PBMCs of ERAP1-001 homozygous donors produced significantly higher levels of HLA-B27 (B), ERAP1 (C), MHC class I free heavy chain or FHCs (D), FHC dimers (E), immunoglobulin heavy-chain binding protein (BiP) (F), CCAAT-enhancer-binding protein homologous protein (CHOP) (G), X-box-binding protein 1 (XBP1) (H), Beclin-1 (I), LC3 I (J), LC3 II) (K), Caspase 1 (L), and IL-1β (M) as compared to ERAP1-002 homozygous donors. ELISA was used to detect IL-23 (p19/p40) in culture media. PBMCs from ERAP1-001 homozygous donors produced significantly higher level of IL-23 p19 (512.1 ± 84.94 versus 241.9 ± 36.40, p = 0.0478) (N) and p40 (740.4 ± 71.25 versus 301.7 ± 85.72, p = 0.0041) (O) than PBMCs of ERAP1-002 homozygous donors (data are shown as means with SD). Data are representative of three experiments. * p < 0.05, ** p < 0.01.
Previous studies indicates that non-conservative ERAP1 SNVs, such as K528R and R725Q substitutions, may impact ERAP1 protein production and aminopeptidase activity [14]. We conducted in vitro experiments to evaluate the effects of ERAP1 variations on ERAP1 expression and aminopeptidase activity. The variant cDNAs of ERAP1-001 and ERAP1-002 were cloned into the bi-cistronic mammalian expression vector pcDNA-Lir-EGFP. Fluorescence microscopy was used to measure the transfection effectiveness of EGFP in CHO cells (Figure 2A). Western blot analysis demonstrated that ERAP1 expression in transfected CHO cells was efficient (Figure 2B). Moreover, cells transfected with the ERAP1-001 variant produced a much greater amount of ERAP1 protein than those transfected with the ERAP1-002 variant (Figure 2C), corresponding with the results obtained from the PBMCs of homozygous ERAP1-001 and ERAP1-002 human individuals (Figure 1C). In addition, CHO cells expressing ERAP1-001 had considerably higher enzyme activity than ERAP1-002-expressing cells (p = 0.0105). (Figure 2D). Our research demonstrated that ERAP1-001 exhibited considerably more ERAP1 protein expression and enzyme activity than ERAP1-002.
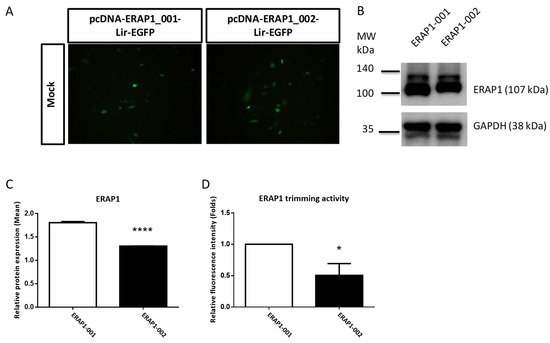
Figure 2.
ERAP1-001 exhibited considerably more ERAP1 protein expression and enzyme activity than ERAP1-002 in transfected cells. (A) Representative images of CHO cells transfected with pcDNA-ERAP1-Lir-EGFP expression constructs. All pictures were taken with a FITC channel (Green) with a 450/490-nm filter set with the same exposure time (900 ms). (B,C) Western blot analysis of cellular lysates from transfected U937 cells with pcDNA-ERAP1-EGFP. Band intensities were quantified and shown as normalized to the loading control. (D) ERAP1 enzyme activity analysis. Enzymatic reactions were performed via a continuous florigenic assay. Data are one representative of three experiments. Data are shown as mean (SD). * p < 0.05; **** p < 0.0001.
3.6. ERAP1 Allelic Variants Affect the Crosstalk of ER Stress (UPR), Autophagy, and Inflammation
Recent evidence suggests that the axis of ER stress, autophagy, and inflammasome plays a crucial role in the etiology of inflammatory illness. U937 cells were transfected with ERAP1 expression constructs in order to determine whether ERAP1 variations impact the interaction between ER stress, autophagy, and inflammation. U937 cells transfected with ERAP1-001 displayed significantly higher levels of HLA-B27, ERAP1 protein, FHCs, FHC dimers, UPR markers (BiP, CHOP and XBP1), and inflammatory markers (Caspase 1 and IL-1) than U937 cells transfected with ERAP1-002. Moreover, U937 cells transfected with ERAP1-001 produced larger quantities of IL-23 in culture supernatants than U937 cells transfected with ERAP1-002. On the other hand, autophagy markers including Beclin 1 (I), LC3 I (J), LC3 II (K), caspase 1 (L), and IL-1β (M) were not significantly different between ERAP1-001 and ERAP1-002 cells.
4. Discussion
As a non-MHC susceptibility gene, ERAP1 was found to interact with HLA-B27 to increase the risk of AS [4,14]. The pathophysiological mechanism behind the impact of ERAP1 allelic variations on AS susceptibility, on the other hand, is unknown. The common ERAP1-001 allelic variant was discovered to be a significant risk factor for AS in Taiwanese in the current investigation. According to our findings, epistasis between ERAP1 allelic variants and HLA-B plays a crucial role in the development of AS in Taiwanese. ERAP1-001 induces a higher level of ERAP1 protein expression, enzyme activity, and IL-23 production than ERAP1-002. Increased ERAP1 enzyme activity appears to be a pathophysiological mechanism for the development of AS in HLA-B27+ individuals; therefore, inhibition of ERAP1 enzyme activity may be a viable treatment option for HLA-B27+ AS patients. In HLA-B27− people, however, ERAP1-002 is a risk factor for AS. The importance of epistasis between ERAP1 variants and HLA-B27+/ HLA-B27− in the development of AS in Taiwanese is demonstrated.
ERAP1 is a multifunctional zinc-metallopeptidase that trims antigen peptides to the proper length for MHC-I molecules to display them. Antigenic epitopes are both produced and destroyed by ERAP1. The length of the peptidome is determined by interactions between the N-terminus of the antigen peptide and the enzyme active site of ERAP1, as well as the C-terminus of the peptide and an ERAP1 regulatory site. By changing the sequence and length of antigenic peptides deposited onto the corresponding MHC Class I molecules, ERAP1 affects a major portion of the immunopeptidome [20]. ERAP1 trims peptide precursors in solution to adjust the immunopeptidome’s optimal fit [21].
Peptidome studies of MHC-bound peptides in HLA-B27 positive cell lines expressing various ERAP1 variants reveal that ERAP1 and HLA-B27 have a functional relationship [22]. The structure, immunological effect, and peptide-trimming property of ERAP1 point to a mechanism or pathway for AS vulnerability that involves HLA-B27 [23,24]. ERAP1 directly affects peptide binding and presentation by HLA-B27, which could be a pathogenic mechanism for ERAP1’s participation in AS [25]. According to in vitro peptide catalysis experiments, ERAP1 risk alleles with higher catalytic activity degrade HLA-B27 epitopes more efficiently, resulting in lower HLA-B27 presentation of the identical peptides [26].
mRNA stability, protein translation, and enzymatic activity may all be affected by combined effects of ERAP1 genetic variations [27,28]. Between ERAP-001 (127R, 276I, 528K, and 730Q) and ERAP-002 (R127P, I276M, K528R, and Q730E), there are four residue differences. The molecular basis of ERAP1 SNVs on N-terminal peptidase function was discovered using crystal structure research [29]. The ERAP1 SNV rs30187A > G (K528R) is found near the substrate pocket’s entrance, which may affect the C-terminus stability and aminopeptidase activity [29]. The R127P and K528R alterations affect ERAP1 conformational transition, whereas the Q730E change in ERAP1’s peptide-binding pocket may affect the peptides processed [30]. ERAP1 with 528R has lower enzymatic activity than ERAP1 with 528K, while ERAP1 with Q730E influences the lengths of antigenic peptides produced [22,25,28,31,32,33]. Therefore, the enzyme activity of ERAP1-002 (127P, 276M, 528R, and 730E) will be reduced. Furthermore, lower surface FHC expression on monocytes and HLA-B27-expressing APCs in AS patients was linked to 528R and 730E. HLA-B27 FHC surface expression was reduced by ERAP1 silencing or inhibition in APCs, as was IL-2 production by KIR3DL2CD3-reporter cells, and Th17 growth and IL-17A secretion by CD4+ T cells from AS patients [34].
In both white and East Asian ancestry populations, ERAP1 has been genetically investigated in AS patients by sequencing [35] and intensive SNV mapping and imputation [4]. There were considerable differences in the presence and distribution of ERAP1 coding SNVs among ethnic populations (Figure S2) [22,35,36,37,38,39]. The 1000 Genomes Project dataset discovered ten missense ERAP1 SNVs, two of which are unusual in Taiwanese (residue positions 12 and 346) [38]. ERAP1 SNV haplotypes, on the other hand, were comparable between the Asian 1000 Genomes Project population and Taiwanese (hap2 = ERAP1-001 plus -004, hap8 = ERAP1-002, hap7 = ERAP1, and hap10 = ERAP1-005, respectively). According to ERAP1 SNV data from the 1000 Genome Project, Reeves et al. detected a number of European-specific ERAP1 SNVs using a cDNA cloning/sequencing technique [37], some of those SNVs may be produced by the cloning step. Based on five ERAP1 SNVs, Robert et al. discovered three ERAP1 haplotypes in the European population (349M/528K/575D/725R/730Q; 349M/528R/575D/725R/730E; 349V/528R/575N/725Q/730E). Those three ERAP1 haplotypes could be deduced both in Europeans and Taiwanese (European 349M/528K/575D/725R/730Q = Taiwanese ERAP1-001 + 004, European 349M/528R/575D/725R/730E = Taiwanese ERAP1-002 + 003 + 006, and European 349V/528R/575N/725Q/730E = Taiwanese ERAP1-005). ERAP1-001 (56E, 127R, 276I, 349M, 528K, 575D, 725R, and 730Q) and ERAP1-002 (56E, 127P, 276M, 349M, 528R, 575D, 725R, and 730E) differ at four residues (R127P, I276M, K528R, and Q730E) and two intron SNVs. Two intron SNVs are thought to alter ERAP1 functions [35,40]. Importantly, we examined 11 ERAP1 SNVs and identified two common allelic variants (haplotypes) in Taiwanese. To the best of our knowledge, for the first time in human subjects, we looked at the functional differences between the two most frequent ERAP1 variations in depth, which differed from previous functional investigations of ERAP1 variants [22,37,40,41].
Reduced ERAP1 mRNA expression in the presence of a coding variant has been linked to Behçet’s Disease (BD) [42]. With longer epitopes, the promiscuous sub-peptidome binding property of HLA-B51 indicates a genetic effect on dysregulated CD8+ T response and aberrant NK cell activation, leading to the development of BD [43]. Other HLA-B alleles have been involved in AS development in European populations [6], therefore ERAP1 epistasis may not be limited to HLA-B27. HLA-B40 increased susceptibility to AS in HLA-B27− AS patients in Taiwanese [44], but not in Caucasian [45]. As shown in Table S5, HLA-B40:01 frequencies (phenotype frequency = 37.1%, allele frequency = 18.55%) among 62 HLA-B27− AS patients were similar to those in general Taiwanese population, indicating that HLA-B40 may not play a unique role in AS. HLA-B27− Taiwanese AS patients have lower ERAP1 enzyme activity, suggesting that decreased ERAP1 enzyme activity may play a role in AS. Understanding the pathophysiology of AS in people will require more research into the link between HLA-B alleles and ERAP1 variations.
Misfolding of HLA-B27 HC promotes ER stress and activation of the UPR. BiP and XBP1 are two UPR indicators that are increased upon the activation of the three major UPR pathways. Compared to cells expressing the ERAP1-002 variant, cells expressing the ERAP1-001 variant produced larger quantities of ERAP1 protein, FHCs, FHC dimers, BiP, XBP1, and IL23. The increased synthesis of BiP and XBP1 in cells expressing ERAP1-001 suggests that the overexpression of MHC class I FHCs and FHC dimers results in the activation of UPR, the elevation of IL-23 levels, and proinflammatory response. The enhanced expression of FHCs and FHC dimers in cells bearing HLA-B27 may explain why the link between ERAP1-001 and AS susceptibility is exclusive to HLA-B27+ people.
Autophagy is a lysosome-dependent degradation mechanism that destroys misfolded proteins and damaged ER. UPR not only enhances cytokine-mediated inflammatory responses leading to disease, but also promotes autophagy. The control of autophagy by reducing ER stress could therefore provide cytoprotection [46]. PBMCs expressing the ERAP1-001 variant displayed significantly higher levels of autophagy markers (Beclin 1, LC3 I, and LC3 II), inflammation markers (caspase 1 and IL-1), and IL-23 (p19 and p40) compared to PBMCs expressing the ERAP1-002 variant (Figure 1). Increased productions of BiP, CHOP, and XBP1 in cells expressing ERAP1-001 suggest that ERAP1-001 may cause the misfolding of HLA-B complexes or HLA-B27 associated with abnormal peptides, resulting in the activation of the UPR [47] and subsequent induction of autophagy (Beclin 1, LC3 I and LC3 II), inflammation (caspase 1 and IL-1β) [48]. Nonetheless, U937 cells transfected with ERAP1-001 displayed comparable levels of autophagy markers (Beclin 1, LC3 I and LC3 II) compared to those with ERAP1-002 (Figure 3). In this regard, we speculated that endogenous ERAP1 may mask the effect of ERAP1-001 and ERAP1-002 variants on autophagy in U937 cells.
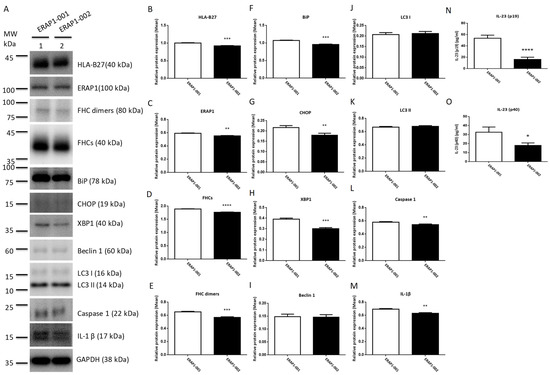
Figure 3.
Effects of ERAP1-001 and ERAP1-002 variants on misfolding proteins, UPR markers, autophagy markers, inflammation markers, and IL-23 in transfected U937 cells. Proteins in cell lysates of transfected U937 cells were detected by Western blot analysis and a panel of proteins from were analyzed (A). GAPDH was used as the loading control where indicated. Band intensities were quantified and normalized to the loading control. Protein levels of HLA-B27 (B), ERAP1 (C), FHCs (D), FHC dimers (E), BiP (F), CHOP (G), XBP1 (H), caspase 1 (L), and IL-1β (M) were significantly higher in ERAP1-001 U937 cells as compared to ERAP1-002 cells. Protein levels of Beclin 1 (I), LC3 I (J), LC3 II (K), caspase 1 (L), and IL-1β (M) were not significantly different between ERAP1-001 and ERAP1-002 cells. Culture media were collected for ELISA to detect IL-23 -p19/p40. U937 cells transfected with ERAP1-001 produced larger quantities of IL-23-p19 subunit (p < 0.0001, 53.78 ± 2.696 versus 16.28 ± 1.964) (N) and IL-23-p40 subunit (p = 0.0158, 32.74 ± 3.288 versus 18.10 ± 1.563) (O) than U937 cells transfected with ERAP1-002 (mean ± SD are shown). Combined data were from three experiments. * p < 0.05; ** p < 0.01, *** p < 0.001, and **** p ≤ 0.0001.
Although the exact mechanism underlying IL-23 overexpression remains unknown, IL-23 was detectable in the peripheral blood and tissues of AS patients. In particular, it is unknown if the overproduction of IL-23 could be linked to the misfolding of HLA-B27 by inducing the UPR [49]. On the other hand, the proof that IL-23 is active at the surface of the intestinal mucosa [50] and the evidence that certain types of bacterial stimuli may alter its intestinal expression [51] suggest the role of microbes in the IL-23 overexpression observed in AS [52]. Multiple roles could be carried out by ERAP1 in the extracellular area [53]. ERAP1 produced into the circulation in response to viral or inflammatory stimuli that boost NO production may function as an acute-phase host defense protein [54]. Exosomes contain ERAP1, TNF-α, IFN-γ, and CCL3, which play a crucial role in the inflammatory process by activating macrophages [55]. The immunomodulation cytokine profiles of the IL-17/IL-23 pathway are affected by the ERAP1 SNPs rs30187, rs2287987 [56], and rs27038 [57]. Emerging immunological and genetic evidence validated the central function of the IL-17A/IL-23 axis, resulting in the continued development of new medications targeting distinct components, such as medicines blocking the p40 subunit of IL-12/IL-23 or the p19 subunit of IL-23 for therapeutic purposes. Our results indicate that the genetic relationship between HLA-B27 and ERAP1 is a critical predictor of IL-23 pathogenesis in AS.
5. Conclusions
The present investigation definitively determined a functional role of ERAP1 allelic variants In AS. Ex vivo and in vitro research demonstrated that ERAP1 allelic variations have a substantial effect on ERAP1 expression and enzyme activity, FHCs, FHC dimers, and UPR markers (BiP, CHOP and XBP1). Two common ERAP1-001 and ERAP1-002 variants are associated with susceptibility to AS in Taiwanese in an HLA-B27-dependent manner.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells11152427/s1, Figure S1: Pair-wise LD patterns with D’ (left side) and r2 (right side) measures of eleven ERAP1 SNPs for healthy controls (A) and AS cases plus normal (B). Figure S2: ERAP1 allelic variants detected among different populations. Figure S3: The original Western blots of Figure 1A, Figure 2B and Figure 3A. Table S1: ERAP1 SNVs and allelic variants (SNV haplotypes) in Taiwanese healthy controls and AS patients. Table S2: Association of ERAP1 SNVs with AS susceptibility in Taiwanese. Table S3: Association of ERAP1 SNVs with HLA-B27 positivity among AS patients. Table S4: Distributions of ERAP1 allelic variants in HLA-B27+ AS patients (AS B27+) and HLA-B27+ healthy controls (Control B27+). Table S5: Distributions of HLA-B alleles in 62 HLA-B27− AS patients.
Author Contributions
Conceptualization, resources, funding acquisition: C.-M.W., J.W. and J.-Y.C.; methodology, data curation, investigation: M.-K.L.; investigation Y.-J.J.W., J.-C.L. and J.-W.Z.; original draft preparation C.-M.W., J.-Y.C. and J.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by funding from the Chang Gung Memorial Hospital (Grant numbers: CMRPG5K0131, CMRPG 5H0022, CMRPG 3J1422 and CMRPG 5I0061) and the Ministry of Science and Technology, Taiwan (Grant numbers: MOST 109-2314-B-182-068-MY3 and 107-2314-B-182-059-MY3).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of Chang Gung Memorial Hospital (protocol code 201902162B0C101).
Informed Consent Statement
Written informed consent has been obtained from the patients to publish this paper.
Data Availability Statement
Not applicable.
Acknowledgments
We greatly appreciate the Shin Chu Blood Donor Center for the collection of samples.
Conflicts of Interest
The study was carried out in accordance with the relevant guidelines and regulations. The authors declare no conflict of interests and any potential financial conflicts of interest that any of the authors may have.
Abbreviations
AS ankylosing spondylitis; HLA-B27 human leukocyte antigen B27; FHCs MHC class I free heavy chain; UPR unfolded protein response; BiP immunoglobulin heavy-chain binding protein; CHOP CCAAT-enhancer-binding protein homologous protein and XBP1 X-box-binding protein 1.
References
- Tan, S.; Wang, R.; Ward, M.M. Syndesmophyte growth in ankylosing spondylitis. Curr. Opin. Rheumatol. 2015, 27, 326–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morin, M.; Hellgren, K.; Frisell, T. Familial aggregation and heritability of ankylosing spondylitis—A Swedish nested case-control study. Rheumatology 2020, 59, 1695–1702. [Google Scholar] [CrossRef] [PubMed]
- Costantino, F.; Breban, M.; Garchon, H.J. Genetics and Functional Genomics of Spondyloarthritis. Front. Immunol. 2018, 9, 2933. [Google Scholar] [CrossRef] [PubMed]
- Cortes, A.; Hadler, J.; Pointon, J.P.; Robinson, P.C.; Karaderi, T.; Leo, P.; Cremin, K.; Pryce, K.; Harris, J.; Lee, S.; et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 2013, 45, 730–738. [Google Scholar]
- Burton, P.R.; Clayton, D.G.; Cardon, L.R.; Craddock, N.; Deloukas, P.; Duncanson, A.; Kwiatkowski, D.P.; McCarthy, M.I.; Ouwehand, W.H.; Samani, N.J.; et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 2007, 39, 1329–1337. [Google Scholar]
- Cortes, A.; Pulit, S.L.; Leo, P.J.; Pointon, J.J.; Robinson, P.C.; Weisman, M.H.; Ward, M.; Gensler, L.S.; Zhou, X.; Garchon, H.J.; et al. Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nat. Commun. 2015, 6, 7146. [Google Scholar] [CrossRef] [Green Version]
- Reveille, J.D.; Sims, A.M.; Danoy, P.; Evans, D.M.; Leo, P.; Pointon, J.J.; Jin, R.; Zhou, X.; Bradbury, L.A.; Appleton, L.H.; et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat. Genet. 2010, 42, 123–127. [Google Scholar]
- Ellinghaus, D.; Jostins, L.; Spain, S.L.; Cortes, A.; Bethune, J.; Han, B.; Park, Y.R.; Raychaudhuri, S.; Pouget, J.G.; Hübenthal, M.; et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 2016, 48, 510–518. [Google Scholar] [CrossRef]
- Serwold, T.; Gonzalez, F.; Kim, J.; Jacob, R.; Shastri, N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature 2002, 419, 480–483. [Google Scholar] [CrossRef]
- Brouwenstijn, N.; Serwold, T.; Shastri, N. MHC class I molecules can direct proteolytic cleavage of antigenic precursors in the endoplasmic reticulum. Immunity 2001, 15, 95–104. [Google Scholar] [CrossRef] [Green Version]
- Serwold, T.; Gaw, S.; Shastri, N. ER aminopeptidases generate a unique pool of peptides for MHC class I molecules. Nat. Immunol. 2001, 2, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Saric, T.; Chang, S.C.; Hattori, A.; York, I.A.; Markant, S.; Rock, K.L.; Tsujimoto, M.; Goldberg, A.L. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat. Immunol. 2002, 3, 1169–1176. [Google Scholar] [CrossRef] [PubMed]
- Hammer, G.E.; Gonzalez, F.; Champsaur, M.; Cado, D.; Shastri, N. The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat. Immunol. 2006, 7, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.M.; Spencer, C.C.; Pointon, J.J.; Su, Z.; Harvey, D.; Kochan, G.; Oppermann, U.; Dilthey, A.; Pirinen, M.; Stone, M.A.; et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 2011, 43, 761–767. [Google Scholar] [CrossRef] [Green Version]
- Reeves, E.; Elliott, T.; James, E.; Edwards, C.J. ERAP1 in the pathogenesis of ankylosing spondylitis. Immunol. Res. 2014, 60, 257–269. [Google Scholar] [CrossRef]
- Chen, J.Y.; Wang, C.M.; Ma, C.C.; Luo, S.F.; Edberg, J.C.; Kimberly, R.P.; Wu, J. Association of a transmembrane polymorphism of Fcgamma receptor IIb (FCGR2B) with systemic lupus erythematosus in Taiwanese patients. Arthritis Rheum. 2006, 54, 3908–3917. [Google Scholar] [CrossRef]
- Evnouchidou, I.; Berardi, M.J.; Stratikos, E. A continuous fluorigenic assay for the measurement of the activity of endoplasmic reticulum aminopeptidase 1 competition kinetics as a tool for enzyme specificity investigation. Anal. Biochem. 2009, 395, 33–40. [Google Scholar] [CrossRef]
- Chen, R.; Yao, L.; Meng, T.; Xu, W. The association between seven ERAP1 polymorphisms and ankylosing spondylitis susceptibility. a meta-analysis involving 8,530 cases and 12,449 controls. Rheumatol. Int. 2012, 32, 909–914. [Google Scholar] [CrossRef]
- Wang, C.M.; Ho, H.H.; Chang, S.W.; Wu, Y.J.; Lin, J.C.; Chang, P.Y.; Wu, J.; Chen, J.Y. ERAP1 genetic variations associated with HLA-B27 interaction and disease severity of syndesmophytes formation in Taiwanese ankylosing spondylitis. Arthritis Res. Ther. 2012, 14, R125. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Batliwala, M.; Bouvier, M. ERAP1 enzyme-mediated trimming and structural analyses of MHC I--bound precursor peptides yield novel insights into antigen processing and presentation. J. Biol. Chem. 2019, 294, 18534–18544. [Google Scholar] [CrossRef] [Green Version]
- Mavridis, G.; Arya, R.; Domnick, A.; Zoidakis, J.; Makridakis, M.; Vlahou, A.; Mpakali, A.; Lelis, A.; Georgiadis, D.; Tampé, R.; et al. A systematic re-examination of processing of MHCI-bound antigenic peptide precursors by endoplasmic reticulum aminopeptidase 1. J. Biol. Chem. 2020, 295, 7193–7210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Medel, N.; Sanz-Bravo, A.; Van Nguyen, D.; Galocha, B.; Gomez-Molina, P.; Martin-Esteban, A.; Alvarez-Navarro, C.; de Castro, J.A. Functional interaction of the ankylosing spondylitis-associated endoplasmic reticulum aminopeptidase 1 polymorphism and HLA-B27 in vivo. Mol. Cell. Proteom. MCP 2012, 11, 1416–1429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin-Esteban, A.; Gomez-Molina, P.; Sanz-Bravo, A.; Lopez de Castro, J.A. Combined effects of ankylosing spondylitis-associated ERAP1 polymorphisms outside the catalytic and peptide-binding sites on the processing of natural HLA-B27 ligands. J. Biol. Chem. 2014, 289, 3978–3990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez-Navarro, C.; Lopez de Castro, J.A. ERAP1 in ankylosing spondylitis: Genetics, biology and pathogenetic role. Curr. Opin. Rheumatol. 2013, 25, 419–425. [Google Scholar] [CrossRef]
- Chen, L.; Fischer, R.; Peng, Y.; Reeves, E.; McHugh, K.; Ternette, N.; Hanke, T.; Dong, T.; Elliott, T.; Shastri, N.; et al. Critical role of endoplasmic reticulum aminopeptidase 1 in determining the length and sequence of peptides bound and presented by HLA-B27. Arthritis Rheumatol. 2014, 66, 284–294. [Google Scholar] [CrossRef]
- Seregin, S.S.; Rastall, D.P.; Evnouchidou, I.; Aylsworth, C.F.; Quiroga, D.; Kamal, R.P.; Godbehere-Roosa, S.; Blum, C.F.; York, I.A.; Stratikos, E.; et al. Endoplasmic reticulum aminopeptidase-1 alleles associated with increased risk of ankylosing spondylitis reduce HLA-B27 mediated presentation of multiple antigens. Autoimmunity 2013, 46, 497–508. [Google Scholar] [CrossRef] [Green Version]
- Costantino, F.; Talpin, A.; Evnouchidou, I.; Kadi, A.; Leboime, A.; Said-Nahal, R.; Bonilla, N.; Letourneur, F.; Leturcq, T.; Ka, Z.; et al. ERAP1 Gene Expression Is Influenced by Nonsynonymous Polymorphisms Associated With Predisposition to Spondyloarthritis. Arthritis Rheumatol. 2015, 67, 1525–1534. [Google Scholar] [CrossRef]
- Sanz-Bravo, A.; Alvarez-Navarro, C.; Martín-Esteban, A.; Barnea, E.; Admon, A.; López de Castro, J.A. Ranking the Contribution of Ankylosing Spondylitis-associated Endoplasmic Reticulum Aminopeptidase 1 (ERAP1) Polymorphisms to Shaping the HLA-B*27 Peptidome. Mol. Cell. Proteom. MCP 2018, 17, 1308–1323. [Google Scholar] [CrossRef] [Green Version]
- Kochan, G.; Krojer, T.; Harvey, D.; Fischer, R.; Chen, L.; Vollmar, M.; von Delft, F.; Kavanagh, K.L.; Brown, M.A.; Bowness, P.; et al. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc. Natl. Acad. Sci. USA 2011, 108, 7745–7750. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Navarro, C.; Lopez de Castro, J.A. ERAP1 structure, function and pathogenetic role in ankylosing spondylitis and other MHC-associated diseases. Mol. Immunol. 2014, 57, 12–21. [Google Scholar] [CrossRef]
- Sanz-Bravo, A.; Campos, J.; Mazariegos, M.S.; Lopez de Castro, J.A. Dominant role of the ERAP1 polymorphism R528K in shaping the HLA-B27 Peptidome through differential processing determined by multiple peptide residues. Arthritis Rheumatol. 2015, 67, 692–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamogiannos, A.; Koumantou, D.; Papakyriakou, A.; Stratikos, E. Effects of polymorphic variation on the mechanism of Endoplasmic Reticulum Aminopeptidase 1. Mol. Immunol. 2015, 67, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Giastas, P.; Mpakali, A.; Papakyriakou, A.; Lelis, A.; Kokkala, P.; Neu, M.; Rowland, P.; Liddle, J.; Georgiadis, D.; Stratikos, E. Mechanism for antigenic peptide selection by endoplasmic reticulum aminopeptidase 1. Proc. Natl. Acad. Sci. USA 2019, 116, 26709–26716. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ridley, A.; Hammitzsch, A.; Al-Mossawi, M.H.; Bunting, H.; Georgiadis, D.; Chan, A.; Kollnberger, S.; Bowness, P. Silencing or inhibition of endoplasmic reticulum aminopeptidase 1 (ERAP1) suppresses free heavy chain expression and Th17 responses in ankylosing spondylitis. Ann. Rheum. Dis. 2016, 75, 916–923. [Google Scholar] [CrossRef] [Green Version]
- Harvey, D.; Pointon, J.J.; Evans, D.M.; Karaderi, T.; Farrar, C.; Appleton, L.H.; Sturrock, R.D.; Stone, M.A.; Oppermann, U.; Brown, M.A.; et al. Investigating the genetic association between ERAP1 and ankylosing spondylitis. Hum. Mol. Genet. 2009, 18, 4204–4212. [Google Scholar] [CrossRef] [Green Version]
- Cinar, M.; Akar, H.; Yilmaz, S.; Simsek, I.; Karkucak, M.; Sagkan, R.I.; Pekel, A.; Erdem, H.; Avci, I.Y.; Acikel, C.; et al. A polymorphism in ERAP1 is associated with susceptibility to ankylosing spondylitis in a Turkish population. Rheumatol. Int. 2013, 33, 2851–2858. [Google Scholar] [CrossRef]
- Reeves, E.; Colebatch-Bourn, A.; Elliott, T.; Edwards, C.J.; James, E. Functionally distinct ERAP1 allotype combinations distinguish individuals with Ankylosing Spondylitis. Proc. Natl. Acad. Sci. USA 2014, 111, 17594–17599. [Google Scholar] [CrossRef] [Green Version]
- Ombrello, M.J.; Kastner, D.L.; Remmers, E.F. Endoplasmic reticulum-associated amino-peptidase 1 and rheumatic disease, genetics. Curr. Opin. Rheumatol. 2015, 27, 349–356. [Google Scholar] [CrossRef]
- Roberts, A.R.; Appleton, L.H.; Cortes, A.; Vecellio, M.; Lau, J.; Watts, L.; Brown, M.A.; Wordsworth, P. ERAP1 association with ankylosing spondylitis is attributable to common genotypes rather than rare haplotype combinations. Proc. Natl. Acad. Sci. USA 2017, 114, 558–561. [Google Scholar] [CrossRef] [Green Version]
- Hanson, A.L.; Cuddihy, T.; Haynes, K.; Loo, D.; Morton, C.J.; Oppermann, U.; Leo, P.; Thomas, G.P.; Le Cao, K.A.; Kenna, T.J.; et al. Genetic Variants in ERAP1 and ERAP2 Associated With Immune-Mediated Diseases Influence Protein Expression and the Isoform Profile. Arthritis Rheumatol. 2018, 70, 255–265. [Google Scholar] [CrossRef] [Green Version]
- Haroon, N.; Tsui, F.W.; Uchanska-Ziegler, B.; Ziegler, A.; Inman, R.D. Endoplasmic reticulum aminopeptidase 1 (ERAP1) exhibits functionally significant interaction with HLA-B27 and relates to subtype specificity in ankylosing spondylitis. Ann. Rheum. Dis. 2012, 71, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Padula, M.C.; Leccese, P.; Lascaro, N.; Carbone, T.; Limongi, A.R.; Radice, R.P.; Padula, A.A.; D’Angelo, S.; Martelli, G. From structure to function for the characterization of ERAP1 active site in Behçet syndrome. A novel polymorphism associated with known gene variations. Mol. Immunol. 2020, 117, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shi, H.; Koftori, D.; Sekine, T.; Nicastri, A.; Ternette, N.; Bowness, P. Identification of an Unconventional Subpeptidome Bound to the Behçet’s Disease-associated HLA-B*51:01 that is Regulated by Endoplasmic Reticulum Aminopeptidase 1 (ERAP1). Mol. Cell. Proteom. MCP 2020, 19, 871–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, J.C.; Sung-Ching, H.W.; Hsu, Y.W.; Wen, Y.F.; Wang, W.C.; Wong, R.H.; Lu, H.F.; Gaalen, F.A.; Chang, W.C. Interaction between HLA-B60 and HLA-B27 as a Better Predictor of Ankylosing Spondylitis in a Taiwanese Population. PLoS ONE 2015, 10, e0137189. [Google Scholar]
- Robinson, W.P.; van der Linden, S.M.; Khan, M.A.; Rentsch, H.U.; Cats, A.; Russell, A.; Thomson, G. HLA-Bw60 increases susceptibility to ankylosing spondylitis in HLA-B27+ patients. Arthritis Rheum. 1989, 32, 1135–1141. [Google Scholar] [CrossRef]
- Chipurupalli, S.; Samavedam, U.; Robinson, N. Crosstalk between ER Stress, Autophagy and Inflammation. Front. Med. 2021, 8, 758311. [Google Scholar] [CrossRef]
- Brown, M.A.; Kenna, T.; Wordsworth, B.P. Genetics of ankylosing spondylitis—Insights into pathogenesis. Nat. Rev. Rheumatol. 2016, 12, 81–91. [Google Scholar] [CrossRef]
- Rahmati, M.; Moosavi, M.A.; McDermott, M.F. ER Stress: A Therapeutic Target in Rheumatoid Arthritis? Trends Pharmacol. Sci. 2018, 39, 610–623. [Google Scholar] [CrossRef]
- DeLay, M.L.; Turner, M.J.; Klenk, E.I.; Smith, J.A.; Sowders, D.P.; Colbert, R.A. HLA-B27 misfolding and the unfolded protein response augment interleukin-23 production and are associated with Th17 activation in transgenic rats. Arthritis Rheum. 2009, 60, 2633–2643. [Google Scholar] [CrossRef]
- Becker, C.; Wirtz, S.; Blessing, M.; Pirhonen, J.; Strand, D.; Bechthold, O.; Frick, J.; Galle, P.R.; Autenrieth, I.; Neurath, M.F. Constitutive p40 promoter activation and IL-23 production in the terminal ileum mediated by dendritic cells. J. Clin. Investig. 2003, 112, 693–706. [Google Scholar] [CrossRef] [Green Version]
- Kinnebrew, M.A.; Buffie, C.G.; Diehl, G.E.; Zenewicz, L.A.; Leiner, I.; Hohl, T.M.; Flavell, R.A.; Littman, D.R.; Pamer, E.G. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity 2012, 36, 276–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodall, J.C.; Wu, C.; Zhang, Y.; McNeill, L.; Ellis, L.; Saudek, V.; Gaston, J.S. Endoplasmic reticulum stress-induced transcription factor, CHOP, is crucial for dendritic cell IL-23 expression. Proc. Natl. Acad. Sci. USA 2010, 107, 17698–17703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goto, Y.; Ogawa, K.; Hattori, A.; Tsujimoto, M. Secretion of endoplasmic reticulum aminopeptidase 1 is involved in the activation of macrophages induced by lipopolysaccharide and interferon-gamma. J. Biol. Chem. 2011, 286, 21906–21914. [Google Scholar] [CrossRef] [Green Version]
- Goto, Y.; Nakamura, T.J.; Ogawa, K.; Hattori, A.; Tsujimoto, M. Acute-phase protein-like properties of endoplasmic reticulum aminopeptidase 1. J. Biochem. 2019, 165, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Ogawa, Y.; Tsumoto, H.; Miura, Y.; Nakamura, T.J.; Ogawa, K.; Akimoto, Y.; Kawakami, H.; Endo, T.; Yanoshita, R.; et al. Contribution of the exosome-associated form of secreted endoplasmic reticulum aminopeptidase 1 to exosome-mediated macrophage activation. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 874–888. [Google Scholar] [CrossRef] [PubMed]
- Babaie, F.; Mohammadi, H.; Hemmatzadeh, M.; Ebrazeh, M.; Torkamandi, S.; Yousefi, M.; Hajaliloo, M.; Rezaiemanesh, A.; Salimi, S.; Salimi, R.; et al. Evaluation of ERAP1 gene single nucleotide polymorphisms in immunomodulation of pro-inflammatory and anti-inflammatory cytokines profile in ankylosing spondylitis. Immunol. Lett. 2020, 217, 31–38. [Google Scholar] [CrossRef]
- Hemmatzadeh, M.; Babaie, F.; Ezzatifar, F.; Mohammadi, F.S.; Ebrazeh, M.; Golabi Aghdam, S.; Hajaliloo, M.; Azizi, G.; Gowhari Shabgah, A.; Shekari, N.; et al. Susceptibility to ERAP1 gene single nucleotide polymorphism modulates the inflammatory cytokine setting in ankylosing spondylitis. Int. J. Rheum. Dis. 2019, 22, 715–724. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).