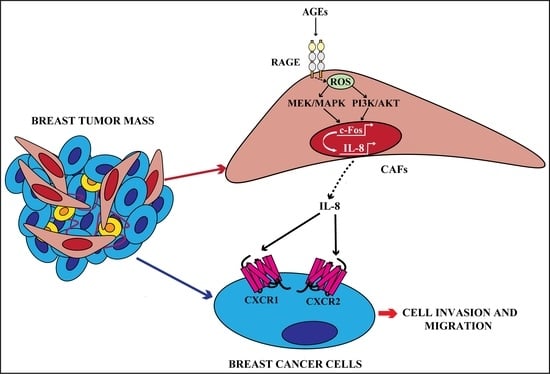
The AGEs/RAGE Transduction Signaling Prompts IL-8/CXCR1/2-Mediated Interaction between Cancer-Associated Fibroblasts (CAFs) and Breast Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analyses
2.2. Reagents
2.3. Cell Cultures
2.4. Gene Expression Studies
2.5. Luciferase Assays and Gene Silencing Experiments
2.6. Western Blot Analysis
2.7. DCFDA Fluorescence Measurement of ROS
2.8. Immunofluorescence Microscopy
2.9. Chromatin Immunoprecipitation (ChIP) Assay
2.10. Conditioned Medium
2.11. Acetone Precipitation of Proteins
2.12. Polarization Assay
2.13. Transwell Migration and Invasion Assays
2.14. Statistical Analysis
3. Results
3.1. AGEs Induce ERK and AKT Phosphorylation in Breast CAFs
3.2. The Activation of AGEs/RAGE Transduction Pathway Up-Regulates IL-8 Levels in CAFs
3.3. c-Fos Is Involved in the Up-Regulation of IL-8 Prompted by AGEs/RAGE Signaling in CAFs
3.4. IL-8/CXCR1/2 Paracrine Activation Promotes the Acquisition of a Migratory and Invasive Phenotype of MDA-MB-231 Breast Cancer Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Vella, V.; De Francesco, E.M.; Lappano, R.; Muoio, M.G.; Manzella, L.; Maggiolini, M.; Belfiore, A. Microenvironmental Determinants of Breast Cancer Metastasis: Focus on the Crucial Interplay Between Estrogen and Insulin/Insulin-Like Growth Factor Signaling. Front. Cell Dev. Biol. 2020, 8, 608412. [Google Scholar] [CrossRef] [PubMed]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Hardefeldt, P.J.; Edirimanne, S.; Eslick, G.D. Diabetes increases the risk of breast cancer: A meta-analysis. Endocr. Relat. Cancer 2012, 19, 793–803. [Google Scholar] [CrossRef]
- Buono, G.; Crispo, A.; Giuliano, M.; de Angelis, C.; Schettini, F.; Forestieri, V.; Lauria, R.; Pensabene, M.; de Laurentiis, M.; Augustin, L.S.A.; et al. Combined effect of obesity and diabetes on early breast cancer outcome: A prospective observational study. Oncotarget 2017, 8, 115709–115717. [Google Scholar] [CrossRef][Green Version]
- Scully, T.; Ettela, A.; LeRoith, D.; Gallagher, E.J. Obesity, Type 2 Diabetes, and Cancer Risk. Front. Oncol. 2021, 10, 615375. [Google Scholar] [CrossRef]
- Park, Y.M.M.; White, A.J.; Nichols, H.B.; O’Brien, K.M.; Weinberg, C.R.; Sandler, D.P. The association between metabolic health, obesity phenotype and the risk of breast cancer. Int. J. Cancer 2017, 140, 2657–2666. [Google Scholar] [CrossRef]
- Lee, H. Obesity-Associated Cancers: Evidence from Studies in Mouse Models. Cells 2022, 11, 1472. [Google Scholar] [CrossRef]
- Vella, V.; Giuliano, M.; Ferlita, A.L.; Pellegrino, M.; Gaudenzi, G.; Alaimo, S.; Massimino, M.; Pulvirenti, A.; Dicitore, A.; Vigneri, P.; et al. Novel mechanisms of tumor promotion by the insulin receptor isoform a in triple-negative breast cancer cells. Cells 2021, 10, 3145. [Google Scholar] [CrossRef]
- Srivastava, S.P.; Goodwin, J.E. Cancer Biology and Prevention in Diabetes. Cells 2020, 9, 1380. [Google Scholar] [CrossRef]
- Boubertakh, B.; Silvestri, C.; Di Marzo, V. Obesity: The Fat Tissue Disease Version of Cancer. Cells 2022, 11, 1872. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef]
- Danforth, D.N. The role of chronic inflammation in the development of breast cancer. Cancers 2021, 13, 3918. [Google Scholar] [CrossRef]
- Aragno, M.; Mastrocola, R. Dietary sugars and endogenous formation of advanced glycation endproducts: Emerging mechanisms of disease. Nutrients 2017, 9, 385. [Google Scholar] [CrossRef]
- Ruiz, H.H.; Ramasamy, R.; Schmidt, A.M. Advanced Glycation End Products: Building on the Concept of the “Common Soil” in Metabolic Disease. Endocrinology 2020, 161, bqz006. [Google Scholar] [CrossRef]
- Rhee, S.Y.; Kim, Y.S. The role of advanced glycation end products in diabetic vascular complications. Diabetes Metab. J. 2018, 42, 188–195. [Google Scholar] [CrossRef]
- Ahmed, N. Advanced glycation endproducts—Role in pathology of diabetic complications. Diabetes Res. Clin. Pract. 2005, 67, 3–21. [Google Scholar] [CrossRef]
- López-Díez, R.; Shekhtman, A.; Ramasamy, R.; Schmidt, A.M. Cellular mechanisms and consequences of glycation in atherosclerosis and obesity. Biochim. Biophys. Acta—Mol. Basis Dis. 2016, 1862, 2244–2252. [Google Scholar] [CrossRef]
- Goldberg, T.; Cai, W.; Peppa, M.; Dardaine, V.; Baliga, B.S.; Uribarri, J.; Vlassara, H. Advanced glycoxidation end products in commonly consumed foods. J. Am. Diet. Assoc. 2004, 104, 1287–1291. [Google Scholar] [CrossRef]
- Twarda-Clapa, A.; Olczak, A.; Białkowska, A.M.; Koziołkiewicz, M. Advanced Glycation End-Products (AGEs): Formation, Chemistry, Classification, Receptors, and Diseases Related to AGEs. Cells 2022, 11, 1312. [Google Scholar] [CrossRef]
- Ott, C.; Jacobs, K.; Haucke, E.; Navarrete Santos, A.; Grune, T.; Simm, A. Role of advanced glycation end products in cellular signaling. Redox Biol. 2014, 2, 411–429. [Google Scholar] [CrossRef]
- Neeper, M.; Schmidt, A.M.; Brett, J.; Yan, S.D.; Wang, F.; Pan, Y.C.E.; Elliston, K.; Stern, D.; Shaw, A. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J. Biol. Chem. 1992, 267, 14998–15004. [Google Scholar] [CrossRef]
- Riehl, A.; Németh, J.; Angel, P.; Hess, J. The receptor RAGE: Bridging inflammation and cancer. Cell Commun. Signal. 2009, 7, 12. [Google Scholar] [CrossRef]
- Nankali, M.; Karimi, J.; Goodarzi, M.T.; Saidijam, M.; Khodadadi, I.; Razavi, A.N.E.; Rahimi, F. Increased Expression of the Receptor for Advanced Glycation End-Products (RAGE) Is Associated with Advanced Breast Cancer Stage. Oncol. Res. Treat. 2016, 39, 622–628. [Google Scholar] [CrossRef]
- Nasser, M.W.; Wani, N.A.; Ahirwar, D.K.; Powell, C.A.; Ravi, J.; Elbaz, M.; Zhao, H.; Padilla, L.; Zhang, X.; Shilo, K.; et al. RAGE Mediates S100A7-Induced Breast Cancer Growth and Metastasis by Modulating the Tumor Microenvironment. Cancer Res. 2015, 75, 974–985. [Google Scholar] [CrossRef]
- Kwak, T.; Drews-Elger, K.; Ergonul, A.; Miller, P.C.; Braley, A.; Hwang, G.H.; Zhao, D.; Besser, A.; Yamamoto, Y.; Yamamoto, H.; et al. Targeting of RAGE-ligand signaling impairs breast cancer cell invasion and metastasis. Oncogene 2017, 36, 1559–1572. [Google Scholar] [CrossRef]
- Muoio, M.G.; Talia, M.; Lappano, R.; Sims, A.H.; Vella, V.; Cirillo, F.; Manzella, L.; Giuliano, M.; Maggiolini, M.; Belfiore, A.; et al. Activation of the s100a7/rage pathway by igf-1 contributes to angiogenesis in breast cancer. Cancers 2021, 13, 621. [Google Scholar] [CrossRef]
- Santolla, M.F.; Talia, M.; Maggiolini, M. S100A4 Is Involved in Stimulatory Effects Elicited by the FGF2/FGFR1 Signaling Pathway in Triple-Negative Breast Cancer (TNBC) Cells. Int. J. Mol. Sci. 2021, 22, 4720. [Google Scholar] [CrossRef]
- Fuentes, E.; Palomo, I.; Rojas, A. Cross-talk between platelet and tumor microenvironment: Role of multiligand/RAGE axis in platelet activation. Blood Rev. 2016, 30, 213–221. [Google Scholar] [CrossRef]
- De Larco, J.E.; Wuertz, B.R.K.; Rosner, K.A.; Erickson, S.A.; Gamache, D.E.; Manivel, J.C.; Furcht, L.T. A potential role for interleukin-8 in the metastatic phenotype of breast carcinoma cells. Am. J. Pathol. 2001, 158, 639–646. [Google Scholar] [CrossRef]
- Bendre, M.S.; Gaddy-Kurten, D.; Mon-Foote, T.; Akel, N.S.; Skinner, R.A.; Nicholas, R.W.; Suva, L.J. Expression of interleukin 8 and not parathyroid hormone-related protein by human breast cancer cells correlates with bone metastasis in vivo. Cancer Res. 2002, 62, 5571–5579. [Google Scholar] [PubMed]
- Bièche, I.; Chavey, C.; Andrieu, C.; Busson, M.; Vacher, S.; Le Corre, L.; Guinebretière, J.M.; Burlinchon, S.; Lidereau, R.; Lazennec, G. CXC chemokines located in the 4q21 region are up-regulated in breast cancer. Endocr. Relat. Cancer 2007, 14, 1039–1052. [Google Scholar] [CrossRef] [PubMed]
- Benoy, I.H.; Salgado, R.; Van Dam, P.; Geboers, K.; Van Marck, E.; Scharpé, S.; Vermeulen, P.B.; Dirix, L.Y. Increased serum interleukin-8 in patients with early and metastatic breast cancer correlates with early dissemination and survival. Clin. Cancer Res. 2004, 10, 7157–7162. [Google Scholar] [CrossRef] [PubMed]
- Nannuru, K.; Sharma, B.; Varney, M.; Singh, R. Role of chemokine receptor CXCR2 expression in mammary tumor growth, angiogenesis and metastasis. J. Carcinog. 2011, 10, 40. [Google Scholar] [CrossRef]
- Ha, H.; Debnath, B.; Neamati, N. Role of the CXCL8-CXCR1/2 axis in cancer and inflammatory diseases. Theranostics 2017, 7, 1543–1588. [Google Scholar] [CrossRef]
- Rossi, D.; Zlotnik, A. The Biology of Chemokines and their Receptors. Annu. Rev. Immunol. 2000, 18, 217–242. [Google Scholar] [CrossRef]
- Yoshimura, T.; Matsushima, K.; Tanaka, S.; Robinson, E.A.; Appella, E.; Oppenheim, J.J.; Leonard, E.J. Purification of a human monocyte-derived neutrophil chemotactic factor that has peptide sequence similarity to other host defense cytokines. Proc. Natl. Acad. Sci. USA 1987, 84, 9233–9237. [Google Scholar] [CrossRef]
- Walz, A.; Peveri, P.; Aschauer, H.; Baggiolini, M. Purification and amino acid sequencing of NAF, a novel neutrophil-activating factor produced by monocytes. Biochem. Biophys. Res. Commun. 1987, 149, 755–761. [Google Scholar] [CrossRef]
- Alfaro, C.; Sanmamed, M.F.; Rodríguez-Ruiz, M.E.; Teijeira, Á.; Oñate, C.; González, Á.; Ponz, M.; Schalper, K.A.; Pérez-Gracia, J.L.; Melero, I. Interleukin-8 in cancer pathogenesis, treatment and follow-up. Cancer Treat. Rev. 2017, 60, 24–31. [Google Scholar] [CrossRef]
- Waugh, D.J.J.; Wilson, C. The interleukin-8 pathway in cancer. Clin. Cancer Res. 2008, 14, 6735–6741. [Google Scholar] [CrossRef]
- Singh, J.K.; Simões, B.M.; Howell, S.J.; Farnie, G.; Clarke, R.B. Recent advances reveal IL-8 signaling as a potential key to targeting breast cancer stem cells. Breast Cancer Res. 2013, 15, 210. [Google Scholar] [CrossRef]
- Singh, J.K.; Simões, B.M.; Clarke, R.B.; Bundred, N.J. Targeting IL-8 signalling to inhibit breast cancer stem cell activity. Expert Opin. Ther. Targets 2013, 17, 1235–1241. [Google Scholar] [CrossRef]
- Tiainen, L.; Hämäläinen, M.; Luukkaala, T.; Tanner, M.; Lahdenperä, O.; Vihinen, P.; Jukkola, A.; Karihtala, P.; Moilanen, E.; Kellokumpu-Lehtinen, P.L. Low Plasma IL-8 Levels During Chemotherapy Are Predictive of Excellent Long-Term Survival in Metastatic Breast Cancer. Clin. Breast Cancer 2019, 19, e522–e533. [Google Scholar] [CrossRef]
- Fernando, R.I.; Castillo, M.D.; Litzinger, M.; Hamilton, D.H.; Palena, C. IL-8 signaling plays a critical role in the epithelial-mesenchymal transition of human carcinoma cells. Cancer Res. 2011, 71, 5296–5306. [Google Scholar] [CrossRef]
- Freund, A.; Chauveau, C.; Brouillet, J.-P.; Lucas, A.; Lacroix, M.; Licznar, A.; Vignon, F.; Lazennec, G. IL-8 expression and its possible relationship with estrogen-receptor-negative status of breast cancer cells. Oncogene 2003, 22, 256–265. [Google Scholar] [CrossRef]
- Lin, Y.; Huang, R.; Chen, L.; Li, S.; Shi, Q.; Jordan, C.; Huang, R.P. Identification of interleukin-8 as estrogen receptor-regulated factor involved in breast cancer invasion and angiogenesis by protein arrays. Int. J. Cancer 2004, 109, 507–515. [Google Scholar] [CrossRef]
- Yao, C.; Lin, Y.; Chua, M.S.; Ye, C.S.; Bi, J.; Li, W.; Zhu, Y.F.; Wang, S.M. Interleukin-8 modulates growth and invasiveness of estrogen receptor-negative breast cancer cells. Int. J. Cancer 2007, 14, 6735–6741. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, Y.; Jia, T.; Sun, Y. Molecular mechanism underlying the tumor-promoting functions of carcinoma-associated fibroblasts. Tumor Biol. 2015, 36, 1385–1394. [Google Scholar] [CrossRef]
- Augsten, M. Cancer-Associated Fibroblasts as Another Polarized Cell Type of the Tumor Microenvironment. Front. Oncol. 2014, 4, 62. [Google Scholar] [CrossRef]
- Lappano, R.; Talia, M.; Cirillo, F.; Rigiracciolo, D.C.; Scordamaglia, D.; Guzzi, R.; Miglietta, A.M.; De Francesco, E.M.; Belfiore, A.; Sims, A.H.; et al. The IL1β-IL1R signaling is involved in the stimulatory effects triggered by hypoxia in breast cancer cells and cancer-associated fibroblasts (CAFs). J. Exp. Clin. Cancer Res. 2020, 39, 153. [Google Scholar] [CrossRef]
- Liu, T.; Zhou, L.; Li, D.; Andl, T.; Zhang, Y. Cancer-associated fibroblasts build and secure the tumor microenvironment. Front. Cell Dev. Biol. 2019, 7, 60. [Google Scholar] [CrossRef]
- Korkaya, H.; Orsulic, S. Editorial: The Tumor Microenvironment: Recent Advances and Novel Therapeutic Approaches. Front. Cell Dev. Biol. 2020, 8, 586176. [Google Scholar] [CrossRef]
- Lan, T.; Chen, L.; Wei, X. Inflammatory cytokines in cancer: Comprehensive understanding and clinical progress in gene therapy. Cells 2021, 10, 100. [Google Scholar] [CrossRef]
- Su, S.; Chen, J.; Yao, H.; Liu, J.; Yu, S.; Lao, L.; Wang, M.; Luo, M.; Xing, Y.; Chen, F.; et al. CD10+GPR77+ Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell 2018, 172, 841–856.e16. [Google Scholar] [CrossRef]
- Lappano, R.; Rigiracciolo, D.C.; Belfiore, A.; Maggiolini, M.; De Francesco, E.M. Cancer associated fibroblasts: Role in breast cancer and potential as therapeutic targets. Expert Opin. Ther. Targets 2020, 24, 559–572. [Google Scholar] [CrossRef]
- Fernández-Nogueira, P.; Fuster, G.; Gutierrez-Uzquiza, Á.; Gascón, P.; Carbó, N.; Bragado, P. Cancer-associated fibroblasts in breast cancer treatment response and metastasis. Cancers 2021, 13, 3146. [Google Scholar] [CrossRef]
- Fridman, E.S.; Ginini, L.; Gil, Z. The Role of Extracellular Vesicles in Metabolic Reprogramming of the Tumor Microenvironment. Cells 2022, 11, 1433. [Google Scholar] [CrossRef]
- Kong, J.; Tian, H.; Zhang, F.; Zhang, Z.; Li, J.; Liu, X.; Li, X.; Liu, J.; Li, X.; Jin, D.; et al. Extracellular vesicles of carcinoma-associated fibroblasts creates a pre-metastatic niche in the lung through activating fibroblasts. Mol. Cancer 2019, 18, 175. [Google Scholar] [CrossRef]
- Boulter, L.; Bullock, E.; Mabruk, Z.; Brunton, V.G. The fibrotic and immune microenvironments as targetable drivers of metastasis. Br. J. Cancer 2021, 124, 27–36. [Google Scholar] [CrossRef]
- Moleirinho, S.; Chang, N.; Sims, A.H.; Tilston-Lünel, A.M.; Angus, L.; Steele, A.; Boswell, V.; Barnett, S.C.; Ormandy, C.; Faratian, D.; et al. KIBRA exhibits MST-independent functional regulation of the Hippo signaling pathway in mammals. Oncogene 2013, 32, 1821–1830. [Google Scholar] [CrossRef]
- Dai, M. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005, 33, e175. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Bolstad, B.M.; Collin, F.; Cope, L.M.; Hobbs, B.; Speed, T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003, 31, e15. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Sims, A.H.; Smethurst, G.J.; Hey, Y.; Okoniewski, M.J.; Pepper, S.D.; Howell, A.; Miller, C.J.; Clarke, R.B. The removal of multiplicative, systematic bias allows integration of breast cancer gene expression datasets—improving meta-analysis and prediction of prognosis. BMC Med. Genom. 2008, 1, 42. [Google Scholar] [CrossRef] [PubMed]
- Pearce, D.A.; Nirmal, A.J.; Freeman, T.C.; Sims, A.H. Continuous biomarker assessment by exhaustive survival analysis. bioRxiv 2017. [Google Scholar] [CrossRef]
- De Francesco, E.M.; Pellegrino, M.; Santolla, M.F.; Lappano, R.; Ricchio, E.; Abonante, S.; Maggiolini, M. GPER mediates activation of HIF1α/VEGF signaling by estrogens. Cancer Res. 2014, 74, 4053–4064. [Google Scholar] [CrossRef]
- Santolla, M.F.; De Francesco, E.M.; Lappano, R.; Rosano, C.; Abonante, S.; Maggiolini, M. Niacin activates the G protein estrogen receptor (GPER)-mediated signalling. Cell. Signal. 2014, 26, 1466–1475. [Google Scholar] [CrossRef]
- Takahashi, A.; de Andrés, M.C.; Hashimoto, K.; Itoi, E.; Oreffo, R.O.C. Epigenetic regulation of interleukin-8, an inflammatory chemokine, in osteoarthritis. Osteoarthr. Cartil. 2015, 23, 1946–1954. [Google Scholar] [CrossRef]
- Santolla, M.F.; Avino, S.; Pellegrino, M.; De Francesco, E.M.; De Marco, P.; Lappano, R.; Vivacqua, A.; Cirillo, F.; Rigiracciolo, D.C.; Scarpelli, A.; et al. SIRT1 is involved in oncogenic signaling mediated by GPER in breast cancer. Cell Death Dis. 2015, 6, e1834. [Google Scholar] [CrossRef]
- Zhang, Y.; Bottinelli, D.; Lisacek, F.; Luban, J.; Strambio-De-Castillia, C.; Varesio, E.; Hopfgartner, G. Optimization of human dendritic cell sample preparation for mass spectrometry-based proteomic studies. Anal. Biochem. 2015, 484, 40–50. [Google Scholar] [CrossRef]
- Fic, E.; Kedracka-Krok, S.; Jankowska, U.; Pirog, A.; Dziedzicka-Wasylewska, M. Comparison of protein precipitation methods for various rat brain structures prior to proteomic analysis. Electrophoresis 2010, 31, 3573–3579. [Google Scholar] [CrossRef]
- De Marco, P.; Lappano, R.; Francesco, E.M.D.; Cirillo, F.; Pupo, M.; Avino, S.; Vivacqua, A.; Abonante, S.; Picard, D.; Maggiolini, M. GPER signalling in both cancer-associated fibroblasts and breast cancer cells mediates a feedforward IL1β/IL1R1 response. Sci. Rep. 2016, 6, 24354. [Google Scholar] [CrossRef]
- Lamers, M.L.; Almeida, M.E.S.; Vicente-Manzanares, M.; Horwitz, A.F.; Santos, M.F. High glucose-mediated oxidative stress impairs cell migration. PLoS ONE 2011, 6, e22865. [Google Scholar] [CrossRef]
- Santolla, M.F.; Vivacqua, A.; Lappano, R.; Rigiracciolo, D.C.; Cirillo, F.; Galli, G.R.; Talia, M.; Brunetti, G.; Miglietta, A.M.; Belfiore, A.; et al. GPER Mediates a Feedforward FGF2/FGFR1 Paracrine Activation Coupling CAFs to Cancer Cells Toward Breast Tumor Progression. Cells 2019, 8, 223. [Google Scholar] [CrossRef]
- Walter, K.R.; Ford, M.E.; Gregoski, M.J.; Kramer, R.M.; Knight, K.D.; Spruill, L.; Nogueira, L.M.; Krisanits, B.A.; Phan, V.; La Rue, A.C.; et al. Advanced glycation end products are elevated in estrogen receptor-positive breast cancer patients, alter response to therapy, and can be targeted by lifestyle intervention. Breast Cancer Res. Treat. 2019, 173, 559–571. [Google Scholar] [CrossRef]
- Hu, P.; Lai, D.; Lu, P.; Gao, J.; He, H. ERK and Akt signaling pathways are involved in advanced glycation end product-induced autophagy in rat vascular smooth muscle cells. Int. J. Mol. Med. 2012, 29, 613–618. [Google Scholar] [CrossRef]
- Palanissami, G.; Paul, S.F.D. RAGE and Its Ligands: Molecular Interplay Between Glycation, Inflammation, and Hallmarks of Cancer—A Review. Horm. Cancer 2018, 9, 295–325. [Google Scholar] [CrossRef]
- Nowotny, K.; Jung, T.; Höhn, A.; Weber, D.; Grune, T. Advanced glycation end products and oxidative stress in type 2 diabetes mellitus. Biomolecules 2015, 5, 194. [Google Scholar] [CrossRef]
- Cepas, V.; Collino, M.; Mayo, J.C.; Sainz, R.M. Redox Signaling and Advanced Glycation Endproducts (AGEs) in Diet-Related Diseases. Antioxidants 2020, 9, 142. [Google Scholar] [CrossRef]
- Rungratanawanich, W.; Qu, Y.; Wang, X.; Essa, M.M.; Song, B.J. Advanced glycation end products (AGEs) and other adducts in aging-related diseases and alcohol-mediated tissue injury. Exp. Mol. Med. 2021, 53, 168–188. [Google Scholar] [CrossRef]
- Garay-Sevilla, M.E.; Gomez-Ojeda, A.; González, I.; Luévano-Contreras, C.; Rojas, A. Contribution of RAGE axis activation to the association between metabolic syndrome and cancer. Mol. Cell. Biochem. 2021, 476, 1555–1573. [Google Scholar] [CrossRef]
- Jundi, K.; Greene, C. Transcription of Interleukin-8: How Altered Regulation Can Affect Cystic Fibrosis Lung Disease. Biomolecules 2015, 5, 1386. [Google Scholar] [CrossRef]
- Khanjani, S.; Terzidou, V.; Johnson, M.R.; Bennett, P.R. NF B and AP-1 drive human myometrial IL8 expression. Mediat. Inflamm. 2012, 2012, 504952. [Google Scholar] [CrossRef]
- Asokan, S.; Bandapalli, O.R. CXCL8 Signaling in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2021, 1302, 25–39. [Google Scholar] [CrossRef]
- Hoffmann, E.; Thiefes, A.; Buhrow, D.; Dittrich-Breiholz, O.; Schneider, H.; Resch, K.; Kracht, M. MEK1-dependent delayed expression of fos-related antigen-1 counteracts c-Fos and p65 NF-κB-mediated interleukin-8 transcription in response to cytokines or growth factors. J. Biol. Chem. 2005, 280, 9706–9718. [Google Scholar] [CrossRef]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Mezawa, Y.; Orimo, A. The roles of tumor- and metastasis-promoting carcinoma-associated fibroblasts in human carcinomas. Cell Tissue Res. 2016, 365, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Hartman, Z.C.; Poage, G.M.; den Hollander, P.; Tsimelzon, A.; Hill, J.; Panupinthu, N.; Zhang, Y.; Mazumdar, A.; Hilsenbeck, S.G.; Mills, G.B.; et al. Growth of Triple-Negative Breast Cancer Cells Relies upon Coordinate Autocrine Expression of the Proinflammatory Cytokines IL-6 and IL-8. Cancer Res. 2013, 73, 3470–3480. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Li, L.; Andrew, S.; Allan, D.; Li, X.; Lee, J.; Ji, G.; Yao, Z.; Gadde, S.; Figeys, D.; et al. An autocrine inflammatory forward-feedback loop after chemotherapy withdrawal facilitates the repopulation of drug-resistant breast cancer cells. Cell Death Dis. 2017, 8, e2932. [Google Scholar] [CrossRef] [PubMed]
- Russo, R.C.; Garcia, C.C.; Teixeira, M.M.; Amaral, F.A. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert Rev. Clin. Immunol. 2014, 10, 593–619. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Koh, M.; Jin, H.; Bae, M.; Lee, S.; Kim, K.M.; Jung, J.; Kim, H.J.; Park, S.Y.; Kim, H.S.; et al. Cancer-associated fibroblasts induce an aggressive phenotypic shift in non-malignant breast epithelial cells via interleukin-8 and S100A8. J. Cell. Physiol. 2021, 236, 7014–7032. [Google Scholar] [CrossRef]
- Ruffini, P.A. The CXCL8-CXCR1/2 axis as a therapeutic target in breast cancer stem-like cells. Front. Oncol. 2019, 9, 40. [Google Scholar] [CrossRef]
- Fousek, K.; Horn, L.A.; Palena, C. Interleukin-8: A chemokine at the intersection of cancer plasticity, angiogenesis, and immune suppression. Pharmacol. Ther. 2021, 219, 107692. [Google Scholar] [CrossRef]
- Jin, K.; Pandey, N.B.; Popel, A.S. Crosstalk between stromal components and tumor cells of TNBC via secreted factors enhances tumor growth and metastasis. Oncotarget 2017, 8, 60210–60222. [Google Scholar] [CrossRef]
- Alassaf, E.; Mueller, A. The role of PKC in CXCL8 and CXCL10 directed prostate, breast and leukemic cancer cell migration. Eur. J. Pharmacol. 2020, 886, 173453. [Google Scholar] [CrossRef]
- Hoffmann, E.; Dittrich-Breiholz, O.; Holtmann, H.; Kracht, M. Multiple control of interleukin-8 gene expression. J. Leukoc. Biol. 2002, 72, 847–855. [Google Scholar] [CrossRef]
- Azenshtein, E.; Meshel, T.; Shina, S.; Barak, N.; Keydar, I.; Ben-Baruch, A. The angiogenic factors CXCL8 and VEGF in breast cancer: Regulation by an array of pro-malignancy factors. Cancer Lett. 2005, 217, 73–86. [Google Scholar] [CrossRef]
- Korkaya, H.; Kim, G.; Davis, A.; Malik, F.; Henry, N.L.; Ithimakin, S.; Quraishi, A.A.; Tawakkol, N.; D’Angelo, R.; Paulson, A.K.; et al. Activation of an IL6 Inflammatory Loop Mediates Trastuzumab Resistance in HER2+ Breast Cancer by Expanding the Cancer Stem Cell Population. Mol. Cell 2012, 47, 570–584. [Google Scholar] [CrossRef]
- Zhai, J.; Shen, J.; Xie, G.; Wu, J.; He, M.; Gao, L.; Zhang, Y.; Yao, X.; Shen, L. Cancer-associated fibroblasts-derived IL-8 mediates resistance to cisplatin in human gastric cancer. Cancer Lett. 2019, 454, 37–43. [Google Scholar] [CrossRef]
- Wang, T.; Notta, F.; Navab, R.; Joseph, J.; Ibrahimov, E.; Xu, J.; Zhu, C.-Q.; Borgida, A.; Gallinger, S.; Tsao, M.-S. Senescent Carcinoma-Associated Fibroblasts Upregulate IL8 to Enhance Prometastatic Phenotypes. Mol. Cancer Res. 2017, 15, 3–14. [Google Scholar] [CrossRef]
- Katanov, C.; Lerrer, S.; Liubomirski, Y.; Leider-Trejo, L.; Meshel, T.; Bar, J.; Feniger-Barish, R.; Kamer, I.; Soria-Artzi, G.; Kahani, H.; et al. Regulation of the inflammatory profile of stromal cells in human breast cancer: Prominent roles for TNF-α and the NF-κB pathway. Stem Cell Res. Ther. 2015, 6, 87. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Spezia, M.; Huang, S.; Yuan, C.; Zeng, Z.; Zhang, L.; Ji, X.; Liu, W.; Huang, B.; Luo, W.; et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018, 5, 77–106. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.S.; Zhao, Z.; Yang, Z.N.; Xu, F.; Lu, H.J.; Zhu, Z.Y.; Shi, W.; Jiang, J.; Yao, P.P.; Zhu, H.P. Risk factors and preventions of breast cancer. Int. J. Biol. Sci. 2017, 13, 1387–1397. [Google Scholar] [CrossRef] [PubMed]
- Seiler, A.; Chen, M.A.; Brown, R.L.; Fagundes, C.P. Obesity, Dietary Factors, Nutrition, and Breast Cancer Risk. Curr. Breast Cancer Rep. 2018, 10, 14–27. [Google Scholar] [CrossRef]
- Bronsveld, H.K.; Jensen, V.; Vahl, P.; De Bruin, M.L.; Cornelissen, S.; Sanders, J.; Auvinen, A.; Haukka, J.; Andersen, M.; Vestergaard, P.; et al. Diabetes and breast cancer subtypes. PLoS ONE 2017, 12, e0170084. [Google Scholar] [CrossRef]
- Boyle, P.; Boniol, M.; Koechlin, A.; Robertson, C.; Valentini, F.; Coppens, K.; Fairley, L.-L.; Boniol, M.; Zheng, T.; Zhang, Y.; et al. Diabetes and breast cancer risk: A meta-analysis. Br. J. Cancer 2012, 107, 1608–1617. [Google Scholar] [CrossRef]
- Harding, J.L.; Andes, L.J.; Gregg, E.W.; Cheng, Y.J.; Weir, H.K.; Bullard, K.M.; Burrows, N.R.; Imperatore, G. Trends in cancer mortality among people with vs without diabetes in the USA, 1988–2015. Diabetologia 2020, 63, 75–84. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, X.; Gu, C.; Xia, J. Diabetes mellitus is associated with breast cancer: Systematic review, meta-analysis, and in silico reproduction. Panminerva Med. 2015, 57, 101–108. [Google Scholar] [PubMed]
- Durrani, I.A.; Bhatti, A.; John, P. The prognostic outcome of ‘type 2 diabetes mellitus and breast cancer’ association pivots on hypoxia-hyperglycemia axis. Cancer Cell Int. 2021, 21, 351. [Google Scholar] [CrossRef]
- Dong, S.; Wang, Z.; Shen, K.; Chen, X. Metabolic Syndrome and Breast Cancer: Prevalence, Treatment Response, and Prognosis. Front. Oncol. 2021, 11, 629666. [Google Scholar] [CrossRef]
- Farhan Mohammed, I.N.; Motawa, I.A.; Abd El Monem Aly, M.; Mohammed Metwally, M.M. Assessment of Interleukin (8) in Type 2 Diabetes Mellitus. Egypt. J. Hosp. Med. 2018, 72, 4403–4406. [Google Scholar] [CrossRef]
- Dabkeviciene, D.; Jonusiene, V.; Zitkute, V.; Zalyte, E.; Grigaitis, P.; Kirveliene, V.; Sasnauskiene, A. The role of interleukin-759 8 (CXCL8) and CXCR2 in acquired chemoresistance of human colorectal carcinoma cells HCT116. Med. Oncol. 2015, 32, 258. [Google Scholar] [CrossRef]
- Imafuji, H.; Matsuo, Y.; Ueda, G.; Omi, K.; Hayashi, Y.; Saito, K.; Tsuboi, K.; Morimoto, M.; Koide, S.; Ogawa, R.; et al. Acquisition of gemcitabine resistance enhances angiogenesis via upregulation of IL-8 production in pancreatic cancer. Oncol. Rep. 2019, 41, 3508–3516. [Google Scholar] [CrossRef]
- Sootichote, R.; Thuwajit, P.; Singsuksawat, E.; Warnnissorn, M.; Yenchitsomanus, P.t.; Ithimakin, S.; Chantharasamee, J.; Thuwajit, C. Compound A attenuates toll-like receptor 4-mediated paclitaxel resistance in breast cancer and melanoma through suppression of IL-8. BMC Cancer 2018, 18, 231. [Google Scholar] [CrossRef]
- Shi, Z.; Yang, W.M.; Chen, L.P.; Yang, D.H.; Zhou, Q.; Zhu, J.; Chen, J.J.; Huang, R.C.; Chen, Z.S.; Huang, R.P. Enhanced chemosensitization in multidrug-resistant human breast cancer cells by inhibition of IL-6 and IL-8 production. Breast Cancer Res. Treat. 2012, 135, 737–747. [Google Scholar] [CrossRef]
- Holmes, W.E.; Lee, J.; Kuang, W.J.; Rice, G.C.; Wood, W.I. Structure and functional expression of a human interleukin-8 receptor. Science 1991, 253, 1278–1280. [Google Scholar] [CrossRef]
- Murphy, P.M.; Tiffany, H.L. Cloning of complementary DNA encoding a functional human interleukin-8 receptor. Science 1991, 253, 1280–1383. [Google Scholar] [CrossRef]
- Kobilka, B.K. G protein coupled receptor structure and activation. Biochim. Biophys. Acta—Biomembr. 2007, 1768, 794–807. [Google Scholar] [CrossRef]
- Stillie, R.; Farooq, S.M.; Gordon, J.R.; Stadnyk, A.W. The functional significance behind expressing two IL-8 receptor types on PMN. J. Leukoc. Biol. 2009, 86, 529–543. [Google Scholar] [CrossRef]
- Castelli, V.; Brandolini, L.; D’Angelo, M.; Giorgio, C.; Alfonsetti, M.; Cocchiaro, P.; Lombardi, F.; Cimini, A.; Allegretti, M. CXCR1/2 Inhibitor Ladarixin Ameliorates the Insulin Resistance of 3T3-L1 Adipocytes by Inhibiting Inflammation and Improving Insulin Signaling. Cells 2021, 10, 2324. [Google Scholar] [CrossRef]
- Chen, L.; Fan, J.; Chen, H.; Meng, Z.; Chen, Z.; Wang, P.; Liu, L. The IL-8/CXCR1 axis is associated with cancer stem cell-like properties and correlates with clinical prognosis in human pancreatic cancer cases. Sci. Rep. 2014, 4, 5911. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, Z.; Wang, Y.; Fu, H.; Wang, Z.; Xie, H.; Zhang, J.; Li, G.; Dai, B.; Ye, D.; et al. High CXC chemokine receptor 1 level represents an independent negative prognosticator in non-metastatic clear-cell renal cell carcinoma patients. Oncoimmunology 2017, 6, e1359450. [Google Scholar] [CrossRef]
- Kemp, D.M.; Pidich, A.; Larijani, M.; Jonas, R.; Lash, E.; Sato, T.; Terai, M.; De Pizzol, M.; Allegretti, M.; Igoucheva, O.; et al. Ladarixin, a dual CXCR1/2 inhibitor, attenuates experimental melanomas harboring different molecular defects by affecting malignant cells and tumor microenvironment. Oncotarget 2017, 8, 14428–14442. [Google Scholar] [CrossRef]
- Brandolini, L.; Cristiano, L.; Fidoamore, A.; Pizzol, M.D.; Giacomo, E.D.; Florio, T.M.; Confalone, G.; Galante, A.; Cinque, B.; Benedetti, E.; et al. Targeting CXCR1 on breast cancer stem cells: Signaling pathways and clinical application modelling. Oncotarget 2015, 6, 43375–43394. [Google Scholar] [CrossRef]
- Wang, R.-X.; Ji, P.; Gong, Y.; Shao, Z.-M.; Chen, S. Value of CXCL8–CXCR1/2 axis in neoadjuvant chemotherapy for triple-negative breast cancer patients: A retrospective pilot study. Breast Cancer Res. Treat. 2020, 181, 561–570. [Google Scholar] [CrossRef]
- Schott, A.F.; Goldstein, L.J.; Cristofanilli, M.; Ruffini, P.A.; McCanna, S.; Reuben, J.M.; Perez, R.P.; Kato, G.; Wicha, M. Phase Ib pilot study to evaluate reparixin in combination with weekly paclitaxel in patients with HER-2–negative metastatic breast cancer. Clin. Cancer Res. 2017, 23, 5358–5365. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santolla, M.F.; Talia, M.; Cirillo, F.; Scordamaglia, D.; De Rosis, S.; Spinelli, A.; Miglietta, A.M.; Nardo, B.; Filippelli, G.; De Francesco, E.M.; et al. The AGEs/RAGE Transduction Signaling Prompts IL-8/CXCR1/2-Mediated Interaction between Cancer-Associated Fibroblasts (CAFs) and Breast Cancer Cells. Cells 2022, 11, 2402. https://doi.org/10.3390/cells11152402
Santolla MF, Talia M, Cirillo F, Scordamaglia D, De Rosis S, Spinelli A, Miglietta AM, Nardo B, Filippelli G, De Francesco EM, et al. The AGEs/RAGE Transduction Signaling Prompts IL-8/CXCR1/2-Mediated Interaction between Cancer-Associated Fibroblasts (CAFs) and Breast Cancer Cells. Cells. 2022; 11(15):2402. https://doi.org/10.3390/cells11152402
Chicago/Turabian StyleSantolla, Maria Francesca, Marianna Talia, Francesca Cirillo, Domenica Scordamaglia, Salvatore De Rosis, Asia Spinelli, Anna Maria Miglietta, Bruno Nardo, Gianfranco Filippelli, Ernestina Marianna De Francesco, and et al. 2022. "The AGEs/RAGE Transduction Signaling Prompts IL-8/CXCR1/2-Mediated Interaction between Cancer-Associated Fibroblasts (CAFs) and Breast Cancer Cells" Cells 11, no. 15: 2402. https://doi.org/10.3390/cells11152402
APA StyleSantolla, M. F., Talia, M., Cirillo, F., Scordamaglia, D., De Rosis, S., Spinelli, A., Miglietta, A. M., Nardo, B., Filippelli, G., De Francesco, E. M., Belfiore, A., Lappano, R., & Maggiolini, M. (2022). The AGEs/RAGE Transduction Signaling Prompts IL-8/CXCR1/2-Mediated Interaction between Cancer-Associated Fibroblasts (CAFs) and Breast Cancer Cells. Cells, 11(15), 2402. https://doi.org/10.3390/cells11152402