Genetically Engineered Pigs as Efficient Salivary Gland Bioreactors for Production of Therapeutically Valuable Human Nerve Growth Factor
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Plasmid Construction
2.3. Genetically Modified Donor Cell Selection
2.4. Somatic Cell Nuclear Transfer
2.5. Observation of EGFP and RFP Expression
2.6. PCR Analysis
2.7. Southern Blot Analysis
2.8. Saliva Collection during Rest Time
2.9. Enzyme-Linked Immunosorbent Assay (ELISA) Analysis
2.10. PC12 Cell Differentiation Assay
2.11. TF1 Cell Proliferation Assay
2.12. Inverse PCR Analysis
2.13. Collection of Saliva by the Surgical and Nonsurgical Methods
2.14. Purification of hNGF
2.15. SDS-PAGE and Western Blot Analysis
2.16. Liquid Chromatography-Mass Spectrometry/Mass Spectrometry (LC-MS/MS) Analysis
2.17. Off-Target Analysis
2.18. Statistical Analysis
3. Results
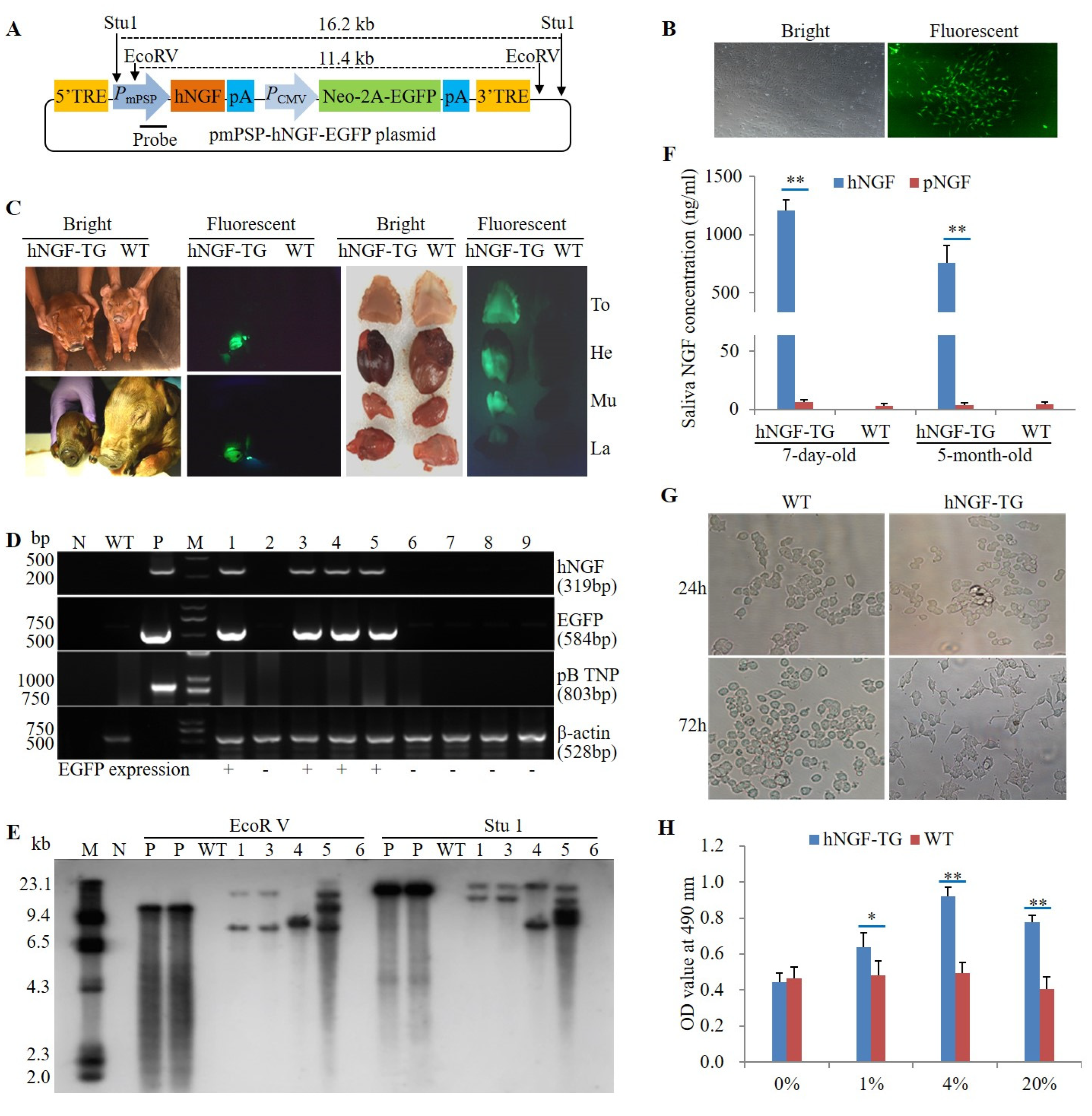
3.1. Production and Identification of F0 Generation hNGF-TG Pigs
3.2. Production and Analysis of F1 Generation hNGF-TG Pigs
3.3. Development of Efficient Methods for Collection of Saliva from hNGF-TG Pigs
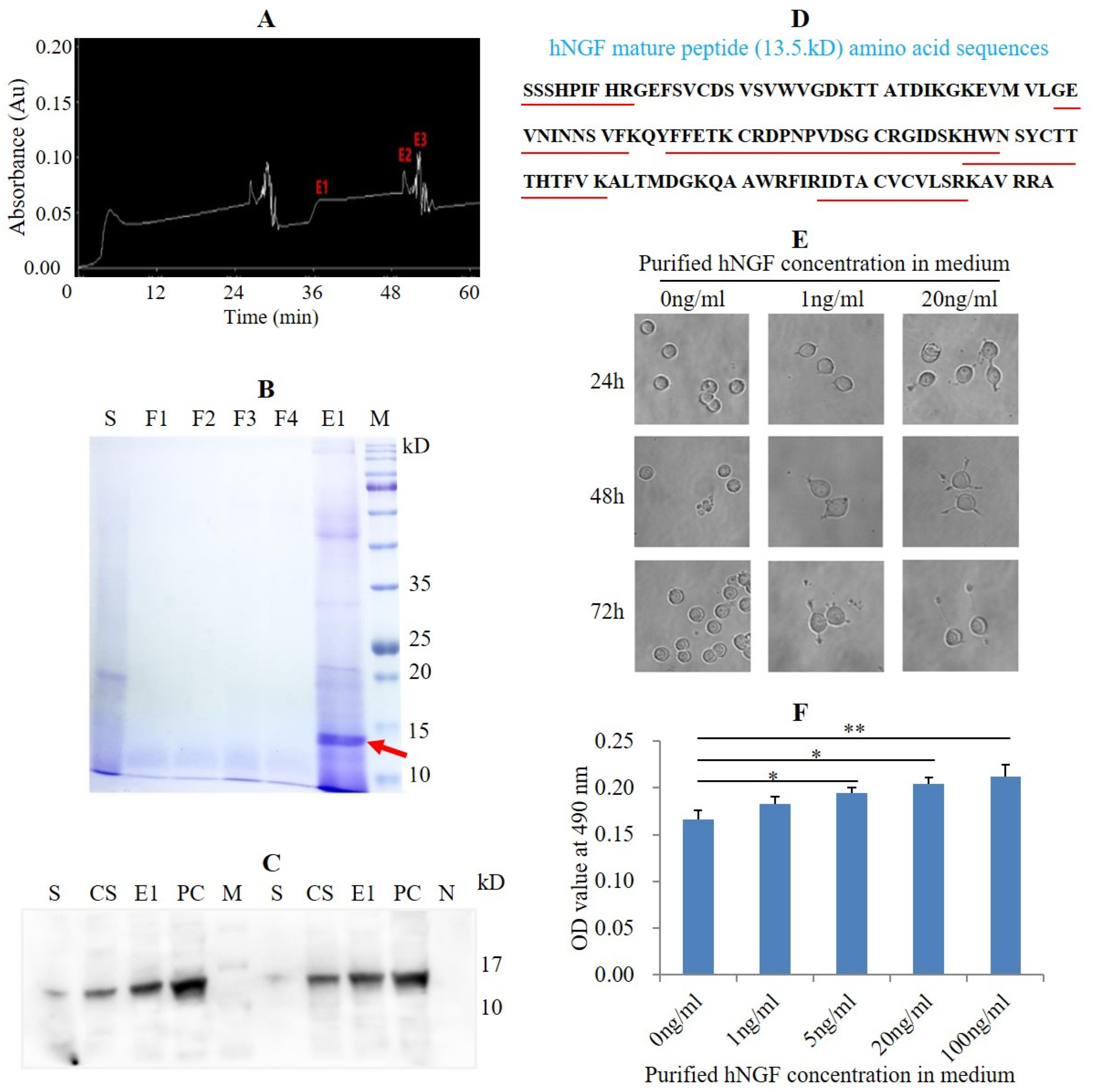
3.4. Purification of hNGF Protein from the Saliva of hNGF-TG Pigs and Bioassay of Purified hNGF
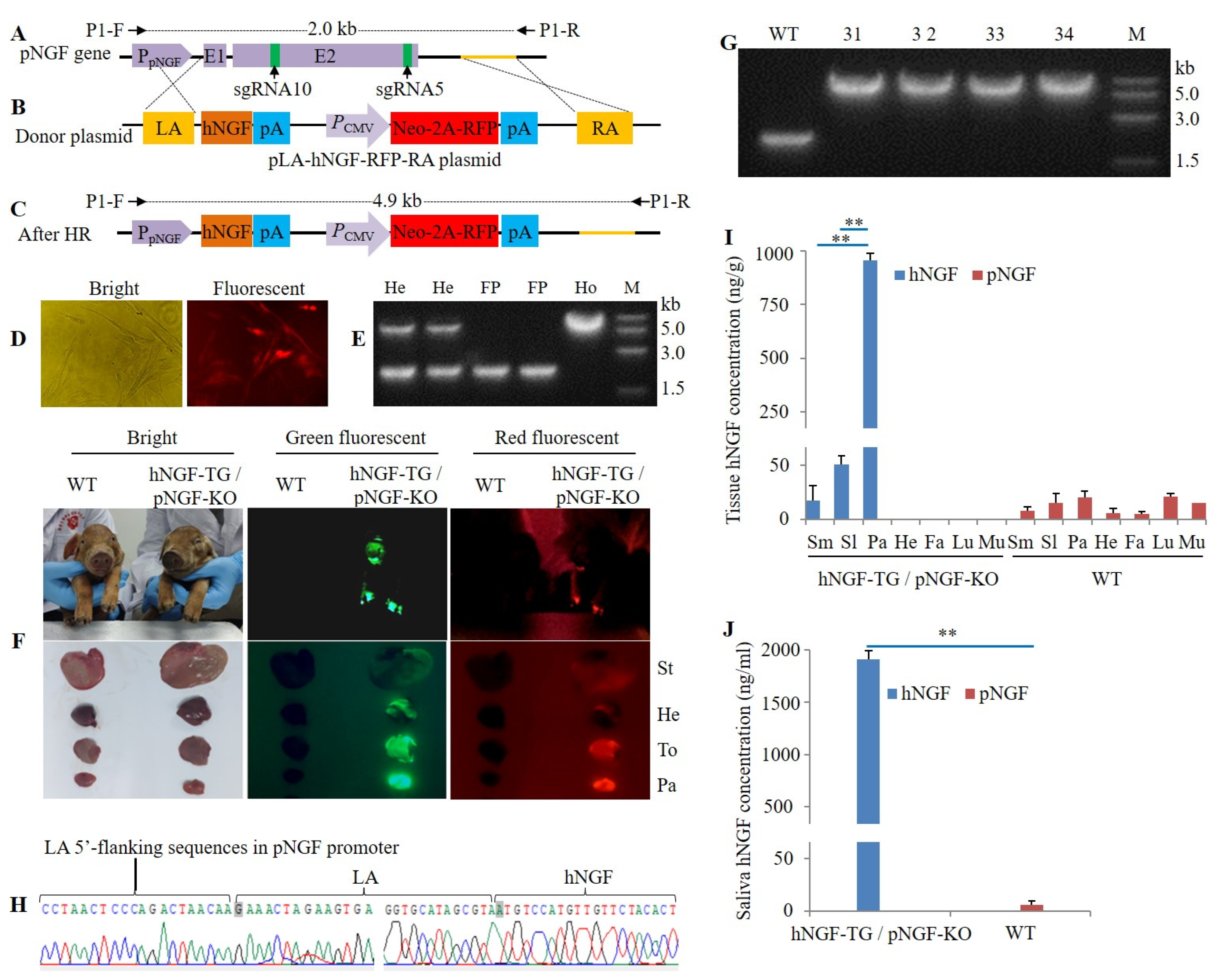
3.5. Production of Double-Transgenic hNGF-TG/ pNGF-KO Pigs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maksimenko, O.; Deykin, A.; Georgiev, P. Use of transgenic animals in biotechnology: Prospects and problems. Acta Naturae 2013, 5, 33–46. [Google Scholar] [CrossRef]
- Bertolini, L.; Meade, H.; Lazzarotto, C.; Martins, L.; Tavares, K.; Bertolini, M.; Murray, J. The transgenic animal platform for biopharmaceutical production. Transgenic Res. 2016, 25, 329–343. [Google Scholar] [CrossRef]
- Demain, A.L.; Vaishnav, P. Production of recombinant proteins by microbes and higher organisms. Biotechnol. Adv. 2009, 27, 297–306. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, S.; Bai, L.; Fan, J.; Liu, E. Expression systems and species used for transgenic animal bioreactors. BioMed Res. Int. 2013, 2013, 580463. [Google Scholar] [CrossRef]
- Sheridan, C. FDA approves ’farmaceutical’ drug from transgenic chickens. Nat. Biotechnol. 2016, 34, 117–119. [Google Scholar] [CrossRef]
- Isenman, L.; Liebow, C.; Rothman, S. The endocrine secretion of mammalian digestive enzymes by exocrine glands. Am. J. Physiol. 1999, 276, E223–E232. [Google Scholar] [CrossRef]
- Humphrey, S.P.; Williamson, R.T. A review of saliva: Normal composition, flow, and function. J. Prosthet. Dent. 2001, 85, 162–169. [Google Scholar] [CrossRef]
- Dawes, C.; Pedersen, A.M.; Villa, A.; Ekström, J.; Proctor, G.B.; Vissink, A.; Aframian, D.; McGowan, R.; Aliko, A.; Narayana, N.; et al. The functions of human saliva: A review sponsored by the World Workshop on Oral Medicine VI. Arch. Oral Biol. 2015, 60, 863–874. [Google Scholar] [CrossRef]
- Maekawa, M.; Beauchemin, K.A.; Christensen, D.A. Effect of concentrate level and feeding management on chewing activities, saliva production, and ruminal pH of lactating dairy cows. J. Dairy Sci. 2002, 85, 1165–1175. [Google Scholar] [CrossRef]
- Golovan, S.P.; Hayes, M.A.; Phillips, J.P.; Forsberg, C.W. Transgenic mice expressing bacterial phytase as a model for phosphorus pollution control. Nat. Biotechnol. 2001, 19, 429–433. [Google Scholar] [CrossRef]
- Salem, A.Z.; López, S.; Ranilla, M.J.; González, J.S. Short- to medium-term effects of consumption of quebracho tannins on saliva production and composition in sheep and goats. J. Anim. Sci. 2013, 91, 1341–1349. [Google Scholar] [CrossRef]
- Norgaard, P.; Grondahl-Nielsen, C.; Grovum, W.L. Technical note: Reversible re-entrant cannulation of the parotid duct in cattle using a new injection anesthesia regimen. J. Anim. Sci. 1996, 74, 1716–1719. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Beal, A.M. Proceedings: A chronic re-entrant parotid duct cannula for long-term salivary collection and replacement in the sheep. J. Physiol. 1974, 242, 22p–24p. [Google Scholar]
- Beal, A.M. The negative correlation between parotid salivary flow and sodium concentration during atropine infusion into conscious sodium-replete sheep. J. Physiol. 1977, 267, 19p–20p. [Google Scholar] [PubMed]
- Cohen, S.; Levi-Montalcini, R. A Nerve Growth-Stimulating Factor Isolated From Snake Venom. Proc. Natl. Acad. Sci. USA 1956, 42, 571–574. [Google Scholar] [CrossRef]
- Weltman, J.K. The 1986 Nobel Prize for Physiology or Medicine awarded for discovery of growth factors: Rita Levi-Montalcini, M.D., and Stanley Cohen, Ph.D. N. Engl. Reg. Allergy Proc. 1987, 8, 47–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, R.; Thrimawithana, T.; Little, P.J.; Xu, J.; Feng, Z.P.; Zheng, W. The nerve growth factor signaling and its potential as therapeutic target for glaucoma. BioMed Res. Int. 2014, 2014, 759473. [Google Scholar] [CrossRef]
- Aloe, L.; Rocco, M.L.; Bianchi, P.; Manni, L. Nerve growth factor: From the early discoveries to the potential clinical use. J. Transl. Med. 2012, 10, 239. [Google Scholar] [CrossRef]
- Cai, J.; Hua, F.; Yuan, L.; Tang, W.; Lu, J.; Yu, S.; Wang, X.; Hu, Y. Potential therapeutic effects of neurotrophins for acute and chronic neurological diseases. BioMed Res. Int. 2014, 2014, 601084. [Google Scholar] [CrossRef]
- Manni, L.; Rocco, M.L.; Bianchi, P.; Soligo, M.; Guaragna, M.; Barbaro, S.P.; Aloe, L. Nerve growth factor: Basic studies and possible therapeutic applications. Growth Factors 2013, 31, 115–122. [Google Scholar] [CrossRef]
- Xu, C.J.; Wang, J.L.; Jin, W.L. The Emerging Therapeutic Role of NGF in Alzheimer’s Disease. Neurochem. Res. 2016, 41, 1211–1218. [Google Scholar] [CrossRef]
- Sheha, H.; Tighe, S.; Hashem, O.; Hayashida, Y. Update On Cenegermin Eye Drops In The Treatment Of Neurotrophic Keratitis. Clin. Ophthalmol. 2019, 13, 1973–1980. [Google Scholar] [CrossRef]
- Schmidt, F.R. Recombinant expression systems in the pharmaceutical industry. Appl. Microbiol. Biotechnol. 2004, 65, 363–372. [Google Scholar] [CrossRef]
- Kang, T.H.; Seong, B.L. Solubility, Stability, and Avidity of Recombinant Antibody Fragments Expressed in Microorganisms. Front. Microbiol. 2020, 11, 1927. [Google Scholar] [CrossRef]
- Xu, L.; Li, Y.; Shi, X.; Han, C.; Tao, L.; Yang, Q.; Rao, C. Expression, purification, and characterization of recombinant mouse nerve growth factor in Chinese hamster ovary cells. Protein Expr. Purif. 2014, 104, 41–49. [Google Scholar] [CrossRef]
- Paoletti, F.; Malerba, F.; Ercole, B.B.; Lamba, D.; Cattaneo, A. A comparative analysis of the structural, functional and biological differences between Mouse and Human Nerve Growth Factor. Biochim. Biophys. Acta 2015, 1854, 187–197. [Google Scholar] [CrossRef]
- Kawaja, M.D.; Smithson, L.J.; Elliott, J.; Trinh, G.; Crotty, A.M.; Michalski, B.; Fahnestock, M. Nerve growth factor promoter activity revealed in mice expressing enhanced green fluorescent protein. J. Comp. Neurol. 2011, 519, 2522–2545. [Google Scholar] [CrossRef]
- Murphy, R.A.; Saide, J.D.; Blanchard, M.H.; Young, M. Nerve growth factor in mouse serum and saliva: Role of the submandibular gland. Proc. Natl. Acad. Sci. USA 1977, 74, 2330–2333. [Google Scholar] [CrossRef]
- Varon, S.; Nomura, J.; Shooter, E.M. Subunit structure of a high-molecular-weight form of the nerve growth factor from mouse submaxillary gland. Proc. Natl. Acad. Sci. USA 1967, 57, 1782–1789. [Google Scholar] [CrossRef]
- Naesse, E.P.; Schreurs, O.; Messelt, E.; Hayashi, K.; Schenck, K. Distribution of nerve growth factor, pro-nerve growth factor, and their receptors in human salivary glands. Eur. J. Oral Sci. 2013, 121, 13–20. [Google Scholar] [CrossRef]
- Zeng, F.; Li, Z.; Zhu, Q.; Dong, R.; Zhao, C.; Li, G.; Li, G.; Gao, W.; Jiang, G.; Zheng, E.; et al. Production of functional human nerve growth factor from the saliva of transgenic mice by using salivary glands as bioreactors. Sci. Rep. 2017, 7, 41270. [Google Scholar] [CrossRef] [PubMed]
- Cadinanos, J.; Bradley, A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007, 35, e87. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.W.; Hossein, M.S.; Bhandari, D.P.; Kim, Y.W.; Kim, J.H.; Park, S.W.; Lee, E.; Park, S.M.; Jeong, Y.I.; Lee, J.Y.; et al. Effects of insulin-transferrin-selenium in defined and porcine follicular fluid supplemented IVM media on porcine IVF and SCNT embryo production. Anim. Reprod. Sci. 2008, 106, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, K.; Suzuki, C.; Itoh, S.; Kikuchi, K.; Iwamura, S.; Rodriguez-Martinez, H. Production of piglets derived from in vitro-produced blastocysts fertilized and cultured in chemically defined media: Effects of theophylline, adenosine, and cysteine during in vitro fertilization. Biol. Reprod. 2003, 69, 2092–2099. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Zhu, Q.; Wu, Z.; Li, Z.; Zeng, F. Effects of gender and age on protein secretion in saliva of transgenic mice expressing human nerve growth factor. Sheng Wu Gong Cheng Xue Bao 2019, 35, 1041–1049. [Google Scholar] [PubMed]
- Coulibaly, S.; Besenfelder, U.; Fleischmann, M.; Zinovieva, N.; Grossmann, A.; Wozny, M.; Bartke, I.; Tögel, M.; Müller, M.; Brem, G. Human nerve growth factor beta (hNGF-beta): Mammary gland specific expression and production in transgenic rabbits. FEBS Lett. 1999, 444, 111–116. [Google Scholar] [CrossRef]
- Fleeman, N.; Mahon, J.; Nevitt, S.; Duarte, R.; Boland, A.; Kotas, E.; Dundar, Y.; McEntee, J.; Ahmad, S. Cenegermin for Treating Neurotrophic Keratitis: An Evidence Review Group Perspective of a NICE Single Technology Appraisal. Pharmacoecon. Open 2019, 3, 453–461. [Google Scholar] [CrossRef]
- Li, Z.; Shi, J.; Liu, D.; Zhou, R.; Zeng, H.; Zhou, X.; Mai, R.; Zeng, S.; Luo, L.; Yu, W.; et al. Effects of donor fibroblast cell type and transferred cloned embryo number on the efficiency of pig cloning. Cell. Reprogram. 2013, 15, 35–42. [Google Scholar] [CrossRef]
- Jang, G.; Park, E.S.; Cho, J.K.; Bhuiyan, M.M.; Lee, B.C.; Kang, S.K.; Hwang, W.S. Preimplantational embryo development and incidence of blastomere apoptosis in bovine somatic cell nuclear transfer embryos reconstructed with long-term cultured donor cells. Theriogenology 2004, 62, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Kurome, M.; Ishikawa, T.; Tomii, R.; Ueno, S.; Shimada, A.; Yazawa, H.; Nagashima, H. Production of transgenic and non-transgenic clones in miniature pigs by somatic cell nuclear transfer. J. Reprod. Dev. 2008, 54, 156–163. [Google Scholar] [CrossRef]
- Lakshmanan, J.; Landel, C.P. Neonatal hyperthyroidism impairs epinephrine-provoked secretion of nerve growth factor and epidermal growth factor in mouse saliva. Pediatr. Res. 1986, 20, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Wallace, L.J.; Partlow, L.M. alpha-Adrenergic regulation of secretion of mouse saliva rich in nerve growth factor. Proc. Natl. Acad. Sci. USA 1976, 73, 4210–4214. [Google Scholar] [CrossRef] [PubMed]
- Eliasson, L.; Carlén, A. An update on minor salivary gland secretions. Eur. J. Oral Sci. 2010, 118, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Abiko, Y.; Saitoh, M. Salivary defensins and their importance in oral health and disease. Curr. Pharm. Des. 2007, 13, 3065–3072. [Google Scholar] [CrossRef] [PubMed]
- Khurshid, Z.; Naseem, M.; Sheikh, Z.; Najeeb, S.; Shahab, S.; Zafar, M.S. Oral antimicrobial peptides: Types and role in the oral cavity. Saudi Pharm. J. 2016, 24, 515–524. [Google Scholar] [CrossRef]
- Amano, O.; Mizobe, K.; Bando, Y.; Sakiyama, K. Anatomy and histology of rodent and human major salivary glands: -overview of the Japan salivary gland society-sponsored workshop. Acta Histochem. Cytochem. 2012, 45, 241–250. [Google Scholar] [CrossRef]





| No. of Cultured (Activated)/Transferred (2-Cell Stage) Cloned Embryos | No. of Total/Farrowed Recipient Sows | No. of Transferred Cloned Embryos Per Recipient | No. of Total Born Pigs (TG/WT) | No. of Survived Pigs (TG/WT) |
|---|---|---|---|---|
| 4650/3296 | 16/3 | 200–220 | 9 (4/5) | 4 (2/2) |
| Collection Methods | Type of Collected Saliva | Volume of Collected Saliva (mL/day/pig) | hNGF/pNGF Concentration (µg/mL) |
|---|---|---|---|
| UPDCB (Surgical) * | Parotid saliva | 512 ± 84 † | 14.2 ± 3.9/0.04 ± 0.02 |
| BLDB (Nonsurgical) ‡ | Oral saliva | 303 ± 76 § | 4.5 ± 0.7/0.02± 0.01 |
| No. of Cultured (Activated)/Transferred (2-Cell Stage) Cloned Embryos | No. of Total/Farrowed Recipient Sows | No. of Transferred Cloned Embryos Per Recipient | Total Number of Born Pigs | Number of Surviving Pigs |
|---|---|---|---|---|
| 1506/1098 | 5/1 | 200–220 | 4 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, F.; Liao, S.; Kuang, Z.; Zhu, Q.; Wei, H.; Shi, J.; Zheng, E.; Xu, Z.; Huang, S.; Hong, L.; et al. Genetically Engineered Pigs as Efficient Salivary Gland Bioreactors for Production of Therapeutically Valuable Human Nerve Growth Factor. Cells 2022, 11, 2378. https://doi.org/10.3390/cells11152378
Zeng F, Liao S, Kuang Z, Zhu Q, Wei H, Shi J, Zheng E, Xu Z, Huang S, Hong L, et al. Genetically Engineered Pigs as Efficient Salivary Gland Bioreactors for Production of Therapeutically Valuable Human Nerve Growth Factor. Cells. 2022; 11(15):2378. https://doi.org/10.3390/cells11152378
Chicago/Turabian StyleZeng, Fang, Sha Liao, Zhe Kuang, Qingchun Zhu, Hengxi Wei, Junsong Shi, Enqin Zheng, Zheng Xu, Sixiu Huang, Linjun Hong, and et al. 2022. "Genetically Engineered Pigs as Efficient Salivary Gland Bioreactors for Production of Therapeutically Valuable Human Nerve Growth Factor" Cells 11, no. 15: 2378. https://doi.org/10.3390/cells11152378
APA StyleZeng, F., Liao, S., Kuang, Z., Zhu, Q., Wei, H., Shi, J., Zheng, E., Xu, Z., Huang, S., Hong, L., Gu, T., Yang, J., Yang, H., Cai, G., Moisyadi, S., Urschitz, J., Li, Z., & Wu, Z. (2022). Genetically Engineered Pigs as Efficient Salivary Gland Bioreactors for Production of Therapeutically Valuable Human Nerve Growth Factor. Cells, 11(15), 2378. https://doi.org/10.3390/cells11152378








