The Impact of Oulema melanopus—Associated Bacteria on the Wheat Defense Response to the Feeding of Their Insect Hosts
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Insects
2.2. Extraction and Purification of RNA
2.3. The RNA-Sequencing and Data Analysis
2.4. Validation of RNA-Sequencing Results by Quantitative Real-Time PCR Analysis
3. Results
3.1. Transcriptomic Profile of the Wheat Plants Subjected to the Treatments Applied
3.1.1. Distribution of DEGs in Wheat Plants Damaged Mechanically and by CLB Larvae Feeding
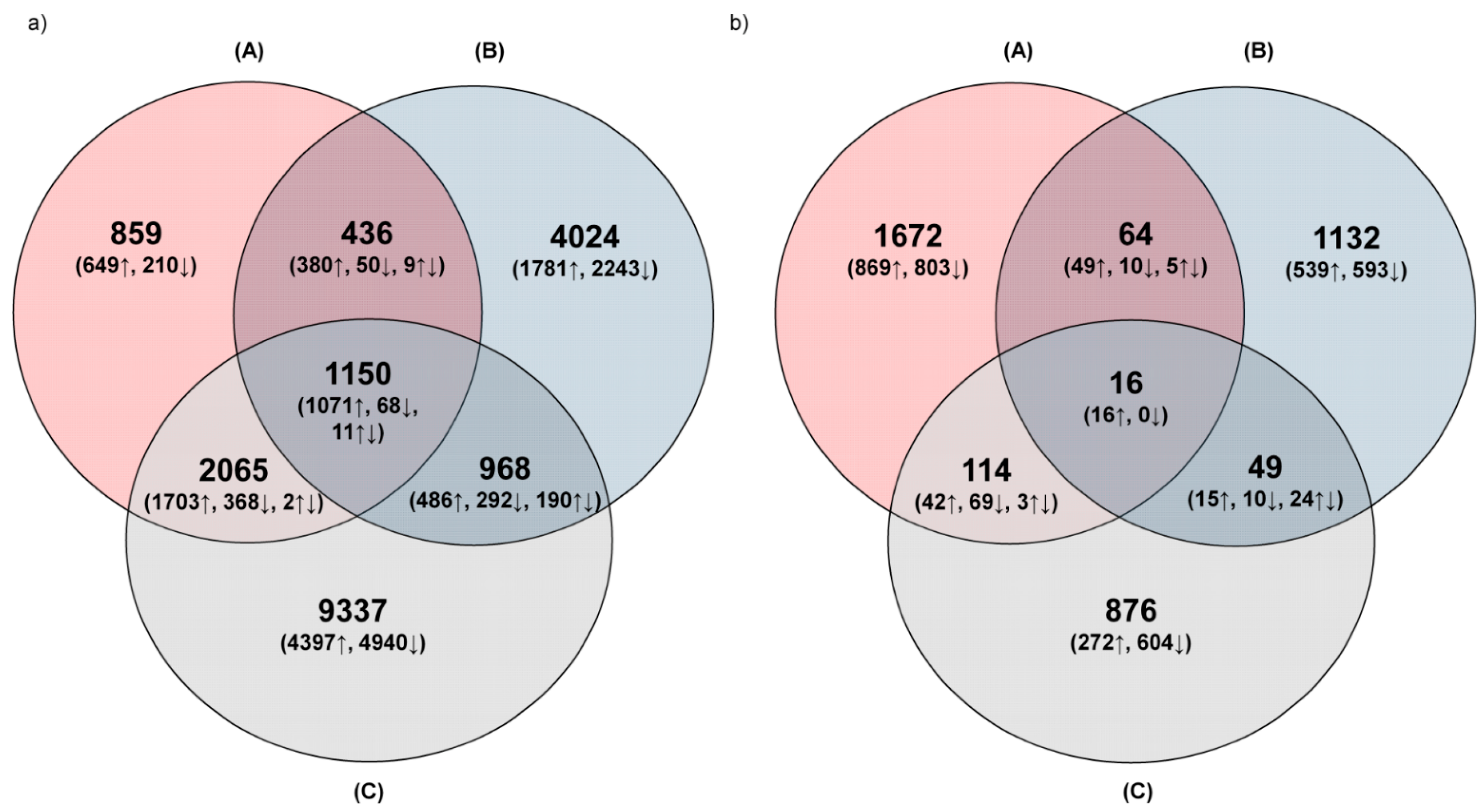
3.1.2. Distribution of Common and Unique DEGs
3.1.3. Gene Ontology Analysis of Common and Unique DEGs for Each Variant of Plant Treatment
3.2. The Effect of Larval-Associated Bacteria on the Plant Defense Response
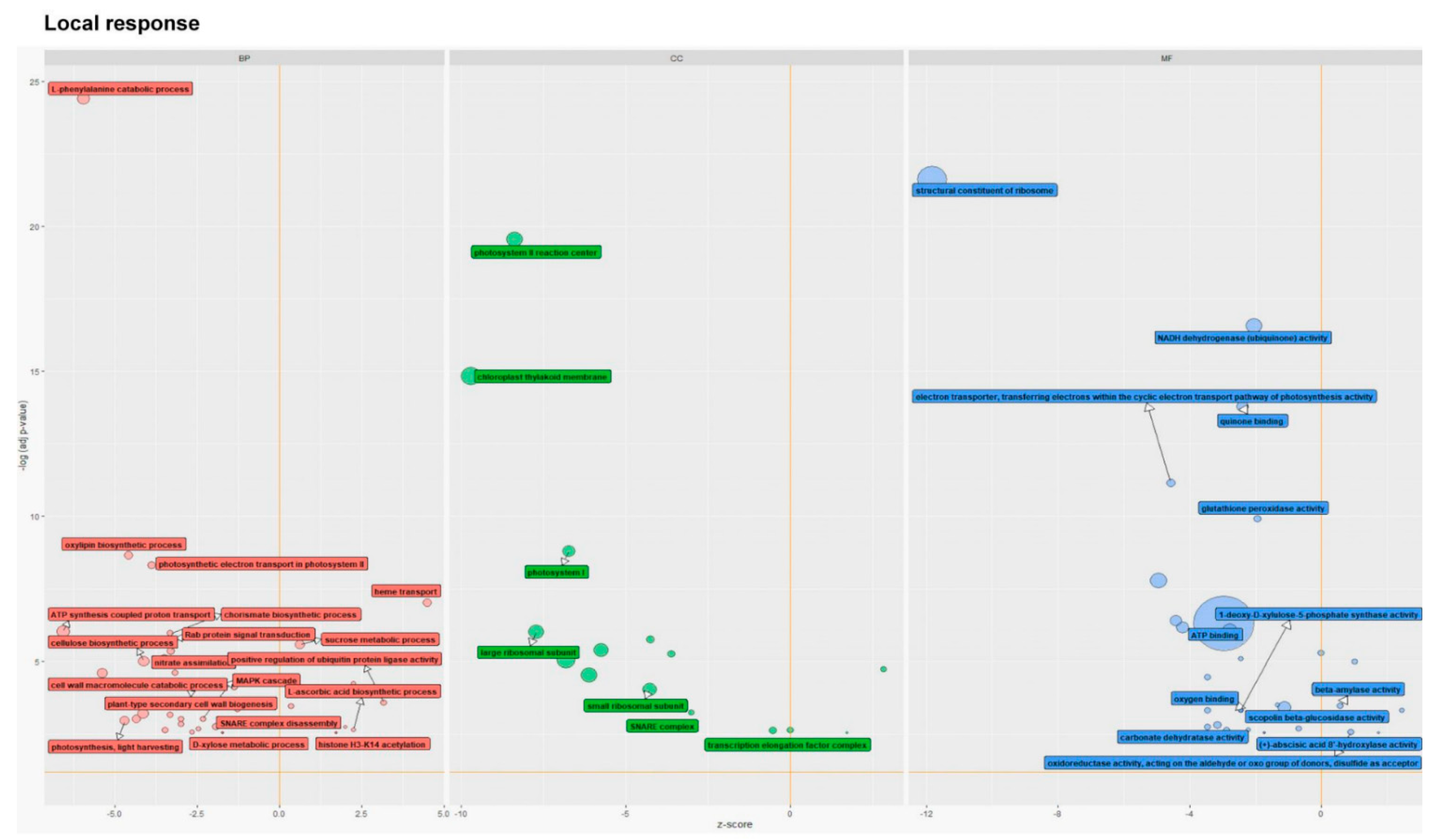
3.2.1. Gene Ontology Overview of Larval-Associated Bacteria-Responsive DEGs
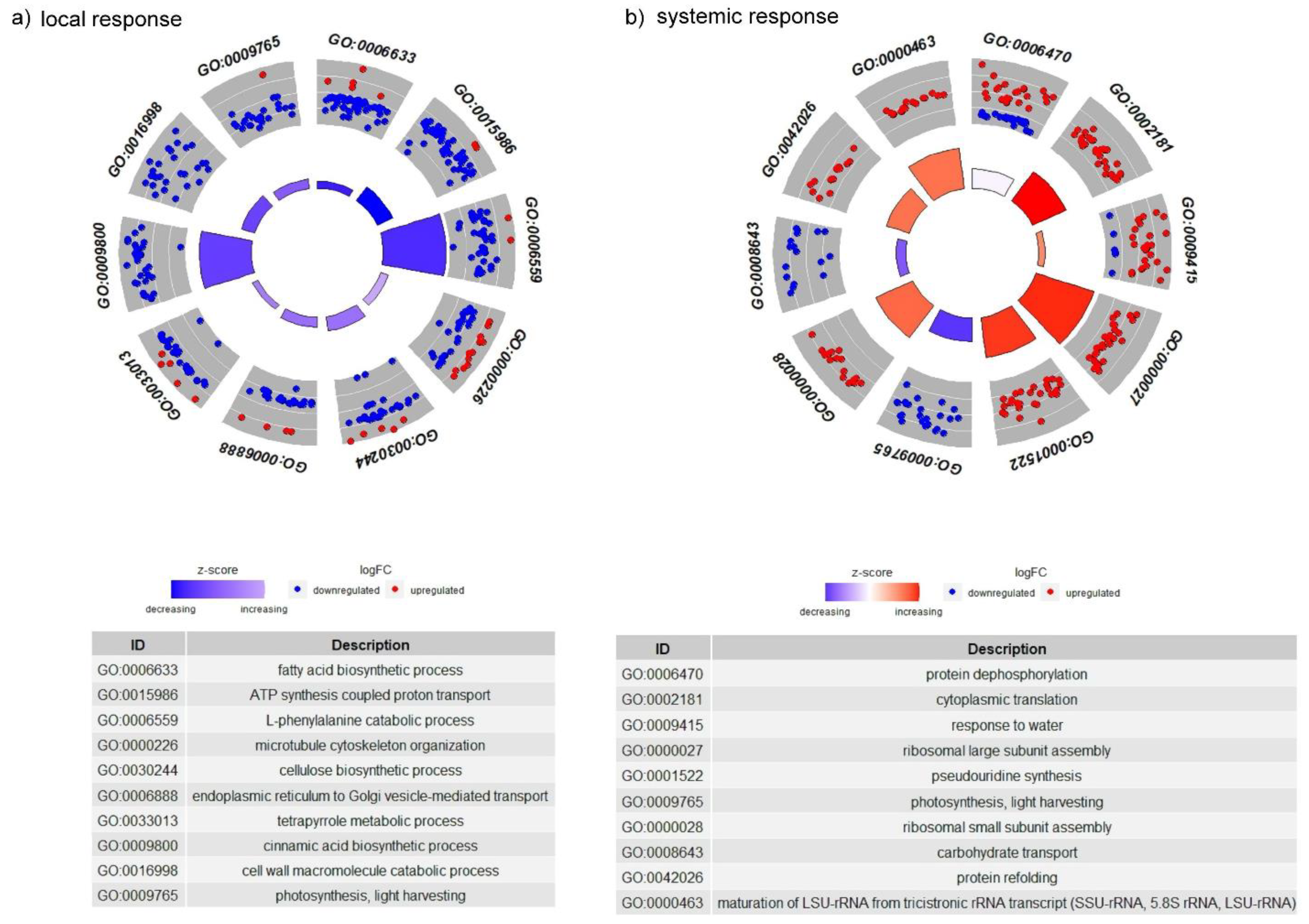
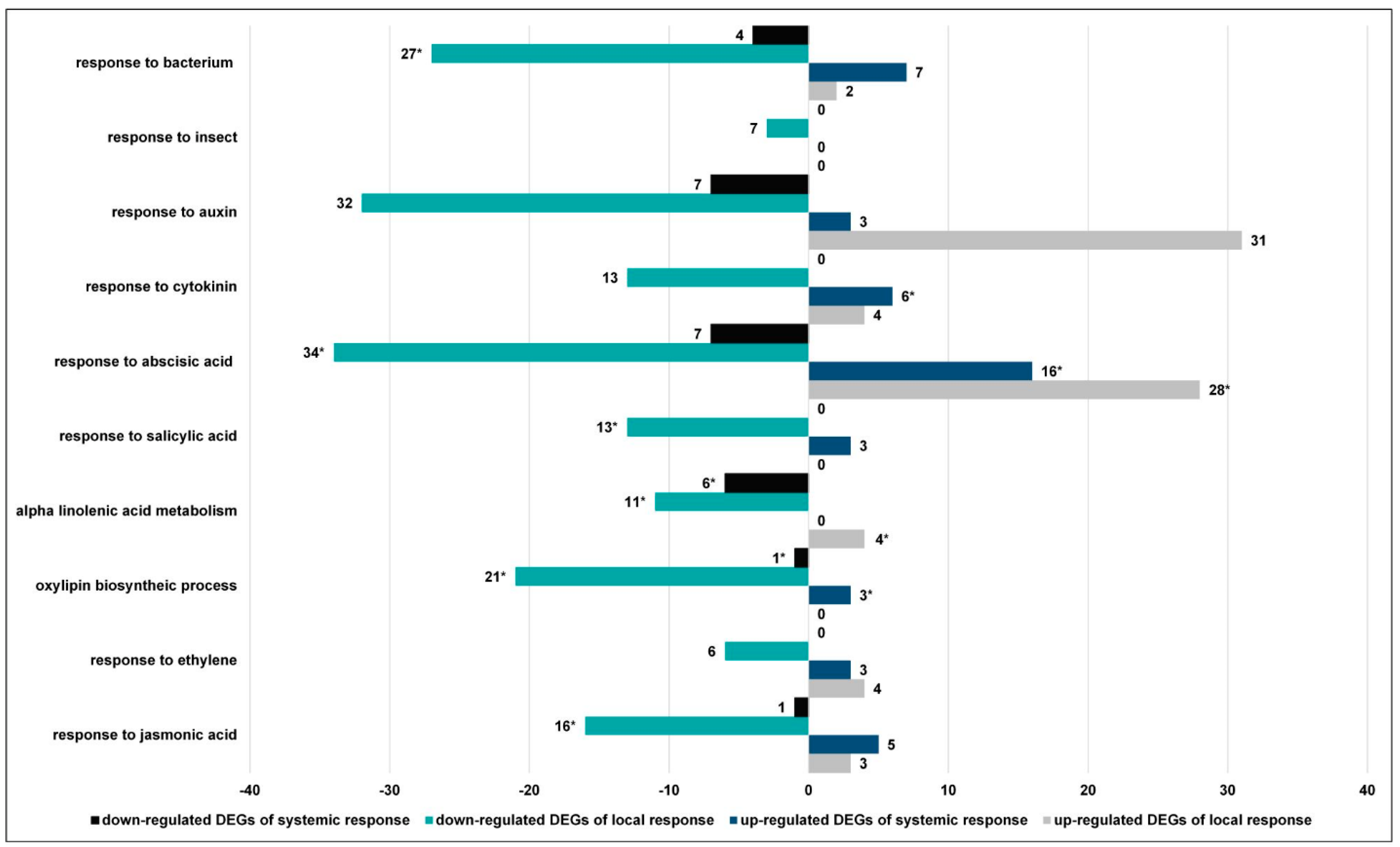
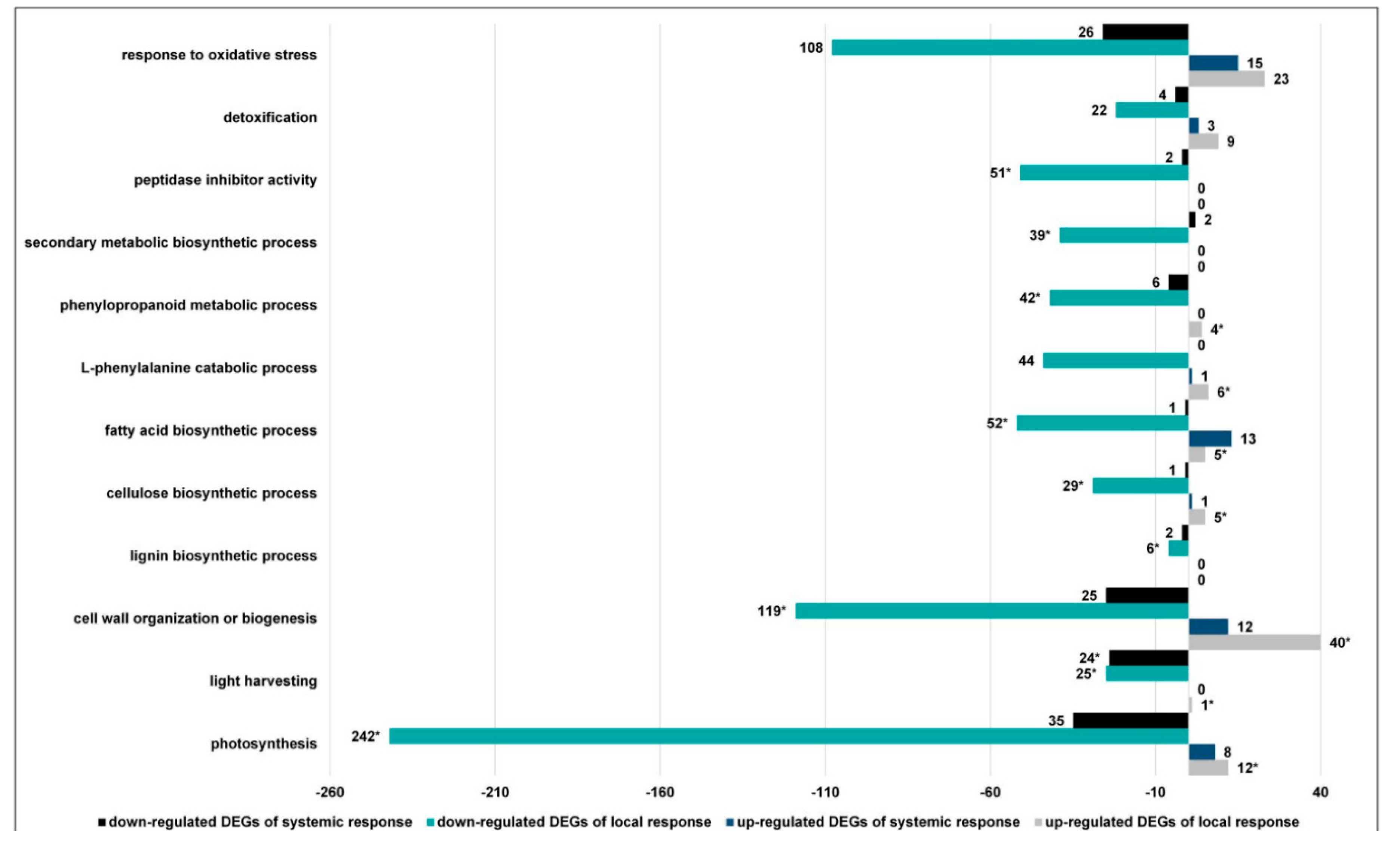
3.2.2. The GO Terms Most Abundantly Represented in the Plants Exposed to Larvae with Natural Intestinal Microflora
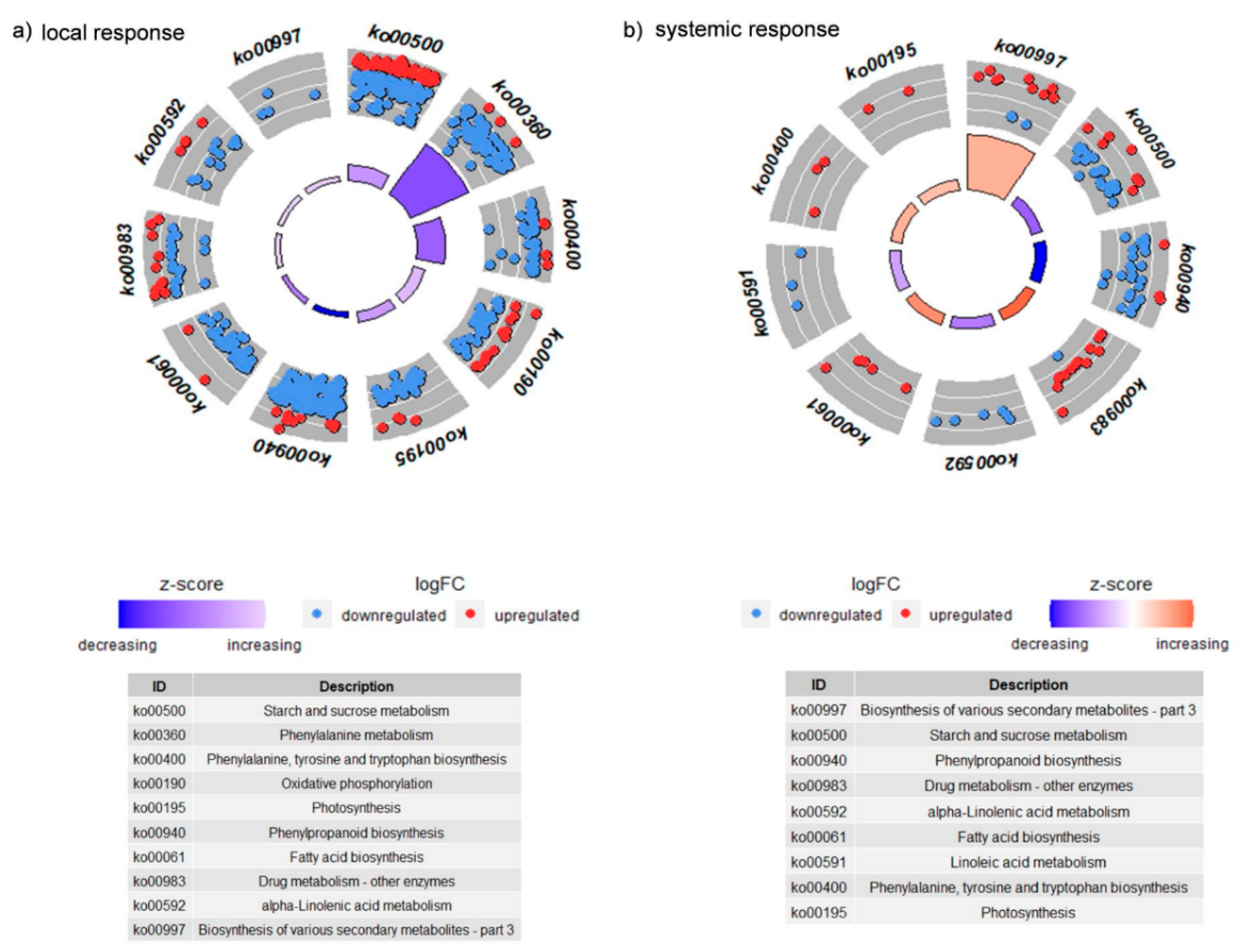
3.2.3. KEGGs Pathways Analysis
3.2.4. CLB Larvae-Associated Bacteria Modulate the Plant Signaling Pathways
3.2.5. CLB Larvae-Associated Bacteria Modulate Processes Associated with the Synthesis of Antifeedant Proteins, Photosynthesis, and Plant Cell Wall Organization
3.2.6. Genes Revealing the Opposite Direction of Expression in Response to Feeding of Larvae with Natural and Reduced Bacterial Flora
3.2.7. Validation of Transcriptome Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qiao, F.; Kong, L.A.; Peng, H.; Huang, W.K.; Wu, D.Q.; Liu, S.M.; Clarke, J.L.; Qiu, D.W.; Peng, D.L. Transcriptional Profiling of Wheat (Triticum aestivum L.) during a Compatible Interaction with the Cereal Cyst Nematode Heterodera avenae. Sci. Rep. 2019, 9, 2184. [Google Scholar] [CrossRef] [PubMed]
- Wielkopolan, B.; Jakubowska, M.; Obrępalska-Stęplowska, A. Beetles as Plant Pathogen Vectors. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, K.; Wielkopolan, B.; Obrępalska-Stęplowska, A. Pantoea ananatis, a New Bacterial Pathogen Affecting Wheat Plants (Triticum L.) in Poland. Pathogens 2020, 9, 1079. [Google Scholar] [CrossRef] [PubMed]
- Elimem, M.; Lahfef, C.; Limem-Sellemi, E. The Emerging Problem in Cereal Crops in North-Eastern Tunisia: The Cereal Leaf Beetles Oulema spp. (Coleoptera; Chrysomelidae; Criocerinae) Dynamic Populations and Infestation Rate. J. Entomol. Zool. Stud. 2022, 10, 9–12. [Google Scholar] [CrossRef]
- Ren, Z.; Liu, J.; Din, G.M.U.; Zhang, H.; Du, Z.; Chen, W.; Liu, T.; Zhang, J.; Zhao, S.; Gao, L. Transcriptome Analysis of Wheat Spikes in Response to Tilletia controversa Kühn Which Cause Wheat Dwarf Bunt. Sci. Rep. 2020, 21567. [Google Scholar] [CrossRef]
- War, A.R.; Taggar, G.K.; Hussain, B.; Taggar, M.S.; Nair, R.M.; Sharma, H.C. Special Issue: Using Non-Model Systems to Explore Plant-Pollinator and Plant-Herbivore Interactions: Plant Defence against Herbivory and Insect Adaptations. AoB Plants 2018, 10. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Anderson, J.P.; Singh, K.B. Plant Defence Responses: What Have We Learnt from Arabidopsis? Funct. Plant Biol. 2005, 32, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Fernández de Bobadilla, M.; Bourne, M.E.; Bloem, J.; Kalisvaart, S.N.; Gort, G.; Dicke, M.; Poelman, E.H. Insect Species Richness Affects Plant Responses to Multi-Herbivore Attack. New Phytol. 2021, 231, 2333–2345. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Varsani, S.; Louis, J. Altering Plant Defenses: Herbivore-Associated Molecular Patterns and Effector Arsenal of Chewing Herbivores. Mol. Plant-Microbe Interact. 2018, 31, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Reymond, P.; Bodenhausen, N.; van Poecke, R.M.P.; Krishnamurthy, V.; Dicke, M.; Farmer, E.E. A Conserved Transcript Pattern in Response to a Specialist and a Generalist Herbivore. Plant Cell 2004, 16, 3132–3147. [Google Scholar] [CrossRef] [PubMed]
- Halitschke, R.; Gase, K.; Hui, D.; Schmidt, D.D.; Baldwin, I.T. Molecular Interactions between the Specialist Herbivore Manduca sexta (Lepidoptera, Sphingidae) and Its Natural Host Nicotiana attenuata. VI. Microarray Analysis Reveals That Most Herbivore-Specific Transcriptional Changes Are Mediated by Fatty Acid-Amino Acid Conjugates. Plant Physiol. 2003, 131, 1894–1902. [Google Scholar] [CrossRef] [PubMed]
- Wielkopolan, B.; Krawczyk, K.; Szabelska-Beręsewicz, A.; Obrępalska-Stęplowska, A. The Structure of the Cereal Leaf Beetle (Oulema melanopus) Microbiome Depends on the Insect’s Developmental Stage, Host Plant, and Origin. Sci. Rep. 2021, 11, 20496. [Google Scholar] [CrossRef]
- Wielkopolan, B.; Obrępalska-Stęplowska, A. Three-Way Interaction among Plants, Bacteria, and Coleopteran Insects. Planta 2016, 244, 313–332. [Google Scholar] [CrossRef] [PubMed]
- Giron, D.; Dedeine, F.; Dubreuil, G.; Huguet, E.; Mouton, L.; Outreman, Y.; Vavre, F.; Simon, J.-C. Influence of Microbial Symbionts on Plant-Insect Interactions. Adv. Bot. Res. 2017, 81, 978. [Google Scholar] [CrossRef]
- Body, M.; Kaiser, W.; Dubreuil, G.; Casas, J.; Giron, D. Leaf-Miners Co-Opt Microorganisms to Enhance Their Nutritional Environment. J. Chem. Ecol. 2013, 39, 969–977. [Google Scholar] [CrossRef]
- Chung, S.H.; Rosa, C.; Scully, E.D.; Peiffer, M.; Tooker, J.F.; Hoover, K.; Luthe, D.S.; Felton, G.W. Herbivore Exploits Orally Secreted Bacteria to Suppress Plant Defenses. Proc. Natl. Acad. Sci. USA 2013, 110, 15728–15733. [Google Scholar] [CrossRef] [PubMed]
- Sugio, A.; Dubreuil, G.; Giron, D.; Simon, J.C. Plant-Insect Interactions under Bacterial Influence: Ecological Implications and Underlying Mechanisms. J. Exp. Bot. 2015, 66, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, N.; Li, W.; Gao, X.; Cha, M.; Qin, L.; Liu, L. Advances in Transcriptomics in the Response to Stress in Plants. Glob. Med. Genet. 2020, 7, 030–034. [Google Scholar] [CrossRef] [PubMed]
- Wielkopolan, B.; Krawczyk, K.; Obrępalska-Stęplowska, A. Gene Expression of Serine and Cysteine Proteinase Inhibitors during Cereal Leaf Beetle Larvae Feeding on Wheat: The Role of Insect-Associated Microorganisms. Arthropod-Plant Interact. 2018, 12, 601–612. [Google Scholar] [CrossRef]
- Jiang, H.; Lei, R.; Ding, S.-W.; Zhu, S. Skewer: A Fast and Accurate Adapter Trimmer for next-Generation Sequencing Paired-End Reads. BMC Bioinformatics 2014, 15, 182. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. Others FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 January 2021).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Oncology (GO) Database and Informatics Resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Walter, W.; Sánchez-Cabo, F.; Ricote, M. GOplot: An R Package for Visually Combining Expression Data with Functional Analysis. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef] [PubMed]
- Giménez, M.J.; Pistón, F.; Atienza, S.G. Identification of Suitable Reference Genes for Normalization of QPCR Data in Comparative Transcriptomics Analyses in the Triticeae. Planta 2011, 233, 163–173. [Google Scholar] [CrossRef]
- Halitschke, R.; Schittko, U.; Pohnert, G.; Boland, W.; Baldwin, I.T. Molecular Interactions between the Specialist Herbivore Manduca sexta (Lepidoptera, Sphingidae) and Its Natural Host Nicotiana attenuata. III. Fatty Acid-Amino Acid Conjugates in Herbivore Oral Secretions Are Necessary and Sufficient for Herbivore-Specific Plant Responses. Plant Physiol. 2001, 125, 711–717. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schittko, U.; Hermsmeier, D.; Baldwin, I.T. Molecular Interactions between the Specialist Herbivore Manduca sexta (Lepidoptera, Sphingidae) and Its Natural Host Nicotiana attenuata. II. Accumulation of Plant MRNAs in Response to Insect-Derived Cues. Plant Physiol. 2001, 125, 701–710. [Google Scholar] [CrossRef] [PubMed] [PubMed Central][Green Version]
- Poelman, E.H.; Kessler, A. Keystone Herbivores and the Evolution of Plant Defenses. Trends Plant Sci. 2016, 21, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth-Defense Tradeoffs in Plants: A Balancing Act to Optimize Fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef]
- Zhang, Y.; Fu, Y.; Wang, Q.; Liu, X.; Li, Q.; Chen, J. Transcriptome Analysis Reveals Rapid Defence Responses in Wheat Induced by Phytotoxic Aphid Schizaphis graminum Feeding. BMC Genomics 2020, 21, 339. [Google Scholar] [CrossRef] [PubMed]
- Alon, M.; Malka, O.; Eakteiman, G.; Elbaz, M.; Moyal Ben Zvi, M.; Vainstein, A.; Morin, S. Activation of the Phenylpropanoid Pathway in Nicotiana Tabacum Improves the Performance of the Whitefly Bemisia tabaci via Reduced Jasmonate Signaling. PLoS ONE 2013, 8, e76619. [Google Scholar] [CrossRef]
- Arena, G.D.; Ramos-González, P.L.; Rogerio, L.A.; Ribeiro-Alves, M.; Casteel, C.L.; Freitas-Astúa, J.; Machado, M.A. Making a Better Home: Modulation of Plant Defensive Response by Brevipalpus mites. Front. Plant Sci. 2018, 9, 1147. [Google Scholar] [CrossRef]
- Wu, J.; Baldwin, I.T. New Insights into Plant Responses to the Attack from Insect Herbivores. Annu. Rev. Genet. 2010, 44, 1–24. [Google Scholar] [CrossRef]
- Rosahl, S. Lipoxygenases in Plants-Their Role in Development and Stress Response. Z Naturforsch C J. Biosci. 1996, 51, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Wager, A.; Browse, J. Social Network: JAZ Protein Interactions Expand Our Knowledge of Jasmonate Signaling. Front. Plant Sci. 2012, 3, 41. [Google Scholar] [CrossRef] [PubMed]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging Insights into the Functions of Pathogenesis-Related Protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Alba, J.M.; Schimmel, B.C.J.; Glas, J.J.; Ataide, L.M.S.; Pappas, M.L.; Villarroel, C.A.; Schuurink, R.C.; Sabelis, M.W.; Kant, M.R. Spider Mites Suppress Tomato Defenses Downstream of Jasmonate and Salicylate Independently of Hormonal Crosstalk. New Phytol. 2015, 205, 828–840. [Google Scholar] [CrossRef]
- Zhang, P.J.; di Li, W.; Huang, F.; Zhang, J.M.; Xu, F.C.; Lu, Y. bin Feeding by Whiteflies Suppresses Downstream Jasmonic Acid Signaling by Eliciting Salicylic Acid Signaling. J. Chem. Ecol. 2013, 39, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Groen, S.C.; Humphrey, P.T.; Chevasco, D.; Ausubel, F.M.; Pierce, N.E.; Whiteman, N.K. Pseudomonas syringae Enhances Herbivory by Suppressing the Reactive Oxygen Burst in Arabidopsis. J. Insect Physiol. 2016, 84, 90–102. [Google Scholar] [CrossRef]
- Kim, D.S.; Hwang, B.K. An Important Role of the Pepper Phenylalanine Ammonia-Lyase Gene (PAL1) in Salicylic Acid-Dependent Signalling of the Defence Response to Microbial Pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zhao, S.; Wang, Z.; Wang, X.; Zhang, X.; Lee, Y.; Xi, J. Quantitative Proteomics Suggests Changes in the Carbohydrate Metabolism of Maize in Response to Larvae of the Belowground Herbivore Holotrichia parallela. PeerJ 2020, 8, e9819. [Google Scholar] [CrossRef]
- Napoleão, T.H.; Albuquerque, L.P.; Santos, N.D.L.; Nova, I.C.V.; Lima, T.A.; Paiva, P.M.G.; Pontual, E.V. Insect Midgut Structures and Molecules as Targets of Plant-Derived Protease Inhibitors and Lectins. Pest Manag. Sci. 2019, 75, 1212–1222. [Google Scholar] [CrossRef] [PubMed]
- Clemente, M.; Corigliano, M.G.; Pariani, S.A.; Sánchez-López, E.F.; Sander, V.A.; Ramos-Duarte, V.A. Plant Serine Protease Inhibitors: Biotechnology Application in Agriculture and Molecular Farming. Int. J. Mol. Sci. 2019, 20, 1345. [Google Scholar] [CrossRef]
- Wielkopolan, B.; Walczak, F.; Podleśny, A.; Nawrot, R.; Obrepalska-Steplowska, A. Identification and Partial Characterization of Proteases in Larval Preparations of the Cereal Leaf Beetle (Oulema melanopus, Chrysomelidae, Coleoptera). Arch. Insect Biochem. Physiol. 2015, 88, 192–202. [Google Scholar] [CrossRef] [PubMed]
- War, A.R.; Paulraj, M.G.; Ahmad, T.; Buhroo, A.A.; Hussain, B.; Ignacimuthu, S.; Sharma, H.C. Mechanisms of Plant Defense against Insect Herbivores. Plant Signal. Behav. 2012, 7, 192–202. [Google Scholar] [CrossRef]
- He, J.; Chen, F.; Chen, S.; Lv, G.; Deng, Y.; Fang, W.; Liu, Z.; Guan, Z.; He, C. Chrysanthemum Leaf Epidermal Surface Morphology and Antioxidant and Defense Enzyme Activity in Response to Aphid Infestation. J. Plant Physiol. 2011, 168, 687–693. [Google Scholar] [CrossRef]
- Heng-Moss, T.; Sarath, G.; Baxendale, F.; Novak, D.; Bose, S.; Ni, X.; Quisenberry, S. Characterization of Oxidative Enzyme Changes in Buffalograsses Challenged by Blissus occiduus. J. Econ. Entomol. 2004, 97, 1086–1095. [Google Scholar] [CrossRef]
- Sethi, A.; McAuslane, H.J.; Rathinasabapathi, B.; Nuessly, G.S.; Nagata, R.T. Enzyme Induction as a Possible Mechanism for Latex-Mediated Insect Resistance in Romaine Lettuce. J. Chem. Ecol. 2009, 35, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Stout, M.J.; Riggio, M.R.; Yang, Y. Direct Induced Resistance in Oryza sativa to Spodoptera frugiperda. Environ. Entomol. 2009, 38, 1174–1181. [Google Scholar] [CrossRef] [PubMed]
- Gulsen, O.; Eickhoff, T.; Heng-Moss, T.; Shearman, R.; Baxendale, F.; Sarath, G.; Lee, D. Characterization of Peroxidase Changes in Resistant and Susceptible Warm-Season Turfgrasses Challenged by Blissus Occiduus. Arthropod-Plant Interact. 2010, 4, 45–55. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Contribution of Phenylpropanoid Metabolism to Plant Development and Plant–Environment Interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef] [PubMed]
- Obrępalska-Stęplowska, A.; Zmienko, A.; Wrzesińska, B.; Goralski, M.; Figlerowicz, M.; Zyprych-Walczak, J.; Siatkowski, I.; Pospieszny, H. The Defense Response of Nicotiana benthamiana to Peanut Stunt Virus Infection in the Presence of Symptom Exacerbating Satellite RNA. Viruses 2018, 10, 449. [Google Scholar] [CrossRef] [PubMed]
- Kerchev, P.I.; Fenton, B.; Foyer, C.H.; Hancock, R.D. Plant Responses to Insect Herbivory: Interactions between Photosynthesis, Reactive Oxygen Species and Hormonal Signalling Pathways. Plant Cell Environ. 2012, 35, 441–453. [Google Scholar] [CrossRef] [PubMed]


| Local Response | |||||
| Up-Regulated DEGs in Response to CLB with Natural Bacterial Flora Down-Regulated DEGs in Response to CLB with a Reduced Bacteria Content | Down-Regulated DEGs in Response to CLB with Natural Bacterial Flora Up-Regulated DEGs in Response to CLB with a Reduced Bacteria Content | ||||
| GO ID | Go Name | # Seq | GO ID | Go Name | # Seq |
| GO:0006470 | protein dephosphorylation | 5 | GO:0071554 | cell wall organization or biogenesis | 6 |
| GO:0009737 | response to abscisic acid | 2 | GO:0006979 | response to oxidative stress | 4 |
| GO:0015979 | photosynthesis | 4 | |||
| GO:0030244 | cellulose biosynthetic process | 3 | |||
| Systemic Response | |||||
| GO:0006470 | protein dephosphorylation | 2 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wielkopolan, B.; Frąckowiak, P.; Wieczorek, P.; Obrępalska-Stęplowska, A. The Impact of Oulema melanopus—Associated Bacteria on the Wheat Defense Response to the Feeding of Their Insect Hosts. Cells 2022, 11, 2342. https://doi.org/10.3390/cells11152342
Wielkopolan B, Frąckowiak P, Wieczorek P, Obrępalska-Stęplowska A. The Impact of Oulema melanopus—Associated Bacteria on the Wheat Defense Response to the Feeding of Their Insect Hosts. Cells. 2022; 11(15):2342. https://doi.org/10.3390/cells11152342
Chicago/Turabian StyleWielkopolan, Beata, Patryk Frąckowiak, Przemysław Wieczorek, and Aleksandra Obrępalska-Stęplowska. 2022. "The Impact of Oulema melanopus—Associated Bacteria on the Wheat Defense Response to the Feeding of Their Insect Hosts" Cells 11, no. 15: 2342. https://doi.org/10.3390/cells11152342
APA StyleWielkopolan, B., Frąckowiak, P., Wieczorek, P., & Obrępalska-Stęplowska, A. (2022). The Impact of Oulema melanopus—Associated Bacteria on the Wheat Defense Response to the Feeding of Their Insect Hosts. Cells, 11(15), 2342. https://doi.org/10.3390/cells11152342







