Transcriptional and Epigenetic Consequences of DMSO Treatment on HepaRG Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Cell Culture
2.3. Real-Time RT-PCR
2.4. Microarray Analysis
2.5. Ingenuity Pathways Analysis
2.6. Gene Set Enrichment Analysis
2.7. Connectivity Map
2.8. Chromatin Immunoprecipitation Followed by Next Generation Sequencing (ChIP-seq)
2.9. ChIP-seq Data Analysis
2.10. Immunoblotting
2.11. Immunocytochemistry
2.12. Extracellular Matrix Deposition Staining
2.13. DNA Synthesis
2.14. Cytochrome P450 Activities
2.15. Statistical Analysis
3. Results
3.1. HepaRG Progenitors Spontaneously Differentiate into Biliary- and Hepatocyte-like Cells
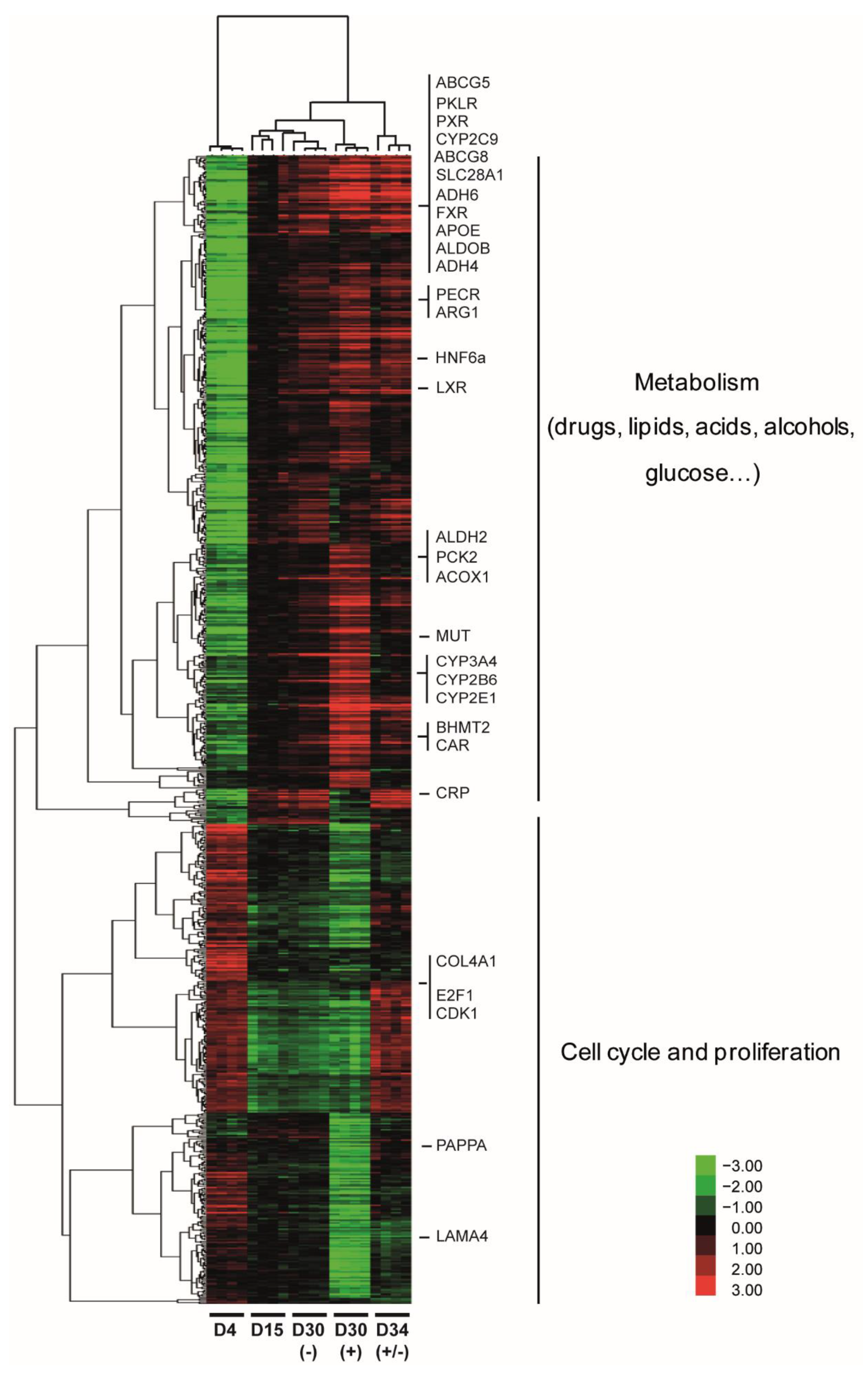
3.2. Pleiotropic Effects of DMSO Enhance Several Hepatic Specific Functions in HepaRG Cells
3.3. DMSO Plays a Role in Matrix and Cytoskeleton Remodeling and Has Anti-Inflammatory Properties
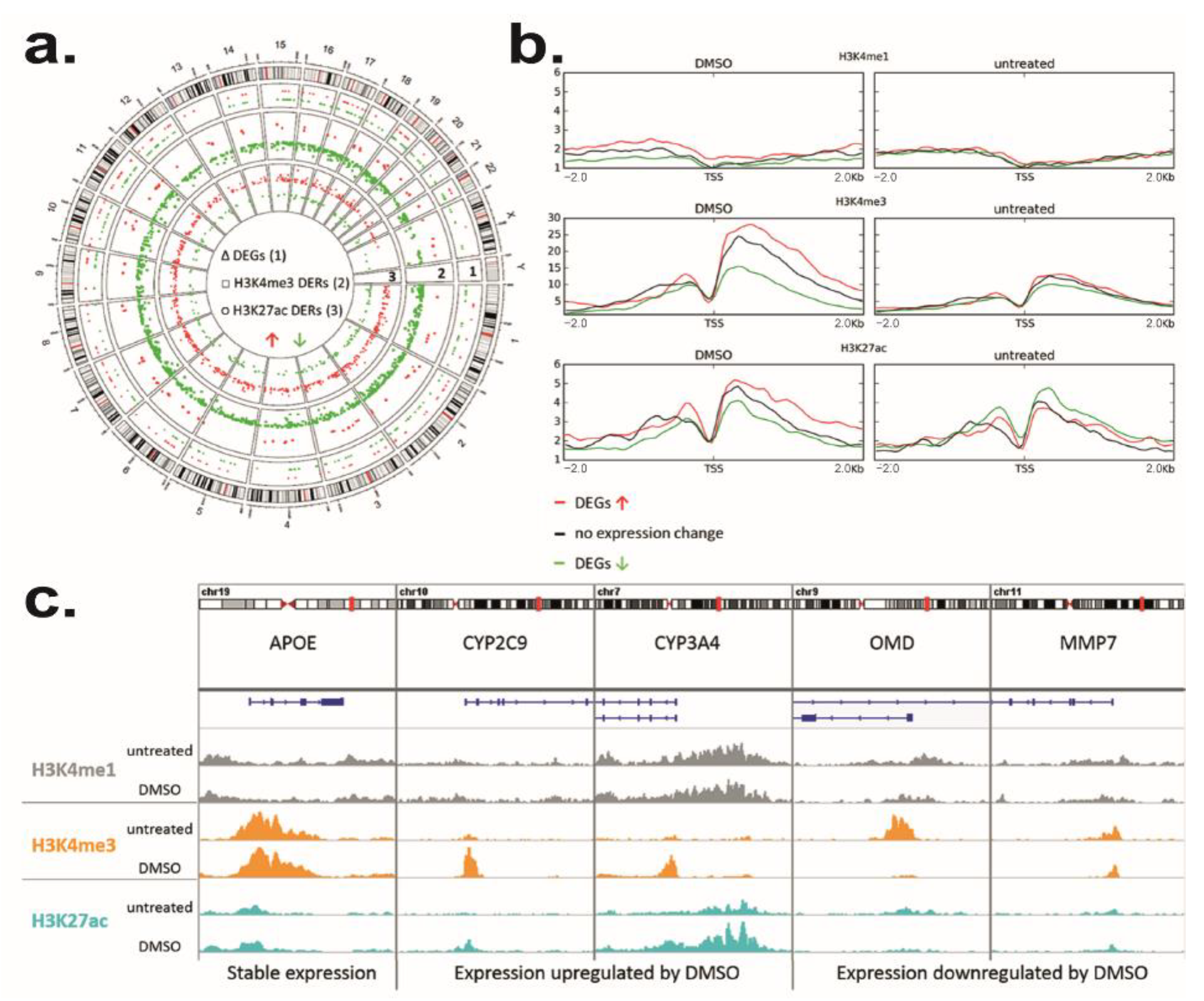
3.4. DMSO Induces Genome-Wide Histone Modification Patterns during HepaRG Differentiation
3.5. Identification of Molecules That Could Mimic or Reverse the DMSO Effect
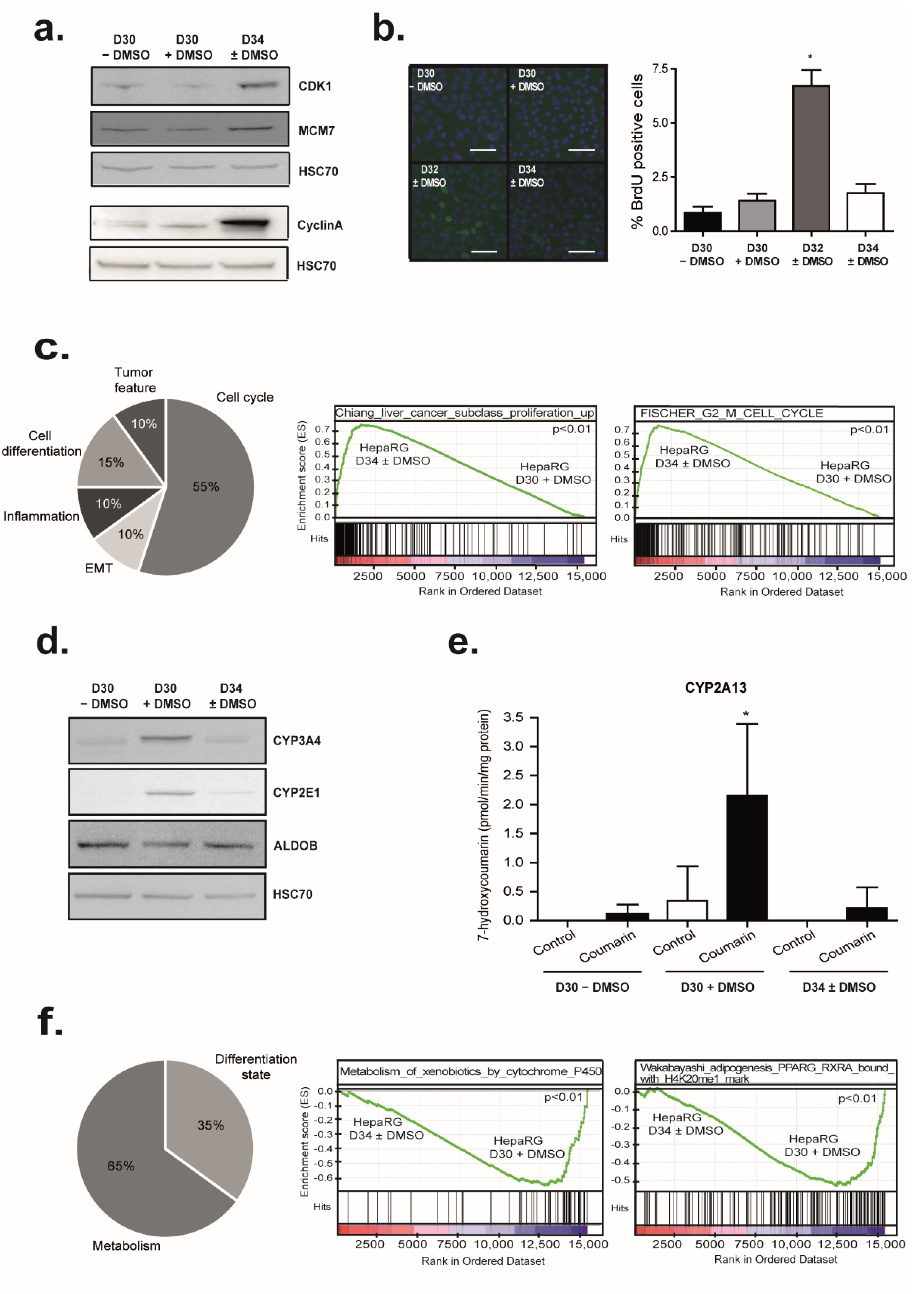
3.6. DMSO Removal Induces Transient Cell Proliferation
3.7. DMSO Removal Differentially Modulates Some Metabolic Functions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guillouzo, A. Liver cell models in in vitro toxicology. Environ. Health Perspect. 1998, 106, 511–532. [Google Scholar] [PubMed]
- Guillouzo, A.; Morel, F.; Fardel, O.; Meunier, B. Use of human hepatocyte cultures for drug metabolism studies. Toxicology 1993, 82, 209–219. [Google Scholar] [CrossRef]
- LeCluyse, E.L. Human hepatocyte culture systems for the in vitro evaluation of cytochrome P450 expression and regulation. Eur. J. Pharm. Sci. 2001, 13, 343–368. [Google Scholar] [CrossRef]
- Guillouzo, A.; Corlu, A.; Aninat, C.; Glaise, D.; Morel, F.; Guguen-Guillouzo, C. The human hepatoma HepaRG cells: A highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem.-Biol. Interact. 2007, 168, 66–73. [Google Scholar] [CrossRef]
- Hart, S.N.; Li, Y.; Nakamoto, K.; Subileau, E.; Steen, D.; Zhong, X. A Comparison of Whole Genome Gene Expression Profiles of HepaRG Cells and HepG2 Cells to Primary Human Hepatocytes and Human Liver Tissues. Drug Metab. Dispos. 2010, 38, 988–994. [Google Scholar] [CrossRef]
- Parmentier, C.; Hendriks, D.F.G.; Heyd, B.; Bachellier, P.; Ingelman-Sundberg, M.; Richert, L. Inter-individual differences in the susceptibility of primary human hepatocytes towards drug-induced cholestasis are compound and time dependent. Toxicol. Lett. 2018, 295, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Rogue, A.; Lambert, C.; Spire, C.; Claude, N.; Guillouzo, A. Interindividual Variability in Gene Expression Profiles in Human Hepatocytes and Comparison with HepaRG Cells. Drug Metab. Dispos. 2012, 40, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Ma, X.; Zou, W.; Wang, C.; Bahbahan, I.S.; Ahuja, T.P.; Tolstikov, V.; Zern, M.A. Differentiation and characterization of metabolically functioning hepatocytes from human embryonic stem cells. Stem Cells 2010, 28, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Touboul, T.; Hannan, N.R.F.; Corbineau, S.; Martinez, A.; Martinet, C.; Branchereau, S.; Mainot, S.; Strick-Marchand, H.; Pedersen, R.; Di Santo, J.; et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 2010, 51, 1754–1765. [Google Scholar] [CrossRef]
- Donato, M.T.; Tolosa, L. Stem-cell derived hepatocyte-like cells for the assessment of drug-induced liver injury. Differentiation 2019, 106, 15–22. [Google Scholar] [CrossRef]
- Ramaswamy, S.; Tonnu, N.; Menon, T.; Lewis, B.M.; Green, K.T.; Wampler, D.; Monahan, P.E.; Verma, I.M. Autologous and Heterologous Cell Therapy for Hemophilia B toward Functional Restoration of Factor IX. Cell Rep. 2018, 23, 1565–1580. [Google Scholar] [CrossRef] [PubMed]
- Tolosa, L.; Caron, J.; Hannoun, Z.; Antoni, M.; López, S.; Burks, D.; Castell, J.V.; Weber, A.; Gomez-Lechon, M.-J.; Dubart-Kupperschmitt, A. Transplantation of hESC-derived hepatocytes protects mice from liver injury. Stem Cell Res. Ther. 2015, 6, 246. [Google Scholar] [CrossRef] [PubMed]
- Camp, J.G.; Sekine, K.; Gerber, T.; Loeffler-Wirth, H.; Binder, H.L.-W.H.; Gac, M.; Kanton, S.; Kageyama, J.; Damm, G.; Seehofer, G.D.D.; et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 2017, 546, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.; Mizuguchi, H. Generation of human pluripotent stem cell-derived hepatocyte-like cells for drug toxicity screening. Drug Metab. Pharmacokinet. 2017, 32, 12–20. [Google Scholar] [CrossRef]
- Gao, X.; Liu, Y. A transcriptomic study suggesting human iPSC-derived hepatocytes potentially offer a better in vitro model of hepatotoxicity than most hepatoma cell lines. Cell Biol. Toxicol. 2017, 33, 407–421. [Google Scholar] [CrossRef]
- Gerets, H.H.J.; Tilmant, K.; Gerin, B.; Chanteux, H.; Depelchin, B.O.; Dhalluin, S.; Atienzar, F.A. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol. Toxicol. 2012, 28, 69–87. [Google Scholar] [CrossRef]
- Liu, Y.; Flynn, T.J.; Xia, M.; Wiesenfeld, P.L.; Ferguson, M.S. Evaluation of CYP3A4 inhibition and hepatotoxicity using DMSO-treated human hepatoma HuH-7 cells. Cell Biol. Toxicol. 2015, 31, 221–230. [Google Scholar] [CrossRef]
- Aninat, C.; Piton, A.; Glaise, D.; Le Charpentier, T.; Langouët, S.; Morel, F.; Guguen-Guillouzo, C.; Guillouzo, A. Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab. Dispos. 2006, 34, 75–83. [Google Scholar] [CrossRef]
- Anthérieu, S.; Chesné, C.; Li, R.; Guguen-Guillouzo, C.; Guillouzo, A. Optimization of the HepaRG cell model for drug metabolism and toxicity studies. Toxicol. In Vitro 2012, 26, 1278–1285. [Google Scholar] [CrossRef]
- Allard, J.; Bucher, S.; Massart, J.; Ferron, P.-J.; Le Guillou, D.; Loyant, R.; Daniel, Y.; Launay, Y.; Buron, N.; Begriche, K.; et al. Drug-induced hepatic steatosis in absence of severe mitochondrial dysfunction in HepaRG cells: Proof of multiple mechanism-based toxicity. Cell Biol. Toxicol. 2021, 37, 151–175. [Google Scholar] [CrossRef]
- Bucher, S.; Jalili, P.; Le Guillou, D.; Begriche, K.; Rondel, K.; Martinais, S.; Zalko, D.; Corlu, A.; Robin, M.-A.; Fromenty, B. Bisphenol a induces steatosis in HepaRG cells using a model of perinatal exposure. Environ. Toxicol. 2017, 32, 1024–1036. [Google Scholar] [CrossRef] [PubMed]
- Kanebratt, K.P.; Andersson, T.B. Evaluation of HepaRG Cells as an in Vitro Model for Human Drug Metabolism Studies. Drug Metab. Dispos. 2008, 36, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Michaut, A.; Le Guillou, D.; Moreau, C.; Bucher, S.; McGill, M.R.; Martinais, S.; Gicquel, T.; Morel, I.; Robin, M.-A.; Jaeschke, H.; et al. A cellular model to study drug-induced liver injury in nonalcoholic fatty liver disease: Application to acetaminophen. Toxicol. Appl. Pharmacol. 2016, 292, 40–55. [Google Scholar] [CrossRef]
- Chen, C.; Soto-Gutierrez, A.; Baptista, P.M.; Spee, B. Biotechnology Challenges to In Vitro Maturation of Hepatic Stem Cells. Gastroenterology 2018, 154, 1258–1272. [Google Scholar] [CrossRef] [PubMed]
- Guguen-Guillouzo, C.; Corlu, A.; Guillouzo, A. Stem cell-derived hepatocytes and their use in toxicology. Toxicology 2010, 270, 3–9. [Google Scholar] [CrossRef]
- Palakkan, A.A.; Nanda, J.; Ross, J.A. Pluripotent stem cells to hepatocytes, the journey so far. Biomed. Rep. 2017, 6, 367–373. [Google Scholar] [CrossRef]
- Padgham, C.R.W.; Paine, A.J.; Phillips, I.R.; Shephard, E.A. Maintenance of total cytochrome P-450 content in rat hepatocyte culture and the abundance of CYP1A2 and CYP2B1/2 mRNAs. Biochem. J. 1992, 285, 929–932. [Google Scholar] [CrossRef][Green Version]
- Isom, H.C.; Secott, T.; Georgoff, I.; Woodworth, C.; Mummaw, J. Maintenance of differentiated rat hepatocytes in primary culture. Proc. Natl. Acad. Sci. USA 1985, 82, 3252–3256. [Google Scholar] [CrossRef]
- Isom, I.; Georgoff, I.; Salditt-Georgieff, M.; Darnell, J.E. Persistence of liver-specific messenger RNA in cultured hepatocytes: Different regulatory events for different genes. J. Cell Biol. 1987, 105, 2877–2885. [Google Scholar] [CrossRef]
- Stoehr, S.A.; Isom, H.C. Gap junction-mediated intercellular communication in a long-term primary mouse hepatocyte culture system. Hepatology 2003, 38, 1125–1135. [Google Scholar] [CrossRef]
- Yoshizawa, T.; Watanabe, S.; Hirose, M.; Miyazaki, A.; Sato, N. Dimethylsulfoxide maintains intercellular communication by preserving the gap junctional protein connexin32 in primary cultured hepatocyte doublets from rats. J. Gastroenterol. Hepatol. 1997, 12, 325–330. [Google Scholar] [CrossRef] [PubMed]
- De La Vega, F.M.; Mendoza-Figureueroa, T. Dimethyl sulfoxide enhances lipid synthesis and secretion by long-term cultures of adult rat hepatocytes. Biochimie 1991, 73, 621–624. [Google Scholar] [CrossRef]
- Su, T.; Waxman, D.J. Impact of dimethyl sulfoxide on expression of nuclear receptors and drug-inducible cytochromes P450 in primary rat hepatocytes. Arch. Biochem. Biophys. 2004, 424, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Gilot, D.; Loyer, P.; Corlu, A.; Glaise, D.; Lagadic-Gossmann, D.; Atfi, A.; Morel, F.; Ichijo, H.; Guguen-Guillouzo, C. Liver protection from apoptosis requires both blockage of initiator caspase activities and inhibition of ASK1/JNK pathway via glutathione S-transferase regulation. J. Biol. Chem. 2002, 277, 49220–49229. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Sainz, B.; Corcoran, P.; Uprichard, S.; Jeong, H. Characterization of increased drug metabolism activity in dimethyl sulfoxide (DMSO)-treated Huh7 hepatoma cells. Xenobiotica 2009, 39, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Sainz, B.; Chisari, F.V. Production of infectious hepatitis C virus by well-differentiated, growth-arrested human hepatoma-derived cells. J. Virol. 2006, 80, 10253–10257. [Google Scholar] [CrossRef]
- Nikolaou, N.; Green, C.J.; Gunn, P.J.; Hodson, L.; Tomlinson, J.W. Optimizing human hepatocyte models for metabolic phenotype and function: Effects of treatment with dimethyl sulfoxide (DMSO). Physiol. Rep. 2016, 4, e12944. [Google Scholar] [CrossRef]
- Alizadeh, E.; Zarghami, N.; Eslaminejad, M.B.; Akbarzadeh, A.; Barzegar, A.; Mohammadi, S.A. The effect of dimethyl sulfoxide on hepatic differentiation of mesenchymal stem cells. Artif. Cells Nanomed. Biotechnol. 2016, 44, 157–164. [Google Scholar] [CrossRef]
- Basma, H.; Soto–Gutiérrez, A.; Yannam, G.R.; Liu, L.; Ito, R.; Yamamoto, T.; Ellis, E.; Carson, S.D.; Sato, S.; Chen, Y.; et al. Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology 2009, 136, 990–999. [Google Scholar] [CrossRef]
- Hay, D.C.; Zhao, D.; Ross, A.; Mandalam, R.; Lebkowski, J.; Cui, W. Direct differentiation of human embryonic stem cells to hepatocyte-like cells exhibiting functional activities. Cloning Stem Cells 2007, 9, 51–62. [Google Scholar] [CrossRef]
- Sivertsson, L.; Synnergren, J.; Jensen, J.; Björquist, P.; Ingelman-Sundberg, M. Hepatic Differentiation and Maturation of Human Embryonic Stem Cells Cultured in a Perfused Three-Dimensional Bioreactor. Stem Cells Dev. 2013, 22, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Chetty, S.; Pagliuca, F.W.; Honore, C.; Kweudjeu, A.; Rezania, A.; Melton, D.A. A simple tool to improve pluripotent stem cell differentiation. Nat. Methods 2013, 10, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Czysz, K.; Minger, S.; Thomas, N. DMSO efficiently down regulates pluripotency genes in human embryonic stem cells during definitive endoderm derivation and increases the proficiency of hepatic differentiation. PLoS ONE 2015, 10, e0117689. [Google Scholar] [CrossRef]
- Kang, M.-H.; You, S.-Y.; Hong, K.; Kim, J.-H. DMSO impairs the transcriptional program for maternal-to-embryonic transition by altering histone acetylation. Biomaterials 2020, 230, 119604. [Google Scholar] [CrossRef]
- Verheijen, M.; Lienhard, M.; Schrooders, Y.; Clayton, O.; Nudischer, R.; Boerno, S.; Timmermann, B.; Selevsek, N.; Schlapbach, R.; Gmuender, H.; et al. DMSO induces drastic changes in human cellular processes and epigenetic landscape in vitro. Sci. Rep. 2019, 9, 4641. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Ke, S.; Chen, J.; Ouyang, N.; Tian, Y. Epigenetic sensitization of pregnane X receptor-regulated gene expression by dimethyl sulfoxide. Toxicol. Lett. 2020, 321, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Gripon, P.; Rumin, S.; Urban, S.; Le Seyec, J.; Glaise, D.; Cannie, I.; Guyomard, C.; Lucas, J.; Trepo, C.; Guguen-Guillouzo, C. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. USA 2002, 99, 15655–15660. [Google Scholar] [CrossRef] [PubMed]
- Cerec, V.; Glaise, D.; Garnier, D.; Morosan, S.; Turlin, B.; Drenou, B.; Gripon, P.; Kremsdorf, D.; Guguen-Guillouzo, C.; Corlu, A. Transdifferentiation of hepatocyte-like cells from the human hepatoma HepaRG cell line through bipotent progenitor. Hepatology 2007, 45, 957–967. [Google Scholar] [CrossRef]
- Dan, Y.Y.; Riehle, K.J.; Lazaro, C.; Teoh, N.; Haque, J.; Campbell, J.S.; Fausto, N. Isolation of multipotent progenitor cells from human fetal liver capable of differentiating into liver and mesenchymal lineages. Proc. Natl. Acad. Sci. USA 2006, 103, 9912–9917. [Google Scholar] [CrossRef]
- Jennen, D.G.J.; Magkoufopoulou, C.; Ketelslegers, H.B.; van Herwijnen, M.H.M.; Kleinjans, J.C.S.; van Delft, J.H.M. Comparison of HepG2 and HepaRG by whole-genome gene expression analysis for the purpose of chemical hazard identification. Toxicol Sci. 2010, 115, 66–79. [Google Scholar] [CrossRef]
- Hoekstra, R.; Nibourg, G.A.; van der Hoeven, T.V.; Ackermans, M.T.; Hakvoort, T.B.; van Gulik, T.M.; Lamers, W.H.; Elferink, R.P.O.; Chamuleau, R.A. The HepaRG cell line is suitable for bioartificial liver application. Int. J. Biochem. Cell Biol. 2011, 43, 1483–1489. [Google Scholar] [CrossRef] [PubMed]
- Troadec, M.-B.; Glaise, D.; Lamirault, G.; Le Cunff, M.; Guérin, E.; Le Meur, N.; Détivaud, L.; Zindy, P.; Leroyer, P.; Guisle, I.; et al. Hepatocyte iron loading capacity is associated with differentiation and repression of motility in the HepaRG cell line. Genomics 2006, 87, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Buick, J.K.; Williams, A.; Gagné, R.; Swartz, C.D.; Recio, L.; Ferguson, S.S.; Yauk, C.L. Flow cytometric micronucleus assay and TGx-DDI transcriptomic biomarker analysis of ten genotoxic and non-genotoxic chemicals in human HepaRGTM cells. Genes Environ. 2020, 42, 5. [Google Scholar] [CrossRef] [PubMed]
- Josse, R.; Rogue, A.; Lorge, E.; Guillouzo, A. An adaptation of the human HepaRG cells to the in vitro micronucleus assay. Mutagenesis 2012, 27, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Savary, C.C.; Jiang, X.; Aubry, M.; Jossé, R.; Kopp-Schneider, A.; Hewitt, P.; Guillouzo, A. Transcriptomic analysis of untreated and drug-treated differentiated HepaRG cells over a 2-week period. Toxicol. In Vitro 2015, 30, 27–35. [Google Scholar] [CrossRef]
- Berriz, G.F.; Beaver, J.E.; Cenik, C.; Tasan, M.; Roth, F.P. Next generation software for functional trend analysis. Bioinformatics 2009, 25, 3043–3044. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.-P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef]
- Marco-Sola, S.; Sammeth, M.; Guigó, R.; Ribeca, P. The GEM mapper: Fast, accurate and versatile alignment by filtration. Nat. Methods 2012, 9, 1185–1188. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Shao, N.-Y.; Liu, X.; Maze, I.; Feng, J.; Nestler, E.J. diffReps: Detecting Differential Chromatin Modification Sites from ChIP-seq Data with Biological Replicates. Mantovani, R.; editor. PLoS ONE 2013, 8, e65598. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed]
- McLean, C.Y.; Bristor, D.; Hiller, M.; Clarke, S.L.; Schaar, B.T.; Lowe, C.B.; Wenger, A.M.; Bejerano, G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010, 28, 495–501. [Google Scholar] [CrossRef]
- Ramírez, F.; Dündar, F.; Diehl, S.; Grüning, B.A.; Manke, T. deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014, 42, W187–W191. [Google Scholar] [CrossRef]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.; Carey, V.J. Software for Computing and Annotating Genomic Ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef] [PubMed]
- Loyer, P.; Glaise, D.; Cariou, S.; Baffet, G.; Meijer, L.; Guguen-Guillouzo, C. Expression and activation of cdks (1 and 2) and cyclins in the cell cycle progression during liver regeneration. J. Biol. Chem. 1994, 269, 2491–2500. [Google Scholar] [CrossRef]
- Corlu, A.; Kneip, B.; Lhadi, C.; Leray, G.; Glaise, D.; Baffet, G.; Bourel, D.; Guguen-Guillouzo, C. A plasma membrane protein is involved in cell contact-mediated regulation of tissue-specific genes in adult hepatocytes. J. Cell Biol. 1991, 115, 505–515. [Google Scholar] [CrossRef]
- Donato, M.T.; Jiménez, N.; Castell, J.V.; Gómez-Lechón, M.J. Fluorescence-based assays for screening nine cytochrome P450 (P450) activities in intact cells expressing individual human P450 enzymes. Drug Metab. Dispos. 2004, 32, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Soucek, P. Novel sensitive high-performance liquid chromatographic method for assay of coumarin 7-hydroxylation. J. Chromatogr. B Biomed. Sci. Appl. 1999, 734, 23–29. [Google Scholar] [CrossRef]
- Iivanainen, A.; Sainio, K.; Sariola, H.; Tryggvason, K. Primary structure and expression of a novel human laminin alpha 4 chain. FEBS Lett. 1995, 365, 183–188. [Google Scholar] [CrossRef]
- Peart, M.J.; Smyth, G.K.; van Laar, R.K.; Bowtell, D.D.; Richon, V.M.; Marks, P.A.; Holloway, A.J.; Johnstone, R.W. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc. Natl. Acad. Sci. USA 2005, 102, 3697–3702. [Google Scholar] [CrossRef] [PubMed]
- Bruzzese, F.; Leone, A.; Rocco, M.; Carbone, C.; Piro, G.; Caraglia, M.; Di Gennaro, E.; Budillon, A. HDAC inhibitor vorinostat enhances the antitumor effect of gefitinib in squamous cell carcinoma of head and neck by modulating ErbB receptor expression and reverting EMT. J. Cell Physiol. 2011, 226, 2378–2390. [Google Scholar] [CrossRef] [PubMed]
- Hasibeder, A.; Venkataramani, V.; Thelen, P.; Radzun, H.-J.; Schweyer, S. Phytoestrogens regulate the proliferation and expression of stem cell factors in cell lines of malignant testicular germ cell tumors. Int. J. Oncol. 2013, 43, 1385–1394. [Google Scholar] [CrossRef] [PubMed]
- Kaimori, A.; Potter, J.J.; Choti, M.; Ding, Z.; Mezey, E.; Koteish, A.A. Histone deacetylase inhibition suppresses the transforming growth factor beta1-induced epithelial-to-mesenchymal transition in hepatocytes. Hepatology 2010, 52, 1033–1045. [Google Scholar] [CrossRef] [PubMed]
- Marks, P.A.; Breslow, R. Dimethyl sulfoxide to vorinostat: Development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 2007, 25, 84–90. [Google Scholar] [CrossRef]
- Leung, Y.Y.; Yao Hui, L.L.; Kraus, V.B. Colchicine—Update on mechanisms of action and therapeutic uses. Semin. Arthritis Rheum. 2015, 45, 341–350. [Google Scholar] [CrossRef]
- Li, J.J.; Lee, S.H.; Kim, D.K.; Jin, R.; Jung, D.-S.; Kwak, S.-J.; Kim, S.H.; Han, S.H.; Lee, J.E.; Moon, S.J.; et al. Colchicine attenuates inflammatory cell infiltration and extracellular matrix accumulation in diabetic nephropathy. Am. J. Physiol.-Ren. Physiol. 2009, 297, F200–F209. [Google Scholar] [CrossRef]
- Garnier, D.; Loyer, P.; Ribault, C.; Guguen-Guillouzo, C.; Corlu, A. Cyclin-dependent kinase 1 plays a critical role in DNA replication control during rat liver regeneration. Hepatology 2009, 50, 1946–1956. [Google Scholar] [CrossRef]
- Ishida, S.; Huang, E.; Zuzan, H.; Spang, R.; Leone, G.; West, M.; Nevins, J.R. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 2001, 21, 4684–4699. [Google Scholar] [CrossRef] [PubMed]
- Chiang, D.Y.; Villanueva, A.; Hoshida, Y.; Peix, J.; Newell, P.; Minguez, B.; LeBlanc, A.C.; Donovan, D.J.; Thung, S.N.; Solé, M.; et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008, 68, 6779–6788. [Google Scholar] [CrossRef]
- Fischer, M.; Grossmann, P.; Padi, M.; DeCaprio, J.A. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 2016, 44, 6070–6086. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, K.-I.; Okamura, M.; Tsutsumi, S.; Nishikawa, N.S.; Tanaka, T.; Sakakibara, I.; Kitakami, J.-I.; Ihara, S.; Hashimoto, Y.; Hamakubo, T.; et al. The peroxisome proliferator-activated receptor gamma/retinoid X receptor alpha heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol. Cell. Biol. 2009, 29, 3544–3555. [Google Scholar] [CrossRef] [PubMed]
- Friend, C.; Scher, W.; Holland, J.G.; Sato, T. Hemoglobin synthesis in murine virus-induced leukemic cells in vitro: Stimulation of erythroid differentiation by dimethyl sulfoxide. Proc. Natl. Acad. Sci. USA 1971, 68, 378–382. [Google Scholar] [CrossRef]
- Tunçer, S.; Gurbanov, R.; Sheraj, I.; Solel, E.; Esenturk, O.; Banerjee, S. Low dose dimethyl sulfoxide driven gross molecular changes have the potential to interfere with various cellular processes. Sci. Rep. 2018, 8, 14828. [Google Scholar] [CrossRef]
- Darling, D.; Tavassoli, M.; Linskens, M.H.; Farzaneh, F. DMSO induced modulation of c-myc steady-state RNA levels in a variety of different cell lines. Oncogene 1989, 4, 175–179. [Google Scholar] [PubMed]
- Santos, N.C.; Figureueira-Coelho, J.; Martins-Silva, J.; Saldanha, C. Multidisciplinary utilization of dimethyl sulfoxide: Pharmacological, cellular, and molecular aspects. Biochem. Pharmacol. 2003, 65, 1035–1041. [Google Scholar] [CrossRef]
- Villa, P.; Arioli, P.; Guaitani, A. Mechanism of maintenance of liver-specific functions by DMSO in cultured rat hepatocytes. Exp. Cell Res. 1991, 194, 157–160. [Google Scholar] [CrossRef]
- Zangar, R.C.; Novak, R.F. Posttranslational elevation of cytochrome P450 3A levels and activity by dimethyl sulfoxide. Arch. Biochem. Biophys. 1998, 353, 1–9. [Google Scholar] [CrossRef]
- Song, Y.M.; Song, S.-O.; Jung, Y.-K.; Kang, E.-S.; Cha, B.-S.; Lee, H.C.; Lee, B.-W. Dimethyl sulfoxide reduces hepatocellular lipid accumulation through autophagy induction. Autophagy 2012, 8, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Pagan, R.; Martín, I.; Llobera, M.; Vilaró, S. Epithelial-mesenchymal transition of cultured rat neonatal hepatocytes is differentially regulated in response to epidermal growth factor and dimethyl sulfoxide. Hepatology 1997, 25, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Cantley, L.C. The phosphoinositide 3-kinase pathway. Science 2002, 296, 1655–1657. [Google Scholar] [CrossRef] [PubMed]
- Dey, N.; Young, B.; Abramovitz, M.; Bouzyk, M.; Barwick, B.; De, P.; Leyland-Jones, B. Differential activation of Wnt-β-catenin pathway in triple negative breast cancer increases MMP7 in a PTEN dependent manner. PLoS ONE 2013, 8, e77425. [Google Scholar] [CrossRef]
- Bufu, T.; Di, X.; Yilin, Z.; Gege, L.; Xi, C.; Ling, W. Celastrol inhibits colorectal cancer cell proliferation and migration through suppression of MMP3 and MMP7 by the PI3K/AKT signaling pathway. Anticancer Drugs 2018, 29, 530–538. [Google Scholar] [CrossRef]
- Hu, H.; Juvekar, A.; Lyssiotis, C.A.; Lien, E.C.; Albeck, J.G.; Oh, D.; Varma, G.; Hung, Y.P.; Ullas, S.; Lauring, J.; et al. Phosphoinositide 3-Kinase Regulates Glycolysis through Mobilization of Aldolase from the Actin Cytoskeleton. Cell 2016, 164, 433–446. [Google Scholar] [CrossRef]
- Nahm, D.-S.; Kim, H.-J.; Mah, J.; Baek, S.-H. In vitro expression of matrix metalloproteinase-1, tissue inhibitor of metalloproteinase-1 and transforming growth factor-beta1 in human periodontal ligament fibroblasts. Eur. J. Orthod. 2004, 26, 129–135. [Google Scholar] [CrossRef][Green Version]
- Demidowich, A.P.; Levine, J.A.; Apps, R.; Cheung, F.K.; Chen, J.; Fantoni, G.; Patel, T.P.; Yanovski, J.A. Colchicine’s effects on metabolic and inflammatory molecules in adults with obesity and metabolic syndrome: Results from a pilot randomized controlled trial. Int. J. Obes. 2020, 44, 1793–1799. [Google Scholar] [CrossRef]
- Lee, S.J.; Kim, Y.G.; Kang, K.W.; Kim, C.W.; Kim, S.G. Effects of colchicine on liver functions of cirrhotic rats: Beneficial effects result from stellate cell inactivation and inhibition of TGF beta1 expression. Chem. Biol. Interact. 2004, 147, 9–21. [Google Scholar] [CrossRef]
- Dvorak, Z.; Ulrichova, J.; Modriansky, M. Role of Microtubules Network in CYP Genes Expression. Curr. Drug Metab. 2005, 6, 545–552. [Google Scholar] [CrossRef]
- Dvořák, Z.; Modriansky, M.; Pichard-Garcia, L.; Balaguer, P.; Vilarem, M.-J.; Ulrichová, J.; Maurel, P.; Pascussi, J.-M. Colchicine down-regulates cytochrome P450 2B6, 2C8, 2C9, and 3A4 in human hepatocytes by affecting their glucocorticoid receptor-mediated regulation. Mol. Pharmacol. 2003, 64, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Pascussi, J.M.; Dvorák, Z.; Gerbal-Chaloin, S.; Assenat, E.; Maurel, P.; Vilarem, M.J. Pathophysiological factors affecting CAR gene expression. Drug Metab. Rev. 2003, 35, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Vrzal, R.; Daujat-Chavanieu, M.; Pascussi, J.-M.; Ulrichova, J.; Maurel, P.; Dvorak, Z. Microtubules-interfering agents restrict aryl hydrocarbon receptor-mediated CYP1A2 induction in primary cultures of human hepatocytes via c-jun-N-terminal kinase and glucocorticoid receptor. Eur. J. Pharmacol. 2008, 581, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, S.-I.; Ishida, S.; Hongo, T.; Ishikawa, Y.; Miyajima, A.; Sawada, J.-I.; Ohno, Y.; Nakazawa, K.; Ozawa, S. Global gene expression changes including drug metabolism and disposition induced by three-dimensional culture of HepG2 cells-Involvement of microtubules. Biochem. Biophys. Res. Commun. 2009, 378, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Roux, A.; Gilbert, S.; Loranger, A.; Marceau, N. Impact of keratin intermediate filaments on insulin-mediated glucose metabolism regulation in the liver and disease association. FASEB J. 2016, 30, 491–502. [Google Scholar] [CrossRef]
- Bays, J.L.; Campbell, H.K.; Heidema, C.; Sebbagh, M.; DeMali, K.A. Linking E-cadherin mechanotransduction to cell metabolism through force-mediated activation of AMPK. Nat. Cell Biol. 2017, 19, 724–731. [Google Scholar] [CrossRef]
- Visavadiya, N.P.; Keasey, M.P.; Razskazovskiy, V.; Banerjee, K.; Jia, C.; Lovins, C.; Wright, G.L.; Hagg, T. Integrin-FAK signaling rapidly and potently promotes mitochondrial function through STAT3. Cell Commun. Signal. 2016, 14, 32. [Google Scholar] [CrossRef]
- Prakash, K.; Fournier, D. Evidence for the implication of the histone code in building the genome structure. Biosystems 2018, 164, 49–59. [Google Scholar] [CrossRef]
- Vanhove, J.; Pistoni, M.; Welters, M.; Eggermont, K.; Vanslembrouck, V.; Helsen, N.; Boon, R.; Najimi, M.; Sokal, E.; Collas, P.; et al. H3K27me3 Does Not Orchestrate the Expression of Lineage-Specific Markers in hESC-Derived Hepatocytes In Vitro. Stem Cell Rep. 2016, 7, 192–206. [Google Scholar] [CrossRef]
- Chiba, T.; Yokosuka, O.; Arai, M.; Tada, M.; Fukai, K.; Imazeki, F.; Kato, M.; Seki, N.; Saisho, H. Identification of genes up-regulated by histone deacetylase inhibition with cDNA microarray and exploration of epigenetic alterations on hepatoma cells. J. Hepatology 2004, 41, 436–445. [Google Scholar] [CrossRef]
- Balint, B.L.; Gabor, P.; Nagy, L. Genome-wide localization of histone 4 arginine 3 methylation in a differentiation primed myeloid leukemia cell line. Immunobiology 2005, 210, 141–152. [Google Scholar] [CrossRef]
- Ancey, P.-B.; Ecsedi, S.; Lambert, M.-P.; Talukdar, F.R.; Cros, M.-P.; Glaise, D.; Narvaez, D.M.; Chauvet, V.; Herceg, Z.; Corlu, A.; et al. TET-Catalyzed 5-Hydroxymethylation Precedes HNF4A Promoter Choice during Differentiation of Bipotent Liver Progenitors. Stem Cell Rep. 2017, 9, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Tashima, T.; Nagatoishi, S.; Sagara, H.; Ohnuma, S.; Tsumoto, K. Osteomodulin regulates diameter and alters shape of collagen fibrils. Biochem. Biophys. Res. Commun. 2015, 463, 292–296. [Google Scholar] [CrossRef]
- Asselah, T.; Bieche, I.; Laurendeau, I.; Paradis, V.; Vidaud, D.; Degott, C.; Martinot, M.; Bedossa, P.; Valla, D.; Vidaud, M.; et al. Liver gene expression signature of mild fibrosis in patients with chronic hepatitis C. Gastroenterology 2005, 129, 2064–2075. [Google Scholar] [CrossRef] [PubMed]
- Quondamatteo, F.; Knittel, T.; Mehde, M.; Ramadori, G.; Herken, R. Matrix metalloproteinases in early human liver development. Histochem. Cell Biol. 1999, 112, 277–282. [Google Scholar] [CrossRef] [PubMed]
- de Abreu Costa, L.; Henrique Fernandes Ottoni, M.; dos Santos, M.G.; Meireles, A.B.; Gomes de Almeida, V.; de Fátima Pereira, W.; Alves de Avelar-Freitas, B.; Eustáquio Alvim Brito-Melo, G. Dimethyl Sulfoxide (DMSO) Decreases Cell Proliferation and TNF-α, IFN-γ, and IL-2 Cytokines Production in Cultures of Peripheral Blood Lymphocytes. Molecules 2017, 22, 1789. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubois-Pot-Schneider, H.; Aninat, C.; Kattler, K.; Fekir, K.; Jarnouen, K.; Cerec, V.; Glaise, D.; Salhab, A.; Gasparoni, G.; Takashi, K.; et al. Transcriptional and Epigenetic Consequences of DMSO Treatment on HepaRG Cells. Cells 2022, 11, 2298. https://doi.org/10.3390/cells11152298
Dubois-Pot-Schneider H, Aninat C, Kattler K, Fekir K, Jarnouen K, Cerec V, Glaise D, Salhab A, Gasparoni G, Takashi K, et al. Transcriptional and Epigenetic Consequences of DMSO Treatment on HepaRG Cells. Cells. 2022; 11(15):2298. https://doi.org/10.3390/cells11152298
Chicago/Turabian StyleDubois-Pot-Schneider, Hélène, Caroline Aninat, Kathrin Kattler, Karim Fekir, Kathleen Jarnouen, Virginie Cerec, Denise Glaise, Abdulrahman Salhab, Gilles Gasparoni, Kubo Takashi, and et al. 2022. "Transcriptional and Epigenetic Consequences of DMSO Treatment on HepaRG Cells" Cells 11, no. 15: 2298. https://doi.org/10.3390/cells11152298
APA StyleDubois-Pot-Schneider, H., Aninat, C., Kattler, K., Fekir, K., Jarnouen, K., Cerec, V., Glaise, D., Salhab, A., Gasparoni, G., Takashi, K., Ishida, S., Walter, J., & Corlu, A. (2022). Transcriptional and Epigenetic Consequences of DMSO Treatment on HepaRG Cells. Cells, 11(15), 2298. https://doi.org/10.3390/cells11152298






