Ssc-MiR-21-5p and Ssc-MiR-615 Regulates the Proliferation and Apoptosis of Leydig Cells by Targeting SOX5
Abstract
:1. Introduction
2. Material and Methods
2.1. Samples Collection
2.2. Cell Culture and Transfection
2.3. Quantitative Real-Time PCR
2.4. Protein Extraction and Western Blot
2.5. EdU Staining Assay
2.6. TUNEL Staining Assay
2.7. Dual-Luciferase Reporter System Assay
2.8. Statistical Analysis
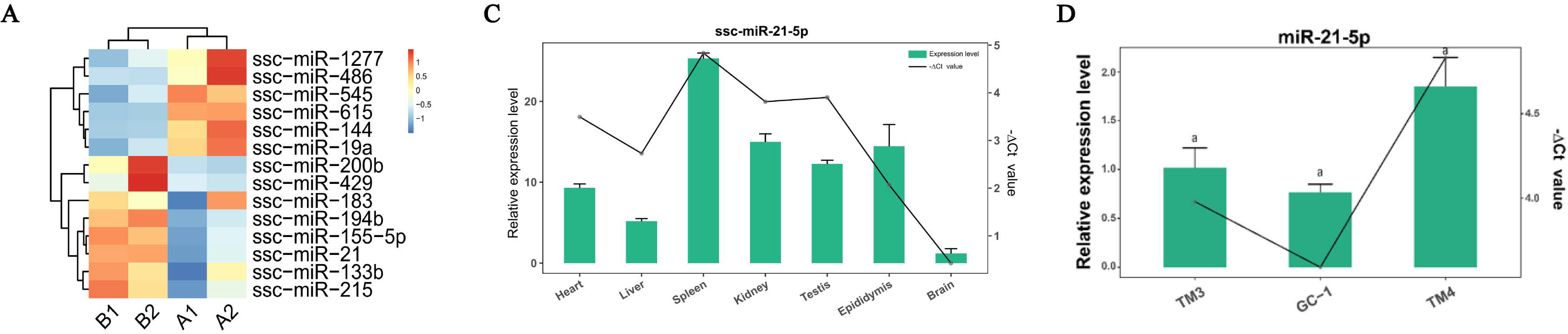
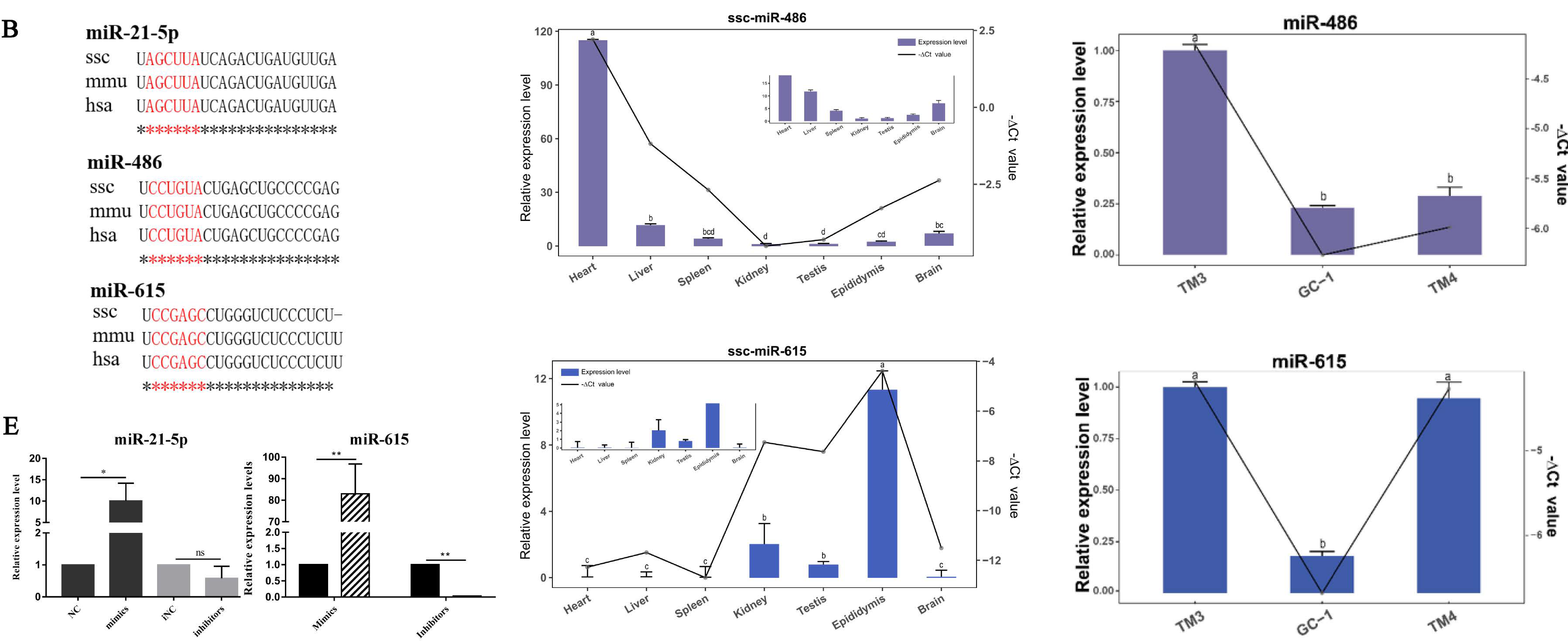
3. Results
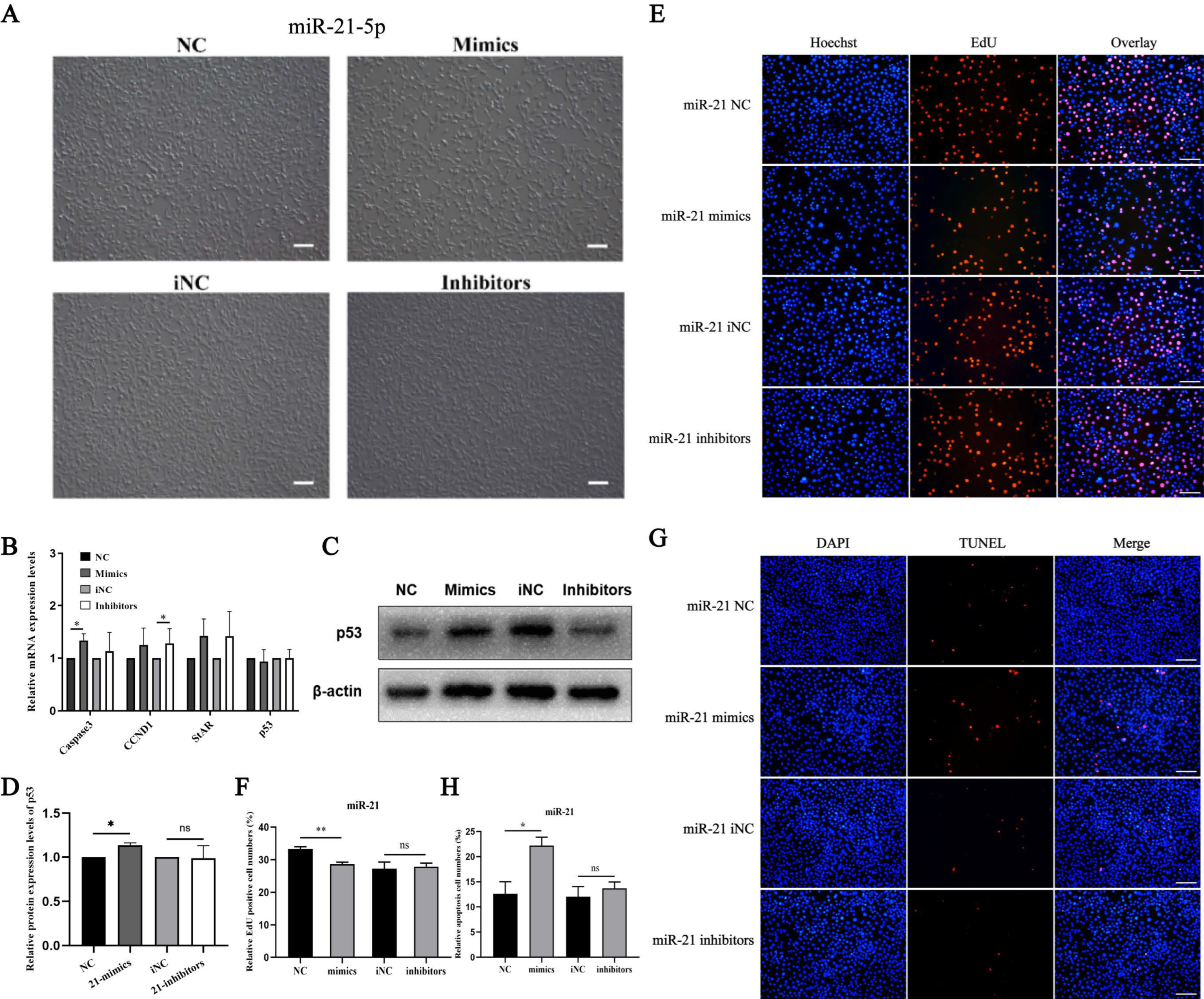
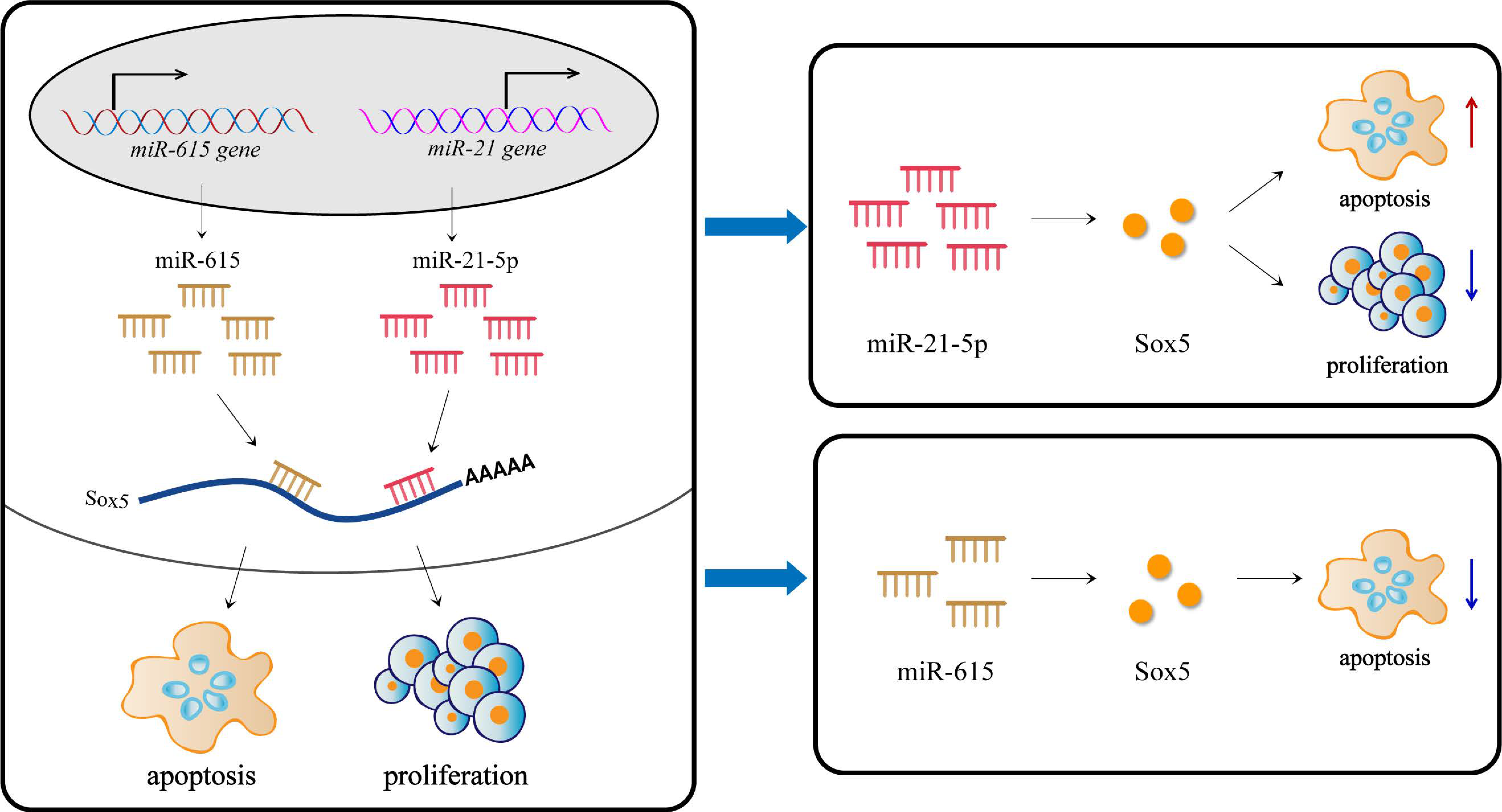
3.1. miR-21-5p Inhibits Proliferation and Promotes Apoptosis
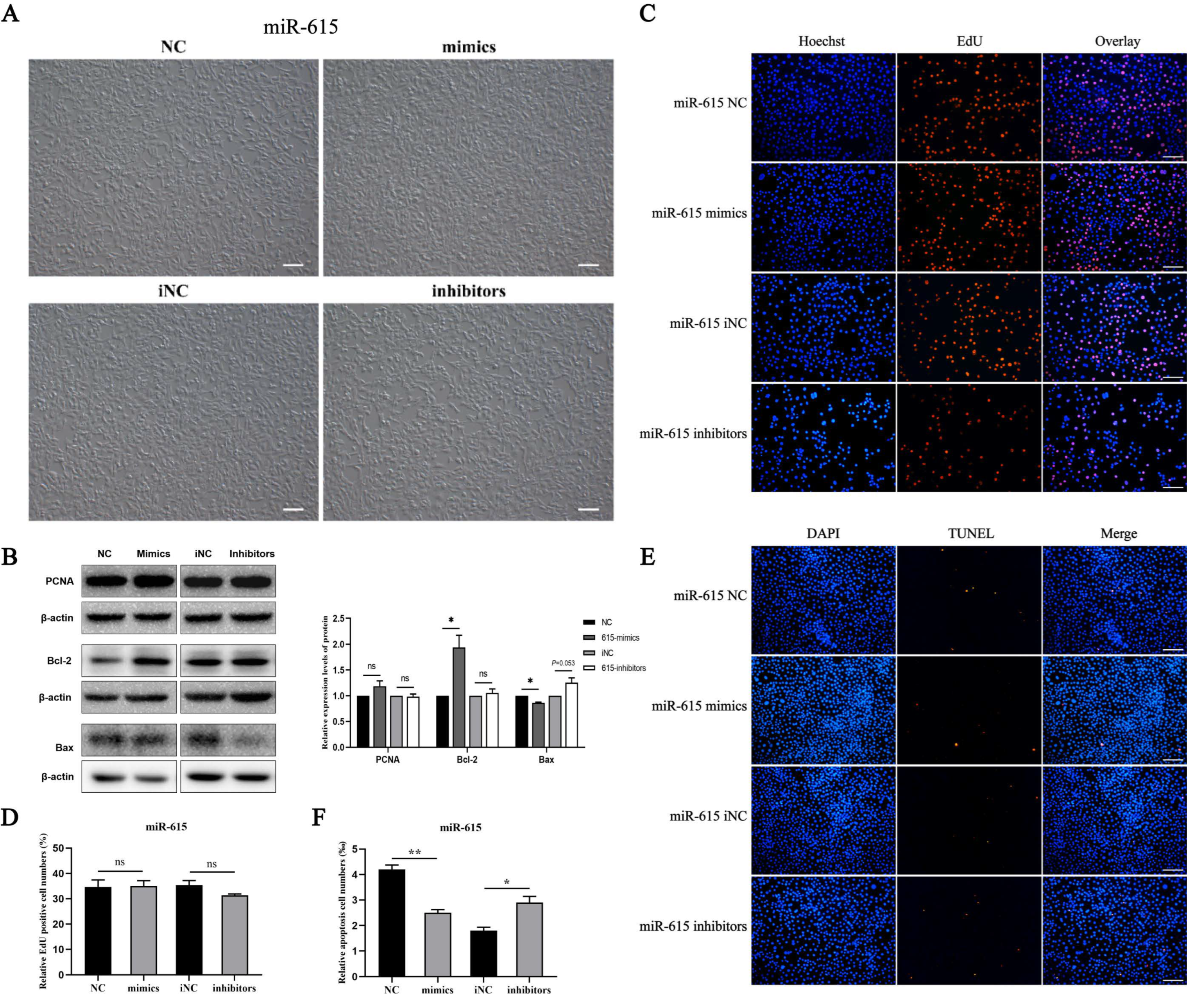
3.2. miR-615 Inhibits Apoptosis but Does Not Affect Proliferation
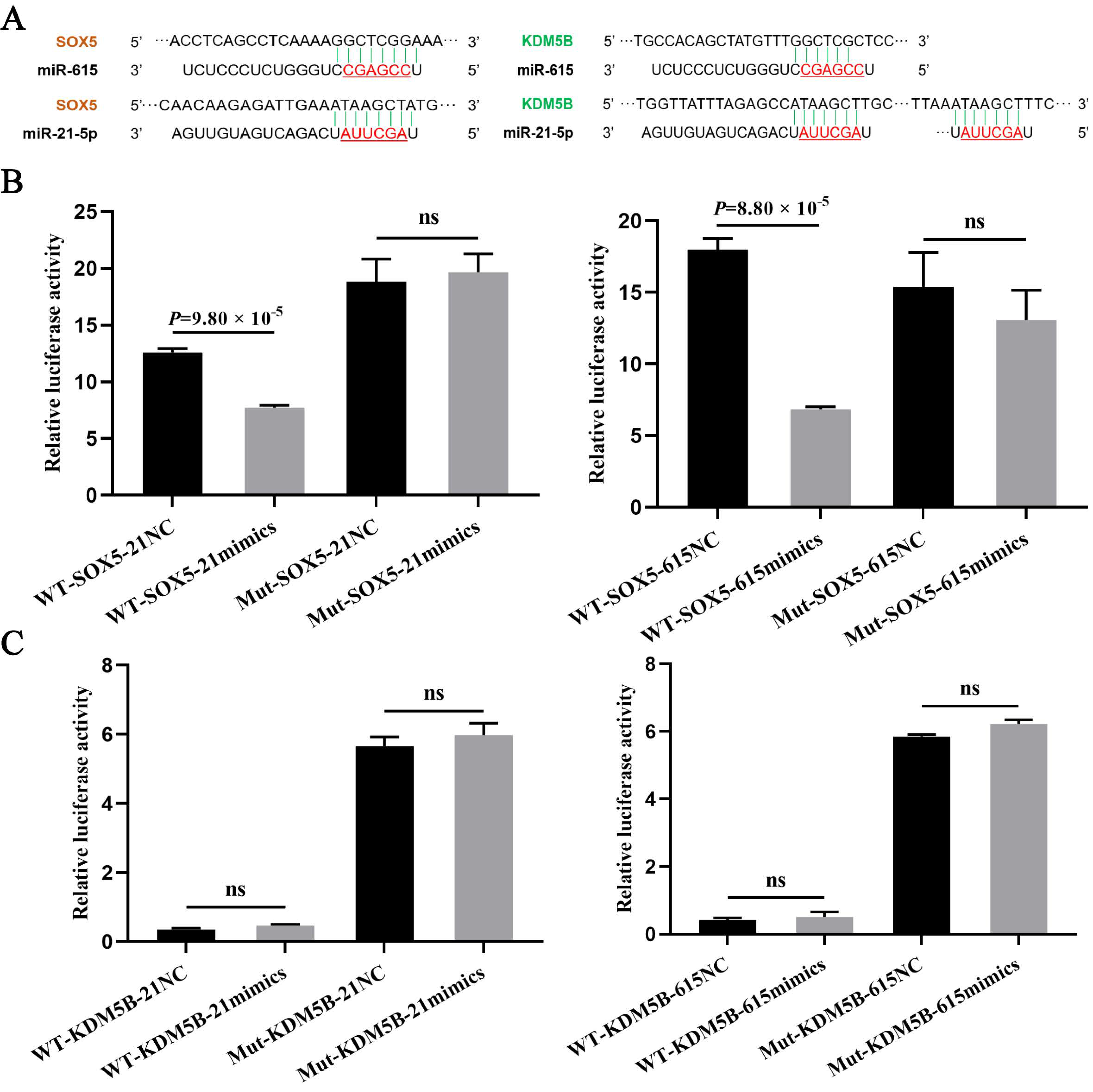
3.3. The Targeting Relationship of Downstream Target Genes of miRNAs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [Green Version]
- Salas-Huetos, A.; James, E.R.; Aston, K.I.; Carrell, D.T.; Jenkins, T.G.; Yeste, M. The role of miRNAs in male human reproduction: A systematic review. Andrology 2020, 8, 7–26. [Google Scholar] [CrossRef] [Green Version]
- Geng, X.J.; Zhao, D.M.; Mao, G.H.; Tan, L. MicroRNA-150 regulates steroidogenesis of mouse testicular Leydig cells by targeting STAR. Reprod. 2017, 154, 229–236. [Google Scholar] [CrossRef]
- Dai, L.; Tsai-Morris, C.H.; Sato, H.; Villar, J.; Kang, J.H.; Zhang, J.; Dufau, M.L. Testis-specific miRNA-469 up-regulated in gonadotropin-regulated testicular RNA helicase (GRTH/DDX25)-null mice silences transition protein 2 and protamine 2 messages at sites within coding region: Implications of its role in germ cell development. J. Biol. Chem. 2011, 286, 44306–44318. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Li, L.; Lin, C.; Hu, M.; Liu, X.; Wang, L.; Le, F.; Jin, F. Decreased miR-149 expression in sperm is correlated with the quality of early embryonic development in conventional in vitro fertilization. Reprod. Toxicol. 2021, 101, 28–32. [Google Scholar] [CrossRef]
- Peak, T.C.; Haney, N.M.; Wang, W.; DeLay, K.J.; Hellstrom, W.J. Stem cell therapy for the treatment of Leydig cell dysfunction in primary hypogonadism. World J. Stem. Cells. 2016, 8, 306–315. [Google Scholar] [CrossRef]
- Teerds, K.J.; Huhtaniemi, I.T. Morphological and functional maturation of Leydig cells: From rodent models to primates. Hum. Reprod. Update 2015, 21, 310–328. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Chen, R.; Ma, L.; Yang, W.; Chen, M.; Zhang, Y.; Yu, S.; Dong, W.; Zeng, W.; Lan, X.; et al. miR-205 Expression Elevated With EDS Treatment and Induced Leydig Cell Apoptosis by Targeting RAP2B via the PI3K/AKT Signaling Pathway. Front. Cell. Dev. Biol. 2020, 8, 448. [Google Scholar] [CrossRef]
- Fu, X.; He, Y.; Wang, X.; Peng, D.; Chen, X.; Li, X.; Wang, Q. Overexpression of miR-21 in stem cells improves ovarian structure and function in rats with chemotherapy-induced ovarian damage by targeting PDCD4 and PTEN to inhibit granulosa cell apoptosis. Stem. Cell. Res. Ther. 2017, 8, 187. [Google Scholar] [CrossRef] [Green Version]
- Hua, R.; Zhang, X.; Li, W.; Lian, W.; Liu, Q.; Gao, D.; Wang, Y.; Lei, M. Ssc-miR-21-5p regulates endometrial epithelial cell proliferation, apoptosis and migration via the PDCD4/AKT pathway. J. Cell. Sci. 2020, 133, jcs248898. [Google Scholar] [CrossRef]
- Niu, Z.; Goodyear, S.M.; Rao, S.; Wu, X.; Tobias, J.W.; Avarbock, M.R.; Brinster, R.L. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA 2011, 108, 12740–12745. [Google Scholar] [CrossRef] [Green Version]
- Li, J.X.; Li, Y.; Xia, T.; Rong, F.Y. miR-21 Exerts Anti-proliferative and Pro-apoptotic Effects in LPS-induced WI-38 Cells via Directly Targeting TIMP3. Cell. Biochem. Biophys. 2021, 79, 781–790. [Google Scholar] [CrossRef]
- Quah, S.; Holland, P.W. The Hox cluster microRNA miR-615: A case study of intronic microRNA evolution. EvoDevo 2015, 6, 31. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Jia, Y.; Jia, L.; Li, T.; Yang, L.; Zhang, G. MicroRNA 615-3p Inhibits the Tumor Growth and Metastasis of NSCLC via Inhibiting IGF2. Oncol. Res. 2019, 27, 269–279. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, J.; Pan, S.; Yang, T.; Sun, X.; Wang, Y.; Shi, X.; Zhao, X.; Guo, J.; Zhang, X. LINC00657 played oncogenic roles in esophageal squamous cell carcinoma by targeting miR-615-3p and JunB. Biomed. Pharmacother. 2018, 108, 316–324. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, G.; Zhang, Z.; Tang, Q.; Zheng, W.; Chen, X.; Chen, F.; Li, Q.; Che, X. Long non-coding RNA HOTTIP promotes renal cell carcinoma progression through the regulation of the miR-615/IGF-2 pathway. Int. J. Oncol. 2018, 53, 2278–2288. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.X.; Tian, Z.G.; Zhu, L.F.; Wu, W.D.; Zhou, S.L.; Zhao, Y.T.; Huang, S. MicroRNA-615-3p promotes the osteoarthritis progression by inhibiting chondrogenic differentiation of bone marrow mesenchymal stem cells. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6212–6220. [Google Scholar] [CrossRef]
- Wang, J.J.; Liu, C.; Shan, K.; Liu, B.H.; Li, X.M.; Zhang, S.J.; Zhou, R.M.; Dong, R.; Yan, B.; Sun, X.H. Circular RNA-ZNF609 regulates retinal neurodegeneration by acting as miR-615 sponge. Theranostics 2018, 8, 3408–3415. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ding, L.; Wang, Y.; Zou, T.B.; Wang, T.; Fu, W.; Lin, Y.; Zhang, X.; Chen, K.; Lei, Y.; et al. MiR-615 Regulates NSC Differentiation In Vitro and Contributes to Spinal Cord Injury Repair by Targeting LINGO-1. Mol. Neurobiol. 2020, 57, 3057–3074. [Google Scholar] [CrossRef]
- Daigle, M.; Roumaud, P.; Martin, L.J. Expressions of Sox9, Sox5, and Sox13 transcription factors in mice testis during postnatal development. Mol. Cell. Biochem. 2015, 407, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Huang, C.; Zhou, J.; Ye, L. Identification of a novel Sox5 transcript in mouse testis. Gene Expr. Patterns. 2021, 41, 119197. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Qin, Y.; Qu, J.; Lu, C.; Wang, Y.; Wu, W.; Song, L.; Wang, S.; Chen, F.; Shen, H.; et al. Evaluation of five candidate genes from GWAS for association with oligozoospermia in a Han Chinese population. PLoS ONE 2013, 8, e80374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, W.; Liu, Y.; Shen, Y.; Yan, Y.; Wang, X.; Yang, D.; Li, L.; Ma, Y.; Tao, D.; Zhang, S.; et al. Genome-wide Loci linked to non-obstructive azoospermia susceptibility may be independent of reduced sperm production in males with normozoospermia. Biol. Reprod. 2015, 92, 41. [Google Scholar] [CrossRef] [Green Version]
- Gu, X.; Li, H.; Chen, X.; Zhang, X.; Mei, F.; Jia, M.; Xiong, C. PEX10, SIRPA-SIRPG, and SOX5 gene polymorphisms are strongly associated with nonobstructive azoospermia susceptibility. J. Assist. Reprod. Genet. 2019, 36, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Li, D.; Wang, L.; Wu, J.; Hu, Y.; Wang, Z.; Chen, Y.; Cao, X.; Jiang, C.; Yan, W.; et al. MicroRNA-449 and microRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway. J. Biol. Chem. 2012, 287, 21686–21698. [Google Scholar] [CrossRef] [Green Version]
| Primer Name | Sequence (5′–3′) | Notes |
|---|---|---|
| miR-21-5p-slp | GTCGTATCCAGTGCAGGGTCCGAGGTATTCG CACTGGATACGACTCAACATC | stem-loop RT-PCR |
| miR-486-slp | GTCGTATCCAGTGCAGGGTCCGAGGTATTCG CACTGGATACGACCTCGGGGC | stem-loop RT-PCR |
| ssc-miR-615-slp | GTCGTATCCAGTGCAGGGTCCGAGGTATTCG CACTGGATACGACAGAGGGAG | stem-loop RT-PCR |
| mmu-miR-615-slp | GTCGTATCCAGTGCAGGGTCCGAGGTATTCG CACTGGATACGACAAGAGGGA | stem-loop RT-PCR |
| q-miR-21-5p-F | ATGGTTCGTGGGTAGCTTATCAGACTGA | qRT-PCR |
| q-miR-486-F | AACTGCAGAATCCTGTACTGAGCTG | qRT-PCR |
| q-miR-615-F | AAGGAAAAAATCCGAGCCTGGGTCTC | qRT-PCR |
| q-miR-U-R | GCAGGGTCCGAGGTATTC | Universal reverse primer of miRNAs for qRT-PCR |
| mus-U6 | F: CGCTTCACGAATTTGCGTGTCAT | qRT-PCR |
| R: GCTTCGGCAGCACATATACTAAAAT | ||
| StAR | F: GGTTCTCAGCTGGAAGACACT | qRT-PCR (146 bp) |
| R: ACCTCGTCCCCATTCTCCTG | ||
| Caspase3 | F: AGCTGGACTGTGGCATTGAG | qRT-PCR (143 bp) |
| R: CCACGACCCGTCCTTTGAAT | ||
| CCND1 | F: CATTCCCTTGACTGCCGAGA | qRT-PCR (177 bp) |
| R: TTGTTCTCATCCGCCTCTGG | ||
| p53 | F: ATGCGGTTCGGGTCCAAAAT | qRT-PCR (154 bp) |
| R: CTAAATGGCAGTCGTTCTCTCC | ||
| β-actin | F: CTCCATCATGAAGTGCGACGT | qRT-PCR (114 bp) |
| R: GTGATCTCCTTCTGCATCCTGTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Q.; Zhang, Y.; Yue, L.; Ren, H.; Pan, C. Ssc-MiR-21-5p and Ssc-MiR-615 Regulates the Proliferation and Apoptosis of Leydig Cells by Targeting SOX5. Cells 2022, 11, 2253. https://doi.org/10.3390/cells11142253
Tang Q, Zhang Y, Yue L, Ren H, Pan C. Ssc-MiR-21-5p and Ssc-MiR-615 Regulates the Proliferation and Apoptosis of Leydig Cells by Targeting SOX5. Cells. 2022; 11(14):2253. https://doi.org/10.3390/cells11142253
Chicago/Turabian StyleTang, Qi, Yanghai Zhang, Linxiu Yue, Hongying Ren, and Chuanying Pan. 2022. "Ssc-MiR-21-5p and Ssc-MiR-615 Regulates the Proliferation and Apoptosis of Leydig Cells by Targeting SOX5" Cells 11, no. 14: 2253. https://doi.org/10.3390/cells11142253
APA StyleTang, Q., Zhang, Y., Yue, L., Ren, H., & Pan, C. (2022). Ssc-MiR-21-5p and Ssc-MiR-615 Regulates the Proliferation and Apoptosis of Leydig Cells by Targeting SOX5. Cells, 11(14), 2253. https://doi.org/10.3390/cells11142253





