Immunocytochemical Analysis of the Wall Ingrowths in the Digestive Gland Transfer Cells in Aldrovanda vesiculosa L. (Droseraceae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Histological and Immunochemical Analysis
2.3. Immunogold Labeling Distribution of AGP, HG, Hemicellulose, and Mannan
3. Results
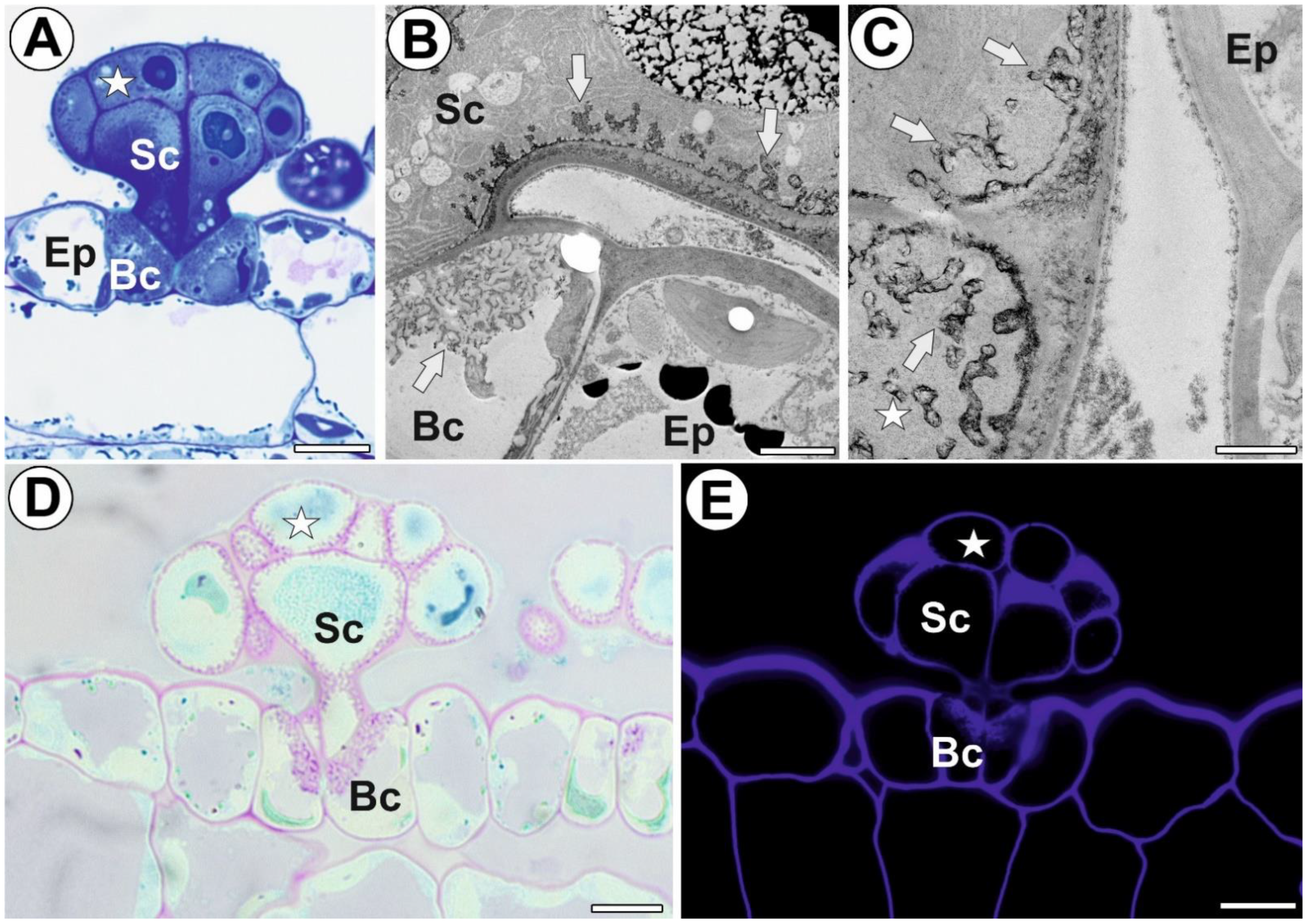
3.1. General Gland Structure and Histochemistry
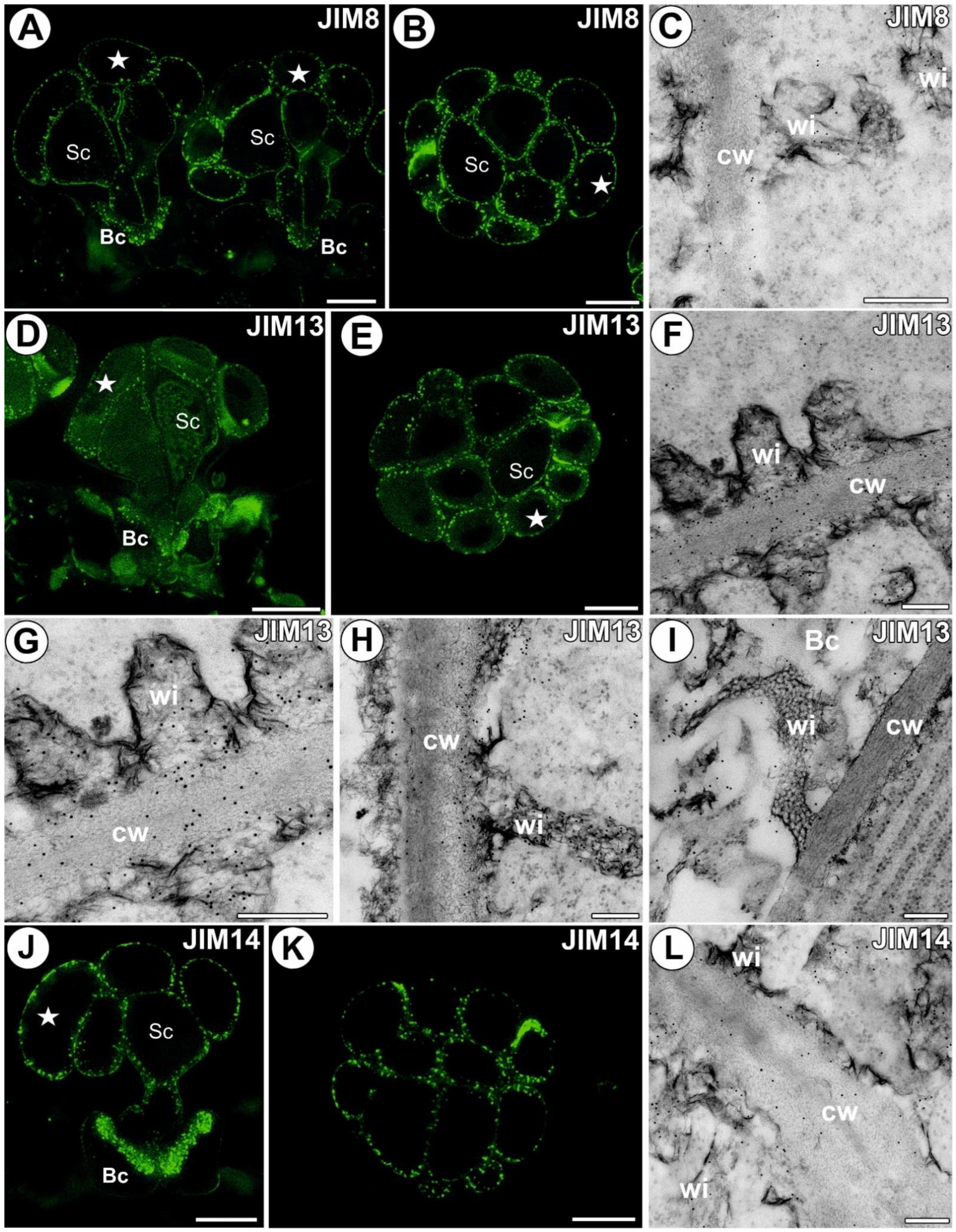
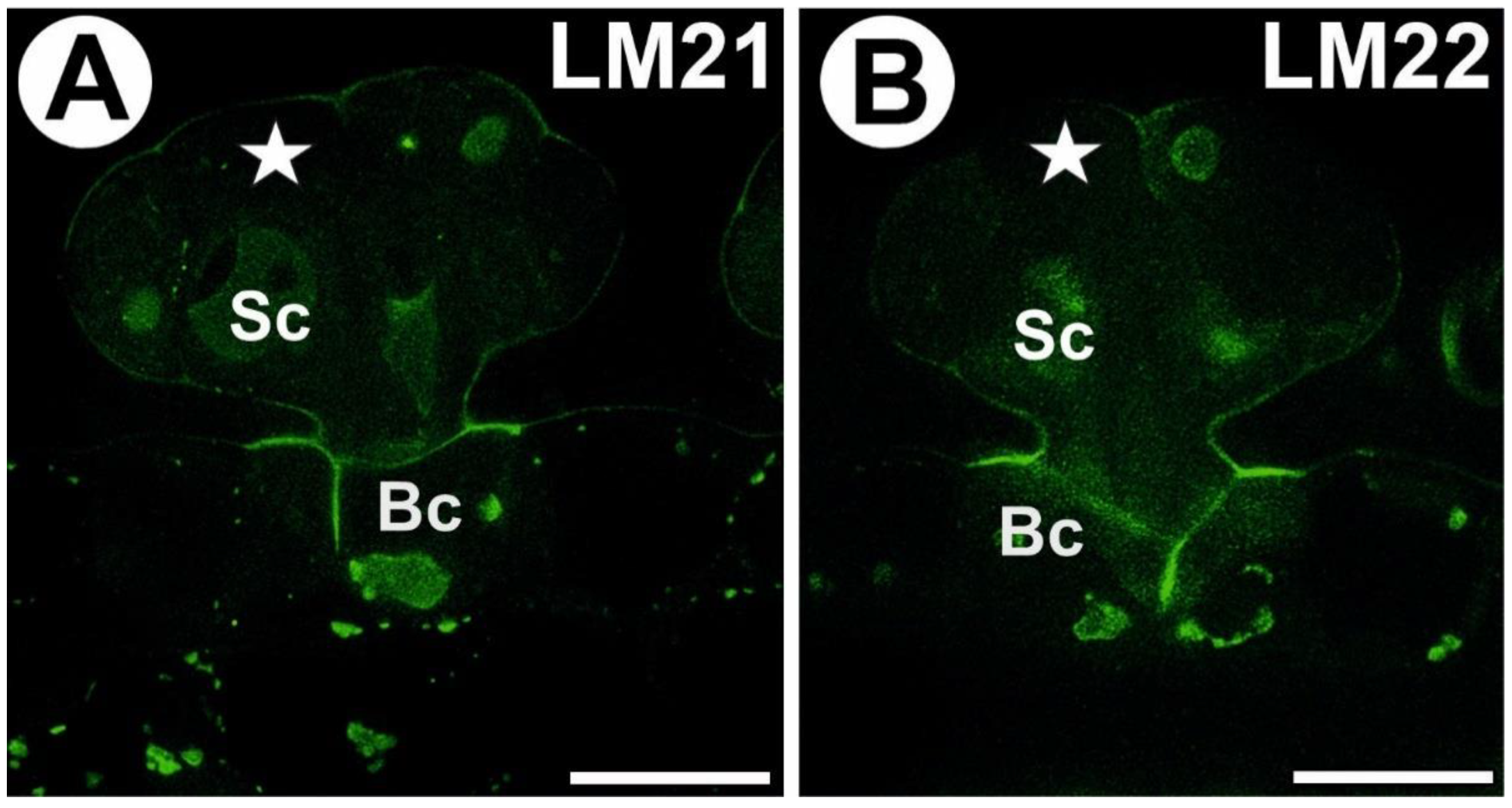
3.2. AGP Distribution
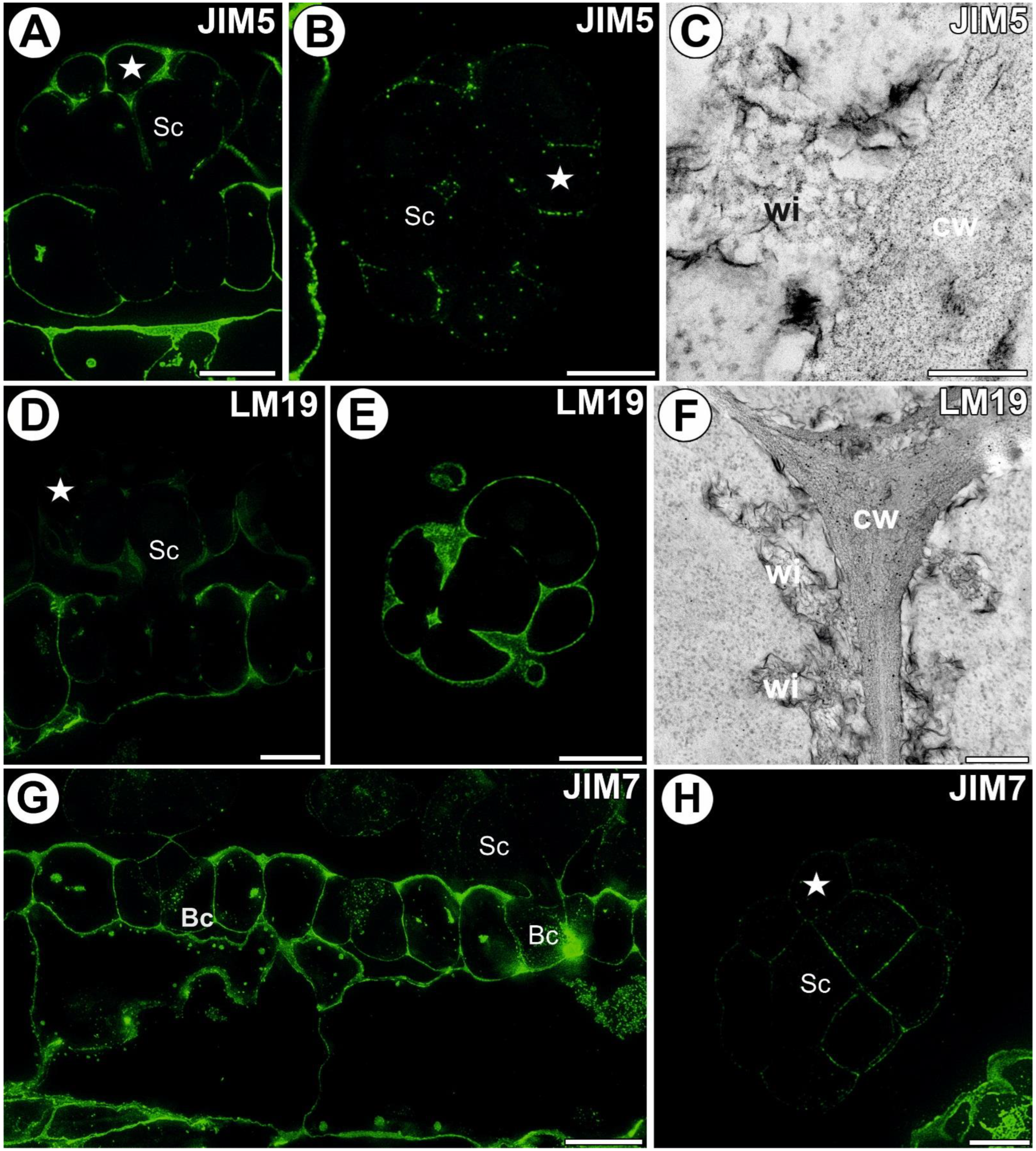
3.3. Homogalacturonan Distribution
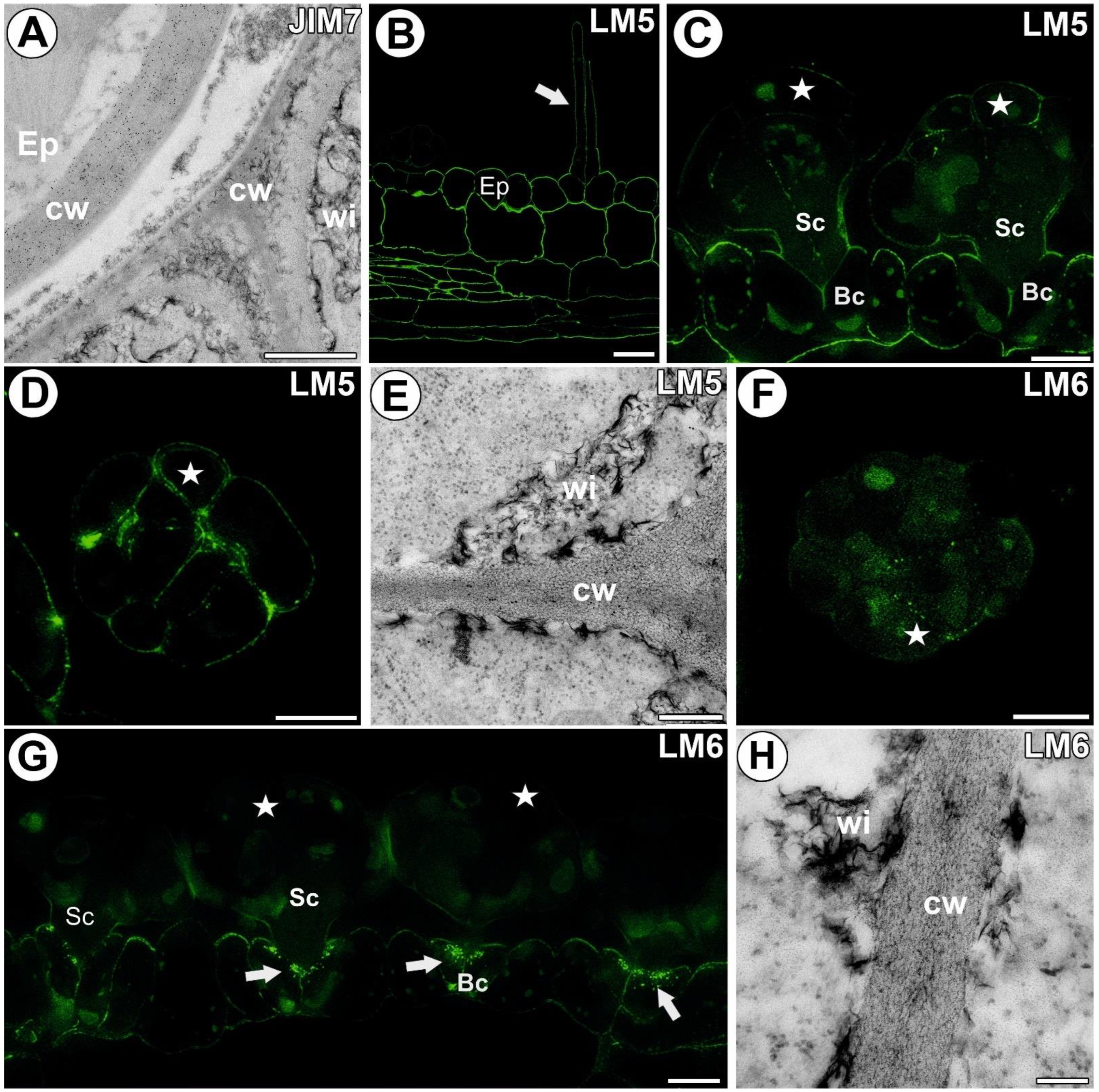
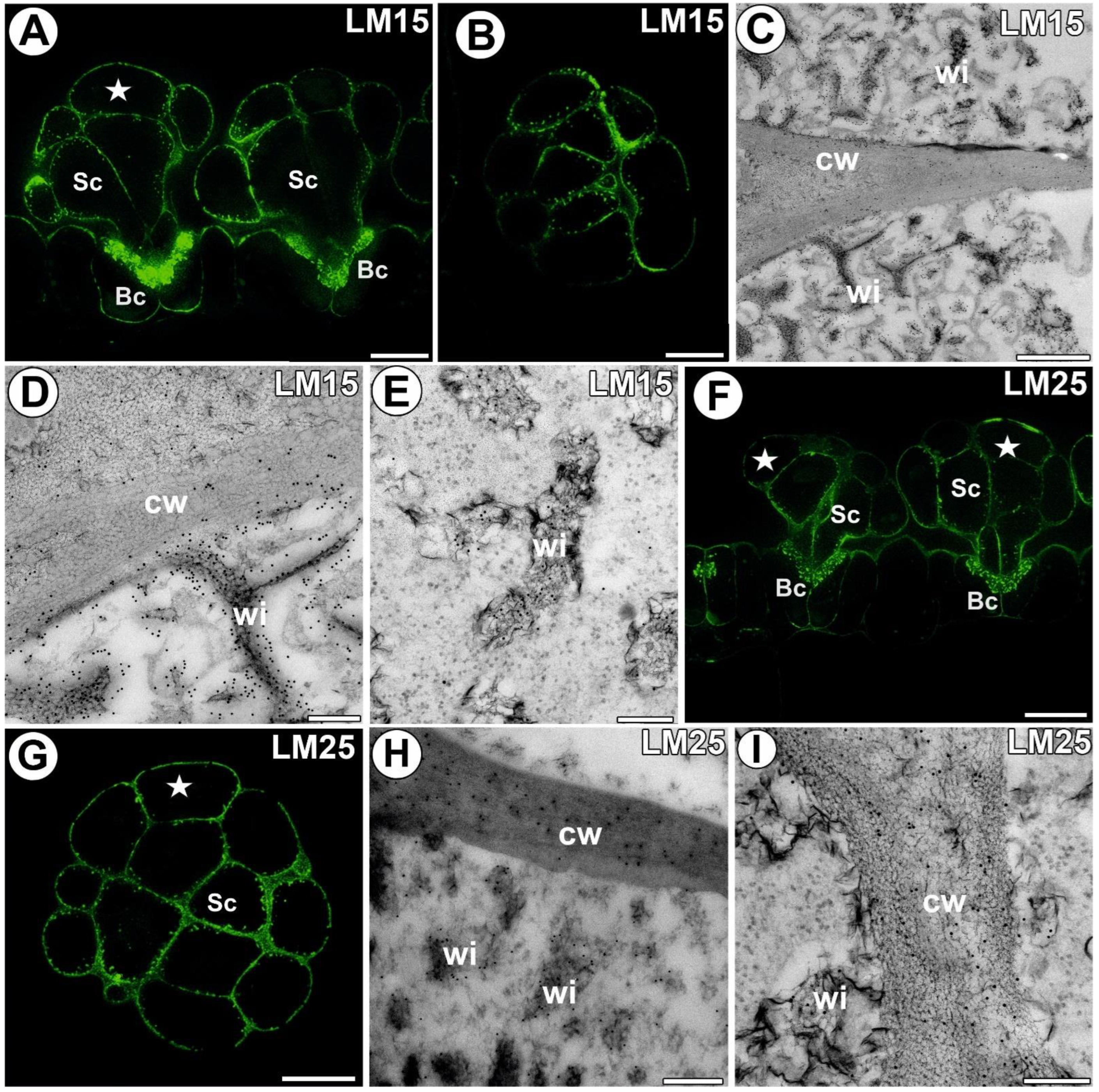
3.4. Hemicellulose and Heteromannan Distribution
3.5. Statistical Analysis
3.5.1. Head Cell Wall
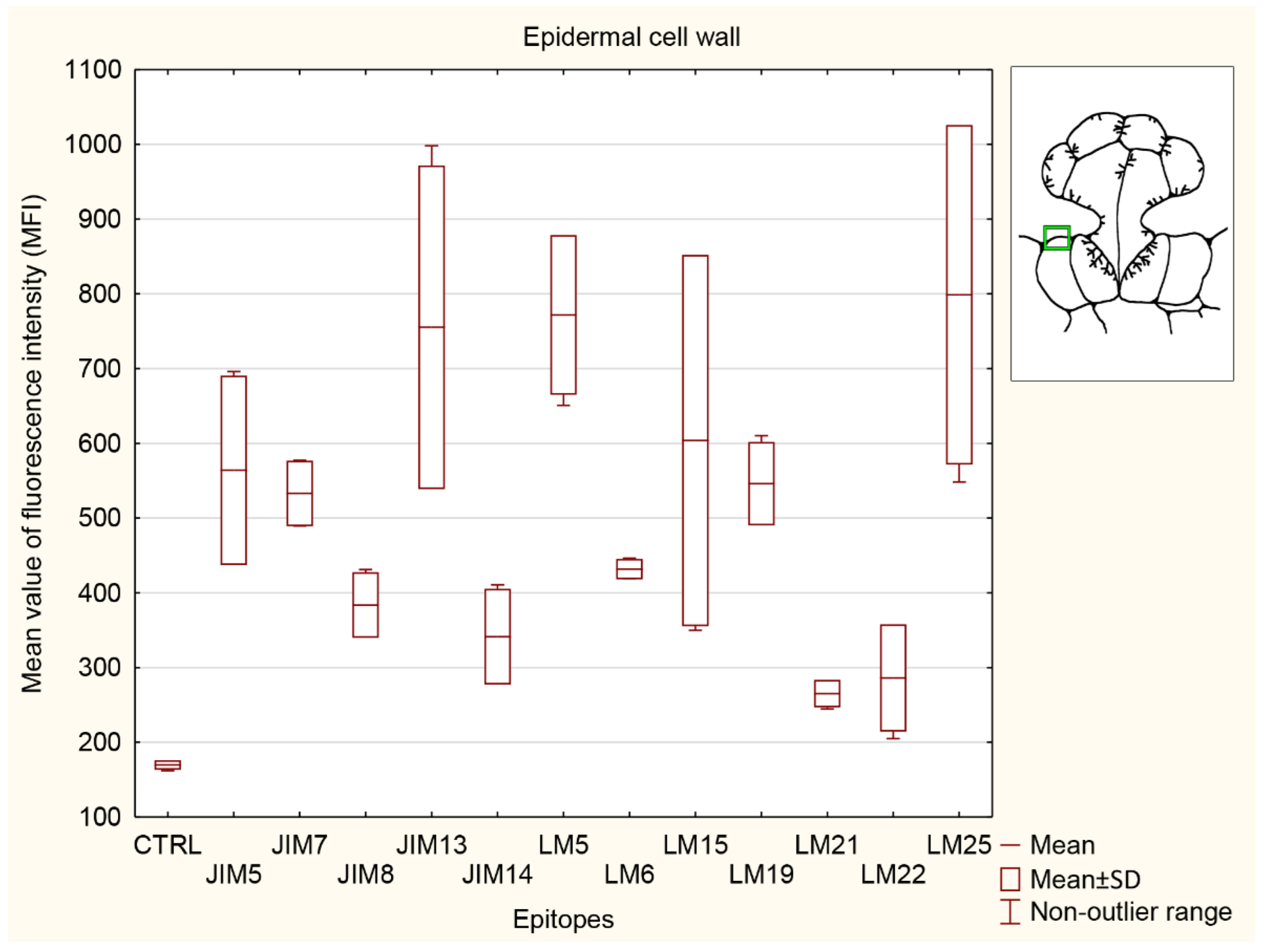
3.5.2. Ordinary Epidermal Cell Wall
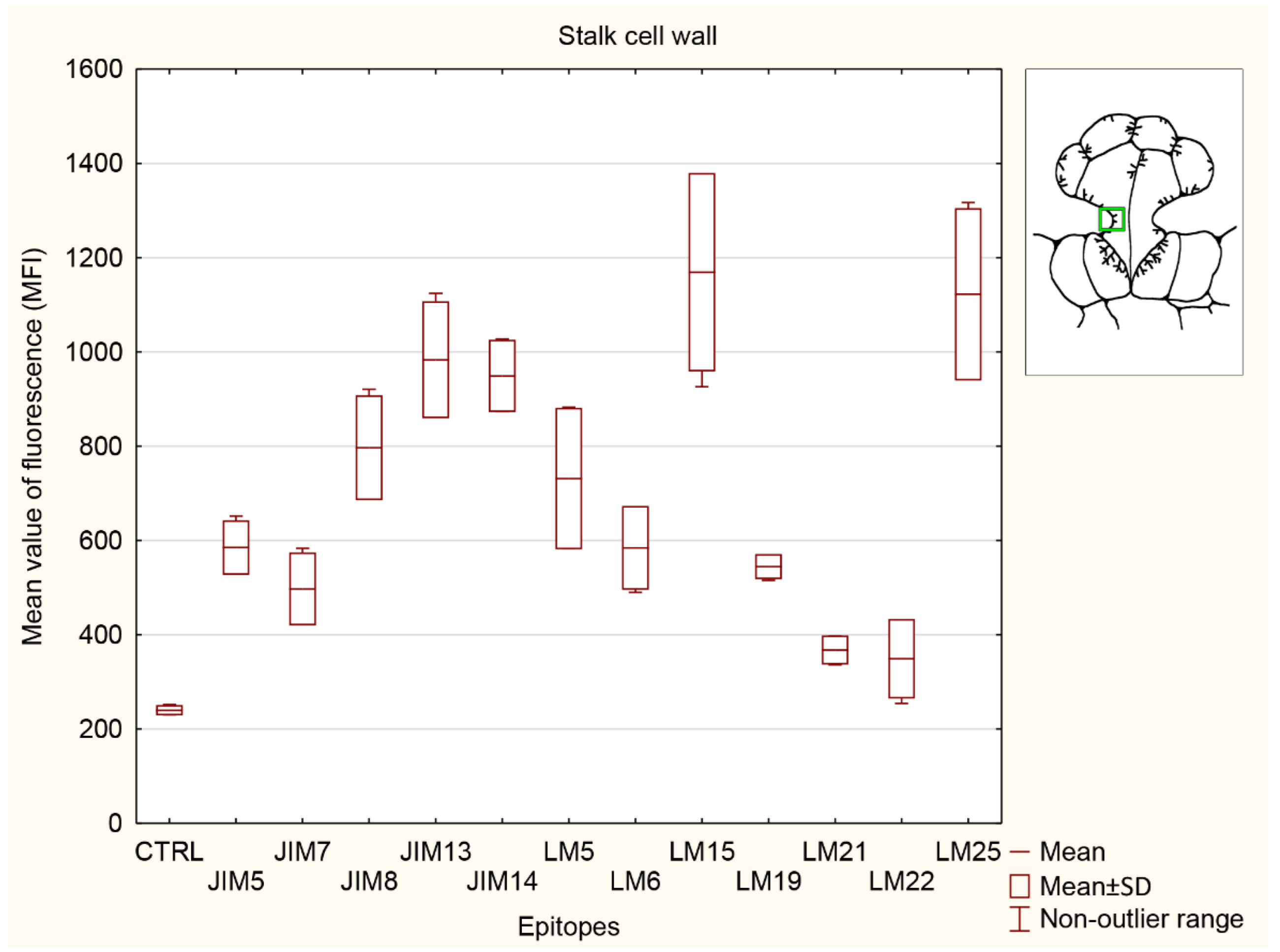
3.5.3. Stalk Cell Wall
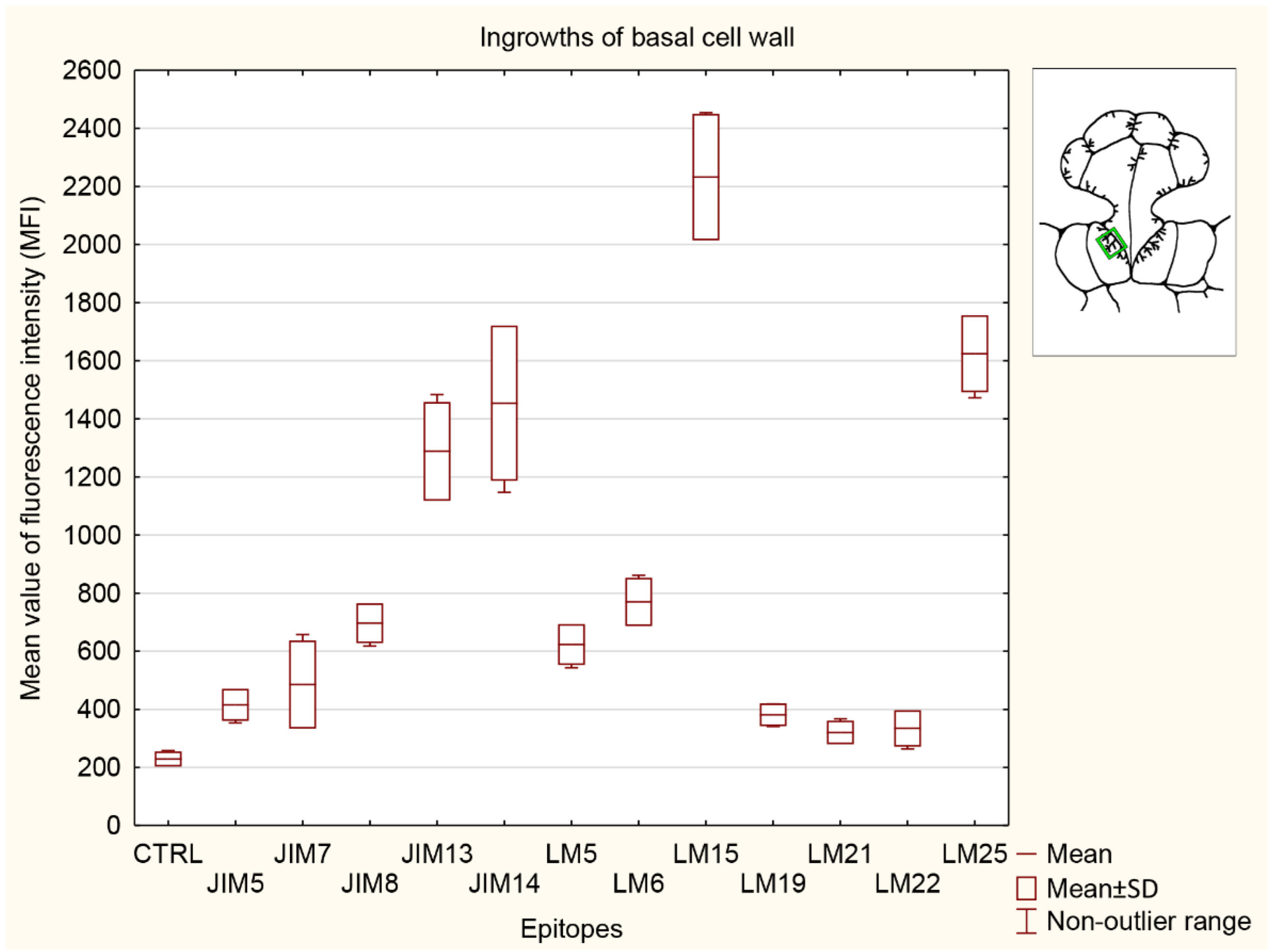
3.5.4. Ingrowths of Basal Cell Wall
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Juniper, B.E.; Robins, R.J.; Joel, D.M. The Carnivorous Plants; Academic Press: London, UK, 1989. [Google Scholar]
- Król, E.; Płachno, B.J.; Adamec, L.; Stolarz, M.; Dziubińska, H.; Trebacz, K. Quite a few reasons for calling carnivores “the most wonderful plants in the world”. Ann. Bot. 2012, 109, 47–64. [Google Scholar] [CrossRef] [PubMed]
- Darnowski, D.; Bauer, U.; Méndez, M.; Horner, J.D.; Płachno, B.J. Prey selection and specialization by carnivorous plants. In Carnivorous Plants: Physiology, Ecology, and Evolution; Oxford University Press: Oxford, UK, 2018; pp. 285–293. ISBN 9780198779841. [Google Scholar]
- Lichtscheidl, I.K.; Lancelle, S.A.; Hepler, P.K. Actin-endoplasmic reticulum complexes in Drosera—Their structural relationship with the plasmalemma, nucleus, and organelles in cells prepared by high pressure freezing. Protoplasma 1990, 155, 116–126. [Google Scholar] [CrossRef]
- Adlassnig, W.; Koller-Peroutka, M.; Bauer, S.; Koshkin, E.; Lendl, T.; Lichtscheidl, I.K. Endocytotic uptake of nutrients in carnivorous plants. Plant J. 2012, 71, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Muravnik, L.E. The ultrastructure of the secretory cells of glandular hairs in two Drosera species as affected by chemical stimulation. Russ. J. Plant Physiol. 2000, 47, 540–548. [Google Scholar]
- Gergely, Z.R.; Martinez, D.E.; Donohoe, B.S.; Mogelsvang, S.; Herder, R.; Andrew Staehelin, L. 3D electron tomographic and biochemical analysis of ER, Golgi and trans Golgi network membrane systems in stimulated Venus flytrap (Dionaea muscipula) glandular cells. J. Biol. Res. 2018, 25, 15. [Google Scholar] [CrossRef]
- Boulogne, C.; Gillet, C.; Hughes, L.; Le Bars, R.; Canette, A.; Hawes, C.R.; Satiat-Jeunemaitre, B. Functional organisation of the endomembrane network in the digestive gland of the Venus flytrap: Revisiting an old story with a new microscopy toolbox. J. Microsc. 2020, 280, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Lichtscheidl, I.; Lancelle, S.; Weidinger, M.; Adlassnig, W.; Koller-Peroutka, M.; Bauer, S.; Krammer, S.; Hepler, P.K. Gland cell responses to feeding in Drosera capensis, a carnivorous plant. Protoplasma 2021, 258, 1291–1306. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.E.; Pickard, B.G. Connections and barriers between cells of Drosera tentacles in relation to their electrophysiology. Planta 1974, 116, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Fineran, B.A.; Gilbertson, J.M. Application of lanthanum and uranyl salts as tracers to demonstrate apoplastic pathways for transport in glands of the carnivorous plant Utricularia monanthos. Eur. J. Cell Biol. 1980, 23, 66–72. [Google Scholar] [PubMed]
- Owen, T.P.; Lennon, K.A.; Santo, M.J.; Anderson, A.N. Pathways for nutrient transport in the pitchers of the carnivorous plant Nepenthes alata. Ann. Bot. 1999, 84, 459–466. [Google Scholar] [CrossRef]
- Scala, J.; Schwab, D.; Simmons, E. The Fine Structure of the Digestive Gland of Venus’s Flytrap. Am. J. Bot. 1968, 55, 649. [Google Scholar] [CrossRef]
- Muravnik, L.E. Morphometrical approach to the secretory activity determination in digestive glands of Aldrovanda vesiculosa (Droseraceae). Bot. Zhurnal 1996, 81, 1–9. [Google Scholar]
- Muravnik, L.E.; Vassilyev, A.E.; Potapova, Y.Y. Ultrastructural aspects of digestive gland functioning in Aldrovanda vesiculosa. Russ. J. Plant Physiol. 1995, 42, 5–13. [Google Scholar]
- Vassilyev, A.E. Dynamics of ultrastructural characters of Drosophyllum lusitanicum Link (Droseraceae) digestive glands during maturation and after stimulation. Taiwania 2005, 50, 167–182. [Google Scholar]
- Atsuzawa, K.; Kanaizumi, D.; Ajisaka, M.; Kamada, T.; Sakamoto, K.; Matsushima, H.; Kaneko, Y. Fine structure of Aldrovanda vesiculosa L: The peculiar lifestyle of an aquatic carnivorous plant elucidated by electron microscopy using cryo-techniques. Microscopy 2020, 69, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Płachno, B.J.; Kapusta, M.; Stolarczyk, P.; Świątek, P. Arabinogalactan Proteins in the Digestive Glands of Dionaea muscipula J.Ellis Traps. Cells 2022, 11, 586. [Google Scholar] [CrossRef] [PubMed]
- Heslop-Harrison, Y.; Heslop-Harrison, J. The digestive glands of Pinguicula: Structure and cytochemistry. Ann. Bot. 1981, 47, 293–319. [Google Scholar] [CrossRef]
- Fineran, B.A. Glandular trichomes in Utricularia: A review of their structure and function. Isr. J. Bot. 1985, 34, 295–330. [Google Scholar] [CrossRef]
- Płachno, B.J.; Kozieradzka-Kiszkurno, M.; Świa̧tek, P. Functional utrastructure of Genlisea (Lentibulariaceae) digestive hairs. Ann. Bot. 2007, 100, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Gunning, B.E.S.; Pate, J.S. “Transfer cells” Plant cells with wall ingrowths, specialized in relation to short distance transport of solutes-Their occurrence, structure, and development. Protoplasma 1969, 68, 107–133. [Google Scholar] [CrossRef]
- Pate, J.S.; Gunning, B.E.S. Transfer cells. Annu. Rev. Plant Physiol. 1972, 23, 173–196. [Google Scholar] [CrossRef]
- Offler, C.E.; McCurdy, D.W.; Patrick, J.W.; Talbot, M.J. Transfer Cells: Cells Specialized for a Special Purpose. Annu. Rev. Plant Biol. 2003, 54, 431–454. [Google Scholar] [CrossRef] [PubMed]
- Offler, C.E.; Patrick, J.W. Transfer cells: What regulates the development of their intricate wall labyrinths? New Phytol. 2020, 228, 427–444. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.; Morris, J.L.; Axe, L.; Taylor, W.A.; Duckett, J.G.; Kenrick, P.; Pressel, S. Earliest record of transfer cells in Lower Devonian plants. New Phytol. 2022, 233, 1456–1465. [Google Scholar] [CrossRef]
- Vaughn, K.C.; Talbot, M.J.; Offler, C.E.; McCurdy, D.W. Wall ingrowths in epidermal transfer cells of Vicia faba cotyledons are modified primary walls marked by localized accumulations of arabinogalactan proteins. Plant Cell Physiol. 2007, 48, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Wardini, T.; Wang, X.D.; Offler, C.E.; Patrick, J.W. Induction of wall ingrowths of transfer cells occurs rapidly and depends upon gene expression in cotyledons of developing Vicia faba seeds. Protoplasma 2007, 231, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Wheeler, S.; Xia, X.; Radchuk, R.; Weber, H.; Offler, C.E.; Patrick, J.W. Differential transcriptional networks associated with key phases of ingrowth wall construction in trans-differentiating epidermal transfer cells of Vicia faba cotyledons. BMC Plant Biol. 2015, 15, 103. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Talbot, M.J.; Offler, C.E.; McCurdy, D.W. Transfer cell wall architecture: A contribution towards understanding localized wall deposition. Protoplasma 2002, 219, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Talbot, M.J.; Wasteneys, G.; McCurdy, D.W.; Offler, C.E. Research note: Deposition patterns of cellulose microfibrils in flange wall ingrowths of transfer cells indicate clear parallels with those of secondary wall thickenings. Funct. Plant Biol. 2007, 34, 307–313. [Google Scholar] [CrossRef]
- Sobczak, M.; Golinowski, W. Structure of cyst nematode feeding sites. In Cell Biology of Plant Nematode Parasitism; Berg, R.H., Taylor, C., Eds.; Springer: Berlin, Germany, 2009; pp. 153–187. [Google Scholar]
- Rodiuc, N.; Vieira, P.; Banora, M.Y.; de Almeida Engler, J. On the track of transfer cell formation by specialized plant-parasitic nematodes. Front. Plant Sci. 2014, 5, 160. [Google Scholar] [CrossRef]
- Bozbuga, R.; Lilley, C.J.; Knox, J.P.; Urwin, P.E. Host-specific signatures of the cell wall changes induced by the plant parasitic nematode, Meloidogyne incognita. Sci. Rep. 2018, 8, 17302. [Google Scholar] [CrossRef]
- Amiard, V.; Demmig-Adams, B.; Mueh, K.E.; Turgeon, R.; Combs, A.F.; Adams, W.W. Role of light and jasmonic acid signaling in regulating foliar phloem cell wall ingrowth development. New Phytol. 2007, 173, 722–731. [Google Scholar] [CrossRef]
- Edwards, J.; Martin, A.P.; Andriunas, F.; Offler, C.E.; Patrick, J.W.; McCurdy, D.W. GIGANTEA is a component of a regulatory pathway determining wall ingrowth deposition in phloem parenchyma transfer cells of Arabidopsis thaliana. Plant J. 2010, 63, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Arun Chinnappa, K.S.; Thu, T.S.N.; Hou, J.; Wu, Y.; McCurdy, D.W. Phloem parenchyma transfer cells in Arabidopsis—An experimental system to identify transcriptional regulators of wall ingrowth formation. Front. Plant Sci. 2013, 4, 102. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Song, W.; Sage, T.; DellaPenna, D. Role of callose synthases in transfer cell wall development in tocopherol deficient Arabidopsis mutants. Front. Plant Sci. 2014, 5, 46. [Google Scholar] [CrossRef] [PubMed]
- Thiel, J.; Weier, D.; Sreenivasulu, N.; Strickert, M.; Weichert, N.; Melzer, M.; Czauderna, T.; Wobus, U.; Weber, H.; Weschke, W. Different hormonal regulation of cellular differentiation and function in nucellar projection and endosperm transfer cells: A microdissection-based transcriptome study of young barley grains. Plant Physiol. 2008, 148, 1436–1452. [Google Scholar] [CrossRef] [PubMed]
- Robert, P.; Jamme, F.; Barron, C.; Bouchet, B.; Saulnier, L.; Dumas, P.; Guillon, F. Change in wall composition of transfer and aleurone cells during wheat grain development. Planta 2011, 233, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, P.; Brewin, N.J. Immunogold localization of callose and other cell wall components in pea nodule transfer cells. Protoplasma 2000, 214, 210–218. [Google Scholar] [CrossRef]
- Ligrone, R.; Vaughn, K.C.; Rascio, N. A cytochemical and immunocytochemical analysis of the wall labyrinth apparatus in leaf transfer cells in Elodea canadensis. Ann. Bot. 2011, 107, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Henry, J.S.; Lopez, R.A.; Renzaglia, K.S. Differential localization of cell wall polymers across generations in the placenta of Marchantia polymorpha. J. Plant Res. 2020, 133, 911–924. [Google Scholar] [CrossRef]
- Henry, J.S.; Renzaglia, K.S. The placenta of physcomitrium patens: Transfer cell wall polymers compared across the three bryophyte groups. Diversity 2021, 13, 378. [Google Scholar] [CrossRef] [PubMed]
- Henry, J.S.; Ligrone, R.; Vaughn, K.C.; Lopez, R.A.; Renzaglia, K.S. Cell wall polymers in the Phaeoceros placenta reflect developmental and functional differences across generations. Bryophyt. Divers. Evol. 2021, 43, 265–283. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.E.; Albert, V.A.; Chase, M.W. Relationships of Droseraceae: A cladistic analysis of RBCL sequence and morphological data. Am. J. Bot. 1994, 81, 1027–1037. [Google Scholar] [CrossRef]
- Cross, A. Aldrovanda: The Waterwheel Plant; Redfern Natural History Productions Ltd.: Poole, UK, 2012; ISBN 190878704X. [Google Scholar]
- Poppinga, S.; Joyeux, M. Different mechanics of snap-trapping in the two closely related carnivorous plants Dionaea muscipula and Aldrovanda vesiculosa. Phys. Rev. E 2011, 84, 041928. [Google Scholar] [CrossRef]
- Adamec, L. Biological flora of Central Europe: Aldrovanda vesiculosa L. Perspect. Plant Ecol. Evol. Syst. 2018, 35, 8–21. [Google Scholar] [CrossRef]
- Płachno, B.J.; Strzemski, M.; Dresler, S.; Adamec, L.; Wojas-Krawczyk, K.; Sowa, I.; Danielewicz, A.; Miranda, V.F.O. A chemometry of Aldrovanda vesiculosa l. (waterwheel, droseraceae) populations. Molecules 2021, 26, 72. [Google Scholar] [CrossRef]
- Knox, J.P.; Day, S.; Roberts, K. A set of cell surface glycoproteins forms an early position, but not cell type, in the root apical meristem of Daucus carota L. Development 1989, 106, 47–56. [Google Scholar] [CrossRef]
- Pennell, R.I.; Knox, J.P.; Scofield, G.N.; Selvendran, R.R.; Roberts, K. A family of abundant plasma membrane-associated glycoproteins related to the arabinogalactan proteins is unique to flowering plants. J. Cell Biol. 1989, 108, 1967–1977. [Google Scholar] [CrossRef]
- Pennell, R.I.; Janniche, L.; Kjellbom, P.; Scofield, G.N.; Peart, J.M.; Roberts, K. Developmental Regulation of a Plasma Membrane Arabinogalactan Protein Epitope in Oilseed Rape Flowers. Plant Cell 1991, 3, 1317–1326. [Google Scholar] [CrossRef]
- Knox, J.P.; Linstead, P.J.; Cooper, J.P.C.; Roberts, K. Developmentally regulated epitopes of cell surface arabinogalactan proteins and their relation to root tissue pattern formation. Plant J. 1991, 1, 317–326. [Google Scholar] [CrossRef]
- Verhertbruggen, Y.; Marcus, S.E.; Haeger, A.; Ordaz-Ortiz, J.J.; Knox, J.P. An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr. Res. 2009, 344, 1858–1862. [Google Scholar] [CrossRef] [PubMed]
- Kerafast. Available online: www.kerafast.com (accessed on 1 December 2021).
- McCartney, L.; Marcus, S.E.; Knox, J.P. Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J. Histochem. Cytochem. 2005, 53, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Marcus, S.E.; Verhertbruggen, Y.; Hervé, C.; Ordaz-Ortiz, J.J.; Farkas, V.; Pedersen, H.L.; Willats, W.G.; Knox, J.P. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 2008, 8, 60. [Google Scholar] [CrossRef]
- Marcus, S.E.; Blake, A.W.; Benians, T.A.S.; Lee, K.J.D.; Poyser, C.; Donaldson, L.; Leroux, O.; Rogowski, A.; Petersen, H.L.; Boraston, A.; et al. Restricted access of proteins to mannan polysaccharides in intact plant cell walls. Plant J. 2010, 64, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Wędzony, M. Fluorescence Microscopy for Botanists; Department of Plant Physiology Monographs 5: Kraków, Poland, 1996; pp. 1–128. [Google Scholar]
- Jauh, G.Y.; Lord, E.M. Localization of pectins and arabinogalactan-proteins in lily (Lilium longiflorum L.) pollen tube and style, and their possible roles in pollination. Planta 1996, 199, 251–261. [Google Scholar] [CrossRef]
- McCurdy, D.W.; Patrick, J.W.; Offler, C.E. Wall ingrowth formation in transfer cells: Novel examples of localized wall deposition in plant cells. Curr. Opin. Plant Biol. 2008, 11, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Showalter, A.M.; Keppler, B.; Lichtenberg, J.; Gu, D.; Welch, L.R. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 2010, 153, 485–513. [Google Scholar] [CrossRef] [PubMed]
- Lamport, D.T.A.; Kieliszewski, M.J.; Showalter, A.M. Salt stress upregulates periplasmic arabinogalactan proteins: Using salt stress to analyse AGP function. New Phytol. 2006, 169, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Lamport, D.T.A.; Várnai, P. Periplasmic arabinogalactan glycoproteins act as a calcium capacitor that regulates plant growth and development. New Phytol. 2013, 197, 58–64. [Google Scholar] [CrossRef]
- Yan, Y.; Takáč, T.; Li, X.; Chen, H.; Wang, Y.; Xu, E.; Xie, L.; Su, Z.; Šamaj, J.; Xu, C. Variable content and distribution of arabinogalactan proteins in banana (Musa spp.) under low temperature stress. Front. Plant Sci. 2015, 6, 353. [Google Scholar] [CrossRef]
- Leszczuk, A.; Szczuka, E.; Zdunek, A. Arabinogalactan proteins: Distribution during the development of male and female gametophytes. Plant Physiol. Biochem. 2019, 135, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Płachno, B.J.; Kapusta, M.; Świątek, P.; Banaś, K.; Miranda, V.F.O.; Bogucka-Kocka, A. Spatio-temporal distribution of cell wall components in the placentas, ovules and female gametophytes of Utricularia during pollination. Int. J. Mol. Sci. 2021, 22, 5622. [Google Scholar] [CrossRef] [PubMed]
- Šamaj, J.; Šamajová, O.; Peters, M.; Baluška, F.; Lichtscheidl, I.; Knox, J.P.; Volkmann, D. Immunolocalization of LM2 arabinogalactan protein epitope associated with endomembranes of plant cells. Protoplasma 2000, 212, 186–196. [Google Scholar] [CrossRef]
- Schulze, W.X.; Sanggaard, K.W.; Kreuzer, I.; Knudsen, A.D.; Bemm, F.; Thøgersen, I.B.; Bräutigam, A.; Thomsen, L.R.; Schliesky, S.; Dyrlund, T.F.; et al. The protein composition of the digestive fluid from the venus flytrap sheds light on prey digestion mechanisms. Mol. Cell. Proteom. 2012, 11, 1306–1319. [Google Scholar] [CrossRef] [PubMed]
- Bemm, F.; Becker, D.; Larisch, C.; Kreuzer, I.; Escalante-Perez, M.; Schulze, W.X.; Ankenbrand, M.; Van De Weyer, A.L.; Krol, E.; Al-Rasheid, K.A.; et al. Venus flytrap carnivorous lifestyle builds on herbivore defense strategies. Genome Res. 2016, 26, 812–825. [Google Scholar] [CrossRef] [PubMed]
- Darvill, A.; Hahn, M.G.; O’Neill, M.A.; York, W.S. Structural Studies of Complex Carbohydrates of Plant Cell Walls; U.S. Department of Energy Office of Scientific and Technical Information: Athens, GA, USA, 2015.
- Liners, F.; Letesson, J.J.; Didembourg, C.; Van Cutsem, P. Monoclonal Antibodies against Pectin: Recognition of a Conformation Induced by Calcium. Plant Physiol. 1989, 91, 1419–1424. [Google Scholar] [CrossRef]
- Bergau, N.; Bennewitz, S.; Syrowatka, F.; Hause, G.; Tissier, A. The development of type VI glandular trichomes in the cultivated tomato Solanum lycopersicum and a related wild species S. habrochaites. BMC Plant Biol. 2015, 15, 289. [Google Scholar] [CrossRef]
- Bowling, A.J.; Maxwell, H.B.; Vaughn, K.C. Unusual trichome structure and composition in mericarps of catchweed bedstraw (Galium aparine). Protoplasma 2008, 233, 223–230. [Google Scholar] [CrossRef]
- Livingston, S.J.; Bae, E.J.; Unda, F.; Hahn, M.G.; Mansfield, S.D.; Page, J.E.; Samuels, A.L. Cannabis Glandular Trichome Cell Walls Undergo Remodeling to Store Specialized Metabolites. Plant Cell Physiol. 2021, 62, 1944–1962. [Google Scholar] [CrossRef]
- Sun, X.; Andrew, I.G.; Harris, P.J.; Hoskin, S.O.; Joblin, K.N.; He, Y. Mapping Pectic-Polysaccharide Epitopes in Cell Walls of Forage Chicory (Cichorium intybus) Leaves. Front. Plant Sci. 2021, 12, 762121. [Google Scholar] [CrossRef]
- Hayashi, T.; Kaida, R. Functions of xyloglucan in plant cells. Mol. Plant 2011, 4, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.B.; Cosgrove, D.J. Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol. 2015, 56, 180–194. [Google Scholar] [CrossRef] [PubMed]

| Epitope/ Type of Cell Wall/Cell | Arabinogalactans | Homogalacturonans | Hemicelluloses | Heteromannans | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| JIM8 | JIM13 | JIM14 | JIM5 | JIM7 | LM5 | LM6 | LM19 | LM15 | LM25 | LM21 | LM22 | |
| Ordinary epidermal cell wall | + | +++ | - | ++ | ++ | +++ | + | ++ | +++ | +++ | - | - |
| Gland head cell wall | +++ | +++ | +++ | + | + | + | + | ++ | ++ | ++ | - | - |
| Gland stalk cell wall | ++ | ++ | ++ | + | + | ++ | ++ | + | +++ | +++ | - | - |
| Ingrowths of gland basal cell wall | + | +++ | +++ | + | + | + | + | - | +++ | +++ | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Płachno, B.J.; Kapusta, M.; Stolarczyk, P.; Świątek, P.; Strzemski, M.; Miranda, V.F.O. Immunocytochemical Analysis of the Wall Ingrowths in the Digestive Gland Transfer Cells in Aldrovanda vesiculosa L. (Droseraceae). Cells 2022, 11, 2218. https://doi.org/10.3390/cells11142218
Płachno BJ, Kapusta M, Stolarczyk P, Świątek P, Strzemski M, Miranda VFO. Immunocytochemical Analysis of the Wall Ingrowths in the Digestive Gland Transfer Cells in Aldrovanda vesiculosa L. (Droseraceae). Cells. 2022; 11(14):2218. https://doi.org/10.3390/cells11142218
Chicago/Turabian StylePłachno, Bartosz J., Małgorzata Kapusta, Piotr Stolarczyk, Piotr Świątek, Maciej Strzemski, and Vitor F. O. Miranda. 2022. "Immunocytochemical Analysis of the Wall Ingrowths in the Digestive Gland Transfer Cells in Aldrovanda vesiculosa L. (Droseraceae)" Cells 11, no. 14: 2218. https://doi.org/10.3390/cells11142218
APA StylePłachno, B. J., Kapusta, M., Stolarczyk, P., Świątek, P., Strzemski, M., & Miranda, V. F. O. (2022). Immunocytochemical Analysis of the Wall Ingrowths in the Digestive Gland Transfer Cells in Aldrovanda vesiculosa L. (Droseraceae). Cells, 11(14), 2218. https://doi.org/10.3390/cells11142218









