Differential Susceptibility of Retinal Neurons to the Loss of Mitochondrial Biogenesis Factor Nrf1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Breeding
2.2. Immunohistochemistry
2.3. Photopic Electroretinography
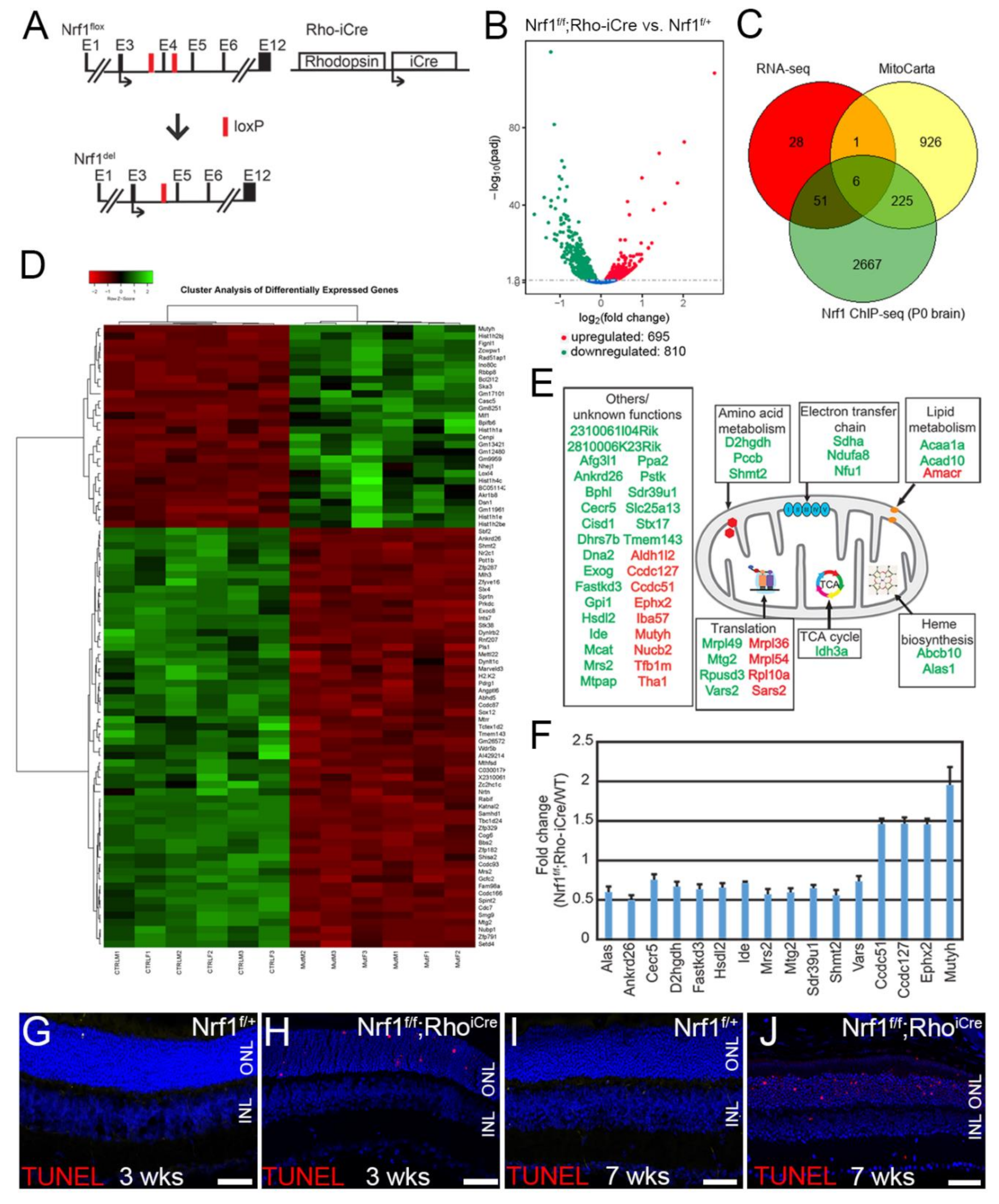
2.4. RNA-Sequencing Analysis
2.5. Quantitative Reverse Transcriptase PCR
2.6. Terminal Deoxynucleotidyl Transferase dUTP Nick-End Labeling (TUNEL) Assay
2.7. Statistical Analysis
3. Results
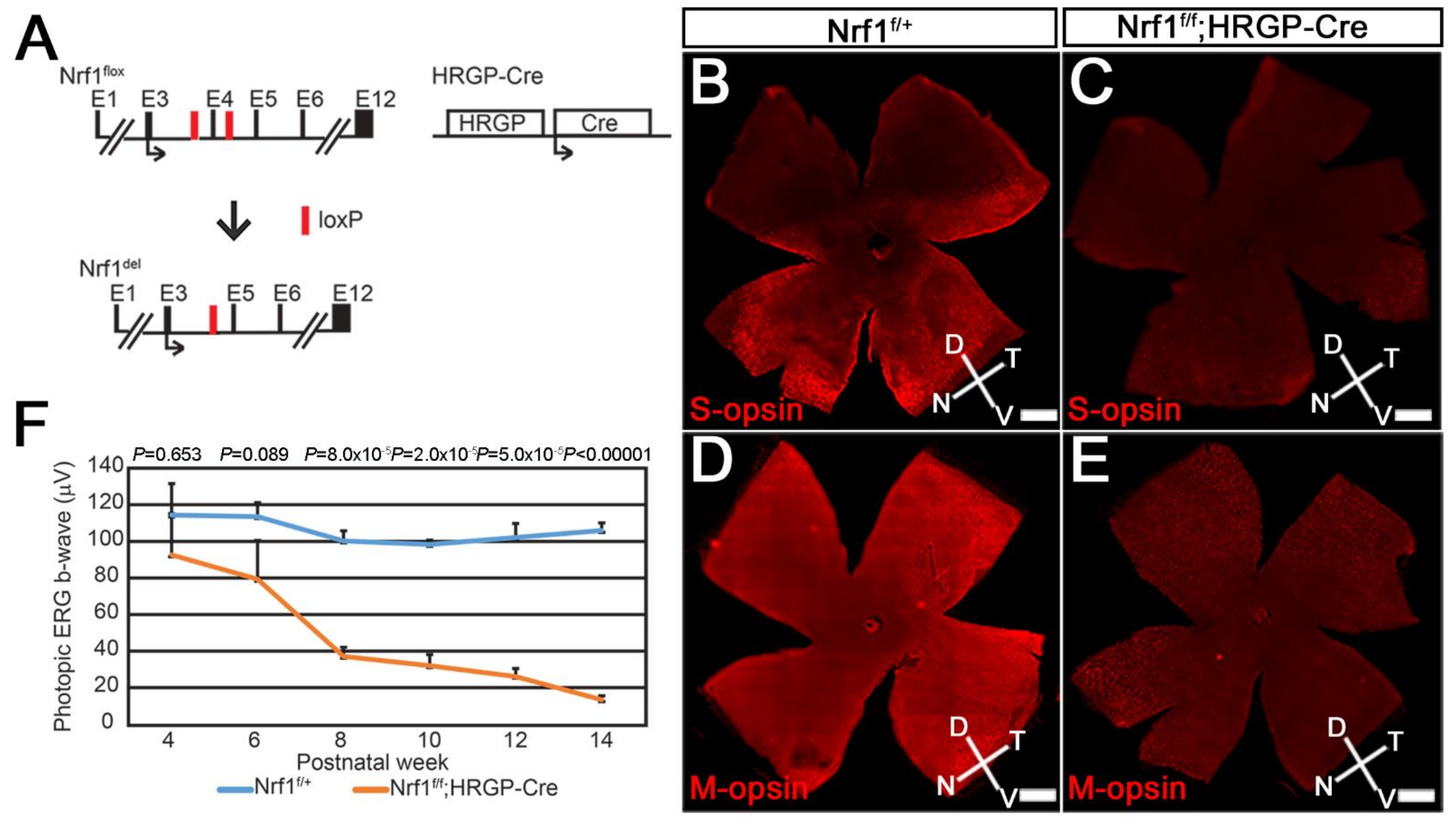
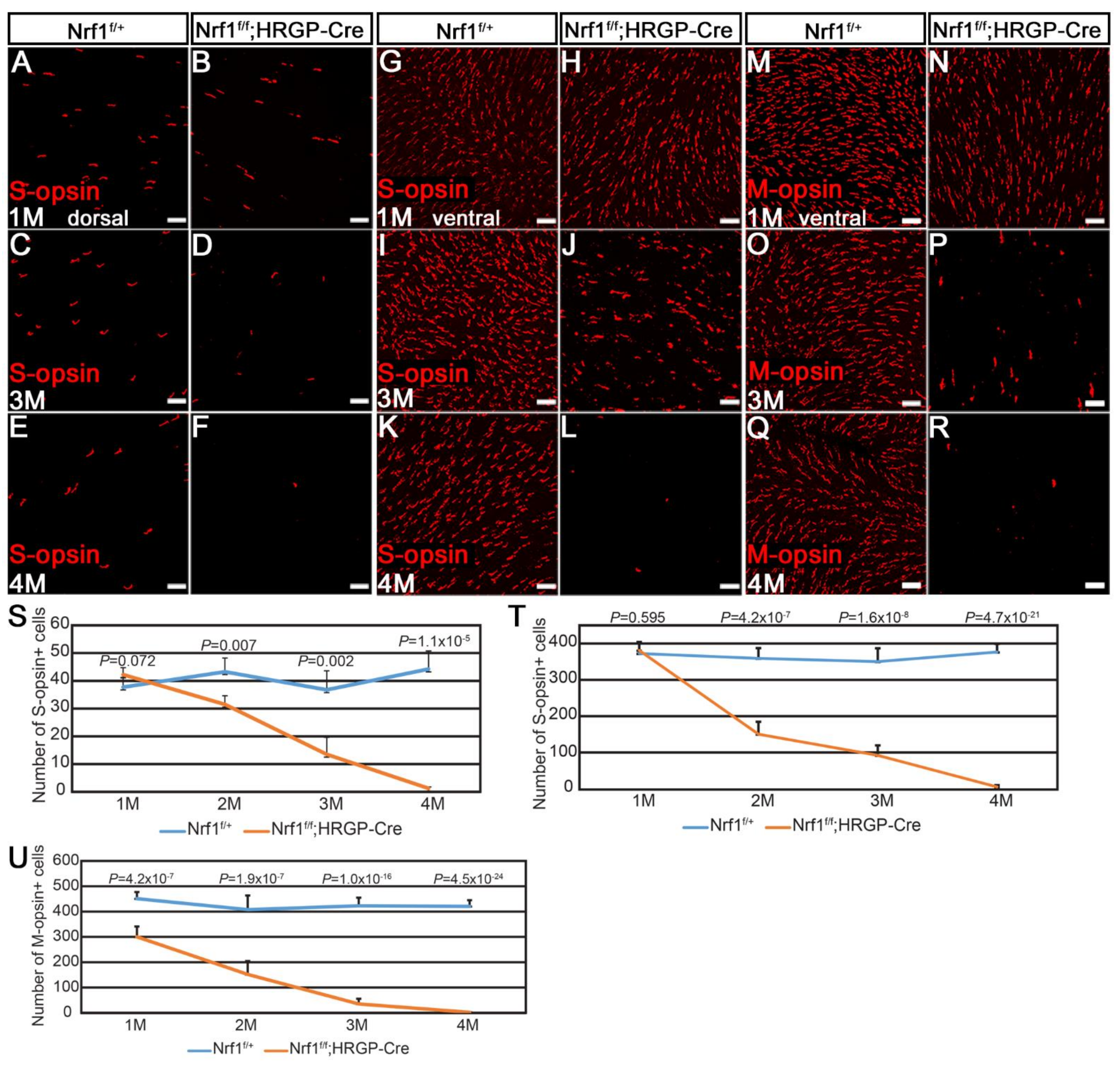
3.1. Progressive Degeneration in Nrf1-Deficient Cone Photoreceptors
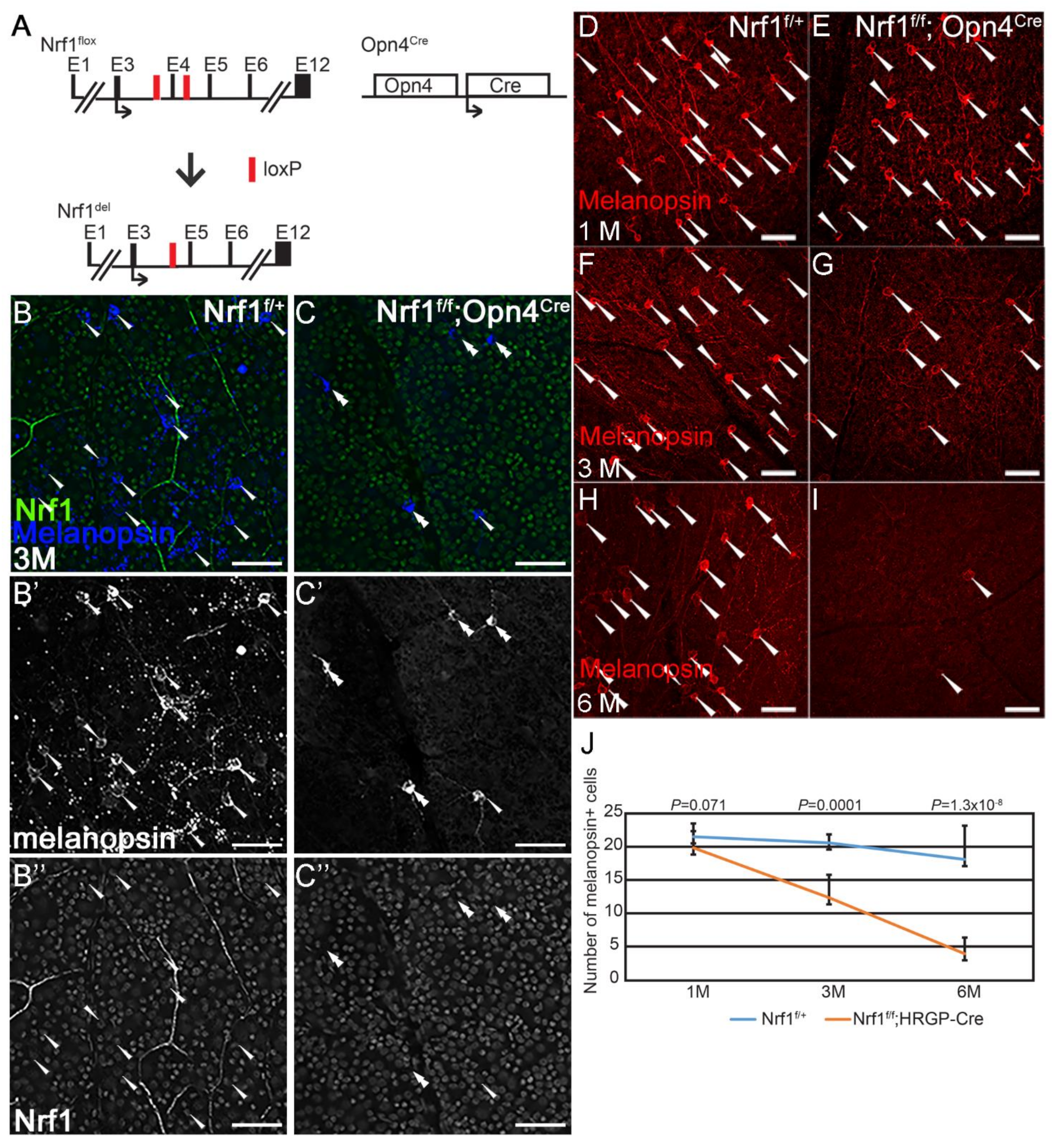
3.2. Slow and Progressive Reduction of Nrf1-Depleted ipRGC
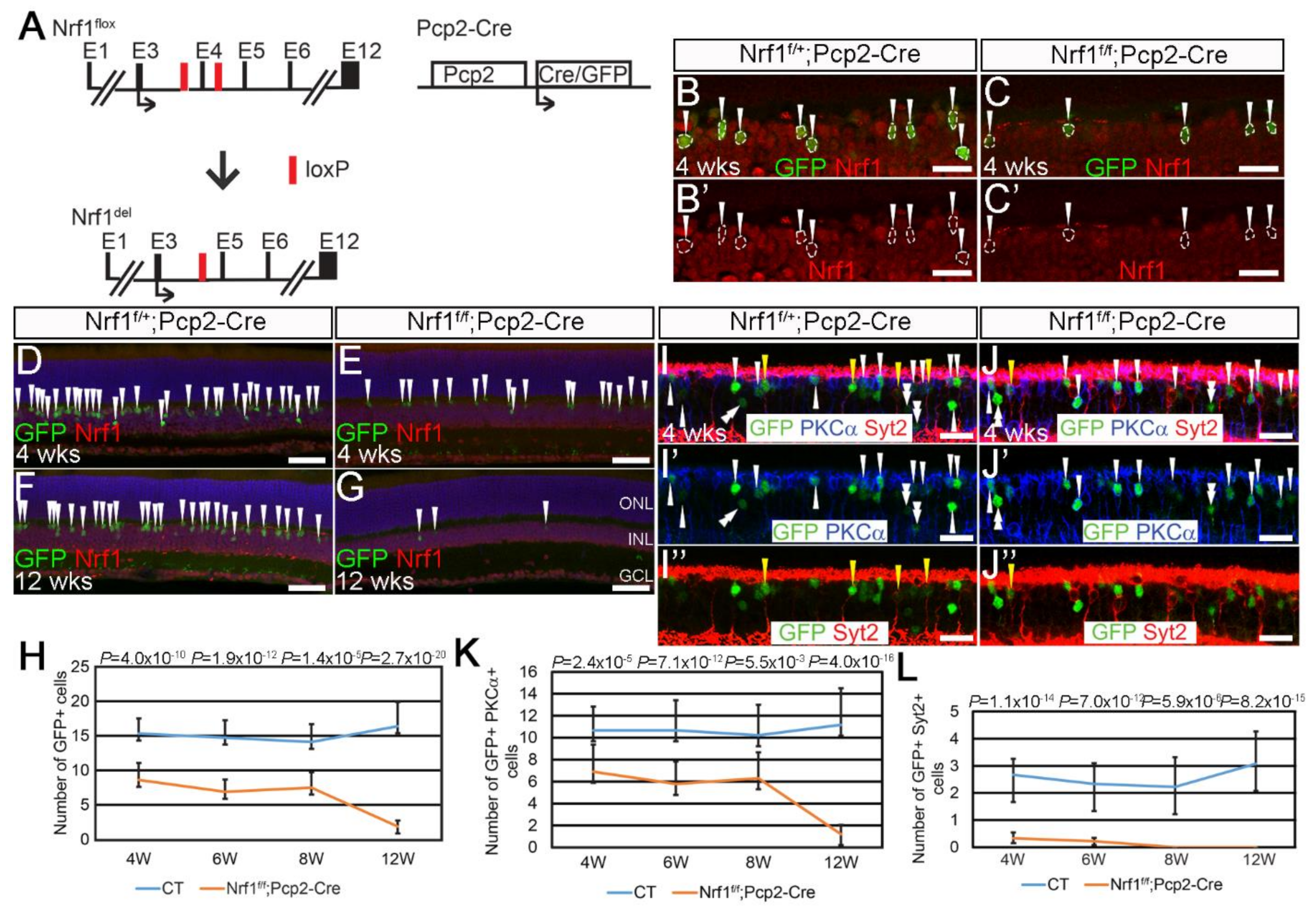
3.3. Highly Sensitive Type 2 and 6 Cone Bipolar Cells to Nrf1 Deletion
3.4. The Most Sensitive Genes in Nrf1-Deficient Rod Photoreceptors
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chan, D.C. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 2006, 22, 79–99. [Google Scholar] [CrossRef] [Green Version]
- Miranda, M.; Bonekamp, N.A.; Kühl, I. Starting the engine of the powerhouse: Mitochondrial transcription and beyond. Biol. Chem. 2022, 403, 779–805. [Google Scholar] [CrossRef] [PubMed]
- Bomba-Warczak, E.; Savas, J.N. Long-lived mitochondrial proteins and why they exist. Trends Cell Biol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Vafai, S.B.; Mootha, V.K. Mitochondrial disorders as windows into an ancient organelle. Nature 2012, 491, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Giarmarco, M.M.; Brock, D.C.; Robbings, B.M.; Cleghorn, W.M.; Tsantilas, K.A.; Kuch, K.C.; Brockerhoff, S.E. Daily mitochondrial dynamics in cone photoreceptors. Proc. Natl. Acad. Sci. USA 2020, 117, 28816–28827. [Google Scholar] [CrossRef]
- Chang, J.Y.; Shi, L.; Ko, M.L.; Ko, G.Y. Circadian Regulation of Mitochondrial Dynamics in Retinal Photoreceptors. J. Biol. Rhythm. 2018, 33, 151–165. [Google Scholar] [CrossRef]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C.; et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Virbasius, C.A.; Virbasius, J.V.; Scarpulla, R.C. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993, 7, 2431–2445. [Google Scholar] [CrossRef]
- Hock, M.B.; Kralli, A. Transcriptional control of mitochondrial biogenesis and function. Annu. Rev. Physiol. 2009, 71, 177–203. [Google Scholar] [CrossRef] [Green Version]
- Spiegelman, B.M. Transcriptional control of mitochondrial energy metabolism through the PGC1 coactivators. Novartis Found. Symp. 2007, 287, 60–63. [Google Scholar]
- Calzone, F.J.; Hoog, C.; Teplow, D.B.; Cutting, A.E.; Zeller, R.W.; Britten, R.J.; Davidson, E.H. Gene regulatory factors of the sea urchin embryo. I. Purification by affinity chromatography and cloning of P3A2, a novel DNA-binding protein. Development 1991, 112, 335–350. [Google Scholar] [CrossRef] [PubMed]
- DeSimone, S.M.; White, K. The Drosophila erect wing gene, which is important for both neuronal and muscle development, encodes a protein which is similar to the sea urchin P3A2 DNA binding protein. Mol. Cell Biol. 1993, 13, 3641–3649. [Google Scholar]
- Becker, T.S.; Burgess, S.M.; Amsterdam, A.H.; Allende, M.L.; Hopkins, N. not really finished is crucial for development of the zebrafish outer retina and encodes a transcription factor highly homologous to human Nuclear Respiratory Factor-1 and avian Initiation Binding Repressor. Development 1998, 125, 4369–4378. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, L.; Engman, H.; Miller, J.B. Coding sequence, chromosomal localization, and expression pattern of Nrf1: The mouse homolog of Drosophila erect wing. Mamm. Genome 2000, 11, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.J.; Scarpulla, R.C. NRF-1: A trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 1990, 4, 1023–1034. [Google Scholar] [CrossRef] [Green Version]
- Satoh, J.; Kawana, N.; Yamamoto, Y. Pathway Analysis of ChIP-Seq-Based NRF1 Target Genes Suggests a Logical Hypothesis of their Involvement in the Pathogenesis of Neurodegenerative Diseases. Gene Regul. Syst. Biol. 2013, 7, 139–152. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Aldinger, K.A.; Cheng, C.V.; Kiyama, T.; Dave, M.; McNamara, H.K.; Zhao, W.; Stafford, J.M.; Descostes, N.; Lee, P.; et al. NRF1 association with AUTS2-Polycomb mediates specific gene activation in the brain. Mol. Cell 2021, 81, 4663–4676.e8. [Google Scholar] [CrossRef]
- Takayama, K.I.; Kosaka, T.; Suzuki, T.; Hongo, H.; Oya, M.; Fujimura, T.; Suzuki, Y.; Inoue, S. Subtype-specific collaborative transcription factor networks are promoted by OCT4 in the progression of prostate cancer. Nat. Commun. 2021, 12, 3766. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, C.; Chen, X.; Takada, M.; Fan, C.; Zheng, X.; Wen, H.; Liu, Y.; Pestell, R.G.; Aird, K.M.; et al. EglN2 associates with the NRF1-PGC1α complex and controls mitochondrial function in breast cancer. EMBO J. 2015, 34, 2953–2970. [Google Scholar] [CrossRef]
- Hossain, M.B.; Ji, P.; Anish, R.; Jacobson, R.H.; Takada, S. Poly(ADP-ribose) Polymerase 1 Interacts with Nuclear Respiratory Factor 1 (NRF-1) and Plays a Role in NRF-1 Transcriptional Regulation. J. Biol. Chem. 2009, 284, 8621–8632. [Google Scholar] [CrossRef] [Green Version]
- Herzig, R.P.; Andersson, U.; Scarpulla, R.C. Dynein light chain interacts with NRF-1 and EWG, structurally and functionally related transcription factors from humans and drosophila. J. Cell Sci. 2000, 113, 4263–4273. [Google Scholar] [CrossRef] [PubMed]
- Kiyama, T.; Long, Y.; Chen, C.K.; Whitaker, C.M.; Shay, A.; Wu, H.; Badea, T.C.; Mohsenin, A.; Parker-Thornburg, J.; Klein, W.H.; et al. Essential Roles of Tbr1 in the Formation and Maintenance of the Orientation-Selective J-RGCs and a Group of OFF-Sustained RGCs in Mouse. Cell Rep. 2019, 27, 900–915.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scarpulla, R.C.; Vega, R.B.; Kelly, D.P. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol. Metab. 2012, 23, 459–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Chen, D.; Sauvé, Y.; McCandless, J.; Chen, Y.J.; Chen, C.K. Rhodopsin-iCre transgenic mouse line for Cre-mediated rod-specific gene targeting. Genesis 2005, 41, 73–80. [Google Scholar] [CrossRef]
- Ecker, J.L.; Dumitrescu, O.N.; Wong, K.Y.; Alam, N.M.; Chen, S.K.; LeGates, T.; Renna, J.M.; Prusky, G.T.; Berson, D.M.; Hattar, S. Melanopsin-expressing retinal ganglion-cell photoreceptors: Cellular diversity and role in pattern vision. Neuron 2010, 67, 49–60. [Google Scholar] [CrossRef] [Green Version]
- Le, Y.Z.; Ash, J.D.; Al-Ubaidi, M.R.; Chen, Y.; Ma, J.X.; Anderson, R.E. Targeted expression of Cre recombinase to cone photoreceptors in transgenic mice. Mol. Vis. 2004, 10, 1011–1018. [Google Scholar]
- Barski, J.J.; Dethleffsen, K.; Meyer, M. Cre recombinase expression in cerebellar Purkinje cells. Genesis 2000, 28, 93–98. [Google Scholar] [CrossRef]
- Lewis, P.M.; Gritli-Linde, A.; Smeyne, R.; Kottmann, A.; McMahon, A.P. Sonic hedgehog signaling is required for expansion of granule neuron precursors and patterning of the mouse cerebellum. Dev. Biol. 2004, 270, 393–410. [Google Scholar] [CrossRef] [Green Version]
- Kiyama, T.; Chen, C.K.; Wang, S.W.; Pan, P.; Ju, Z.; Wang, J.; Takada, S.; Klein, W.H.; Mao, C.A. Essential roles of mitochondrial biogenesis regulator Nrf1 in retinal development and homeostasis. Mol. Neurodegener. 2018, 13, 56. [Google Scholar] [CrossRef]
- Applebury, M.L.; Antoch, M.P.; Baxter, L.C.; Chun, L.L.; Falk, J.D.; Farhangfar, F.; Kage, K.; Krzystolik, M.G.; Lyass, L.A.; Robbins, J.T. The murine cone photoreceptor: A single cone type expresses both S and M opsins with retinal spatial patterning. Neuron 2000, 27, 513–523. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.V.; Weick, M.; Demb, J.B. Spectral and temporal sensitivity of cone-mediated responses in mouse retinal ganglion cells. J. Neurosci. 2011, 31, 7670–7681. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Breuninger, T.; Euler, T. Chromatic coding from cone-type unselective circuits in the mouse retina. Neuron 2013, 77, 559–571. [Google Scholar] [CrossRef] [Green Version]
- Nadal-Nicolas, F.M.; Kunze, V.P.; Ball, J.M.; Peng, B.T.; Krishnan, A.; Zhou, G.; Dong, L.; Li, W. True S-cones are concentrated in the ventral mouse retina and wired for color detection in the upper visual field. eLife 2020, 9, e56840. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.M.; Chen, S.K.; Hattar, S. Intrinsically photosensitive retinal ganglion cells: Many subtypes, diverse functions. Trends Neurosci. 2011, 34, 572–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong-Riley, M.T. Energy metabolism of the visual system. Eye Brain 2010, 2, 99–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Q.; Ivanova, E.; Ganjawala, T.H.; Pan, Z.H. Cre-mediated recombination efficiency and transgene expression patterns of three retinal bipolar cell-expressing Cre transgenic mouse lines. Mol. Vis. 2013, 19, 1310–1320. [Google Scholar]
- Pagliarini, D.J.; Calvo, S.E.; Chang, B.; Sheth, S.A.; Vafai, S.B.; Ong, S.E.; Walford, G.A.; Sugiana, C.; Boneh, A.; Chen, W.K.; et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008, 134, 112–123. [Google Scholar] [CrossRef] [Green Version]
- Calvo, S.E.; Clauser, K.R.; Mootha, V.K. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016, 44, D1251–D1257. [Google Scholar] [CrossRef] [Green Version]
- LaVail, M.M. Rod outer segment disk shedding in rat retina: Relationship to cyclic lighting. Science 1976, 194, 1071–1074. [Google Scholar] [CrossRef]
- Okawa, H.; Sampath, A.P.; Laughlin, S.B.; Fain, G.L. ATP consumption by mammalian rod photoreceptors in darkness and in light. Curr. Biol. 2008, 18, 1917–1921. [Google Scholar] [CrossRef] [Green Version]
- Niven, J.E.; Laughlin, S.B. Energy limitation as a selective pressure on the evolution of sensory systems. J. Exp. Biol. 2008, 211, 1792–1804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ait-Ali, N.; Fridlich, R.; Millet-Puel, G.; Clerin, E.; Delalande, F.; Jaillard, C.; Blond, F.; Perrocheau, L.; Reichman, S.; Byrne, L.C.; et al. Rod-derived cone viability factor promotes cone survival by stimulating aerobic glycolysis. Cell 2015, 161, 817–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, N.; Zhang, Z.; Keung, J.; Youn, S.B.; Ishibashi, M.; Tian, L.M.; Marshak, D.W.; Solessio, E.; Umino, Y.; Fahrenfort, I.; et al. Molecular and functional architecture of the mouse photoreceptor network. Sci. Adv. 2020, 6, eaba7232. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Yu, A.; Murray, K.; Cortopassi, G. Bipolar cell reduction precedes retinal ganglion neuron loss in a complex 1 knockout mouse model. Brain Res. 2017, 1657, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Kim, S.J.; Sohn, Y.I.; Paik, S.S.; Caplette, R.; Simonutti, M.; Moon, K.H.; Lee, E.J.; Min, K.W.; Kim, M.J.; et al. Mitochondrial Protection by Exogenous Otx2 in Mouse Retinal Neurons. Cell Rep. 2015, 13, 990–1002. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.T.; Prochiantz, A.; Kim, J.W. Donating Otx2 to support neighboring neuron survival. BMB Rep. 2016, 49, 69–70. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.D.; Starikovskaya, E.; Derbeneva, O.; Hosseini, S.; Allen, J.C.; Mikhailovskaya, I.E.; Sukernik, R.I.; Wallace, D.C. The role of mtDNA background in disease expression: A new primary LHON mutation associated with Western Eurasian haplogroup J. Hum. Genet. 2002, 110, 130–138. [Google Scholar] [CrossRef]
- Delettre, C.; Lenaers, G.; Griffoin, J.M.; Gigarel, N.; Lorenzo, C.; Belenguer, P.; Pelloquin, L.; Grosgeorge, J.; Turc-Carel, C.; Perret, E.; et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 2000, 26, 207–210. [Google Scholar] [CrossRef]
- Alexander, C.; Votruba, M.; Pesch, U.E.; Thiselton, D.L.; Mayer, S.; Moore, A.; Rodriguez, M.; Kellner, U.; Leo-Kottler, B.; Auburger, G.; et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 2000, 26, 211–215. [Google Scholar] [CrossRef]
- Siliprandi, R.; Canella, R.; Carmignoto, G.; Schiavo, N.; Zanellato, A.; Zanoni, R.; Vantini, G. N-methyl-D-aspartate-induced neurotoxicity in the adult rat retina. Vis. Neurosci. 1992, 8, 567–573. [Google Scholar] [CrossRef]
- Lipton, S.A.; Rosenberg, P.A. Excitatory amino acids as a final common pathway for neurologic disorders. N. Engl. J. Med. 1994, 330, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Shaw, P.J. Excitatory amino acid receptors, excitotoxicity, and the human nervous system. Curr. Opin. Neurol. Neurosurg. 1993, 6, 414–422. [Google Scholar] [PubMed]
- Yu-Wai-Man, P.; Griffiths, P.G.; Gorman, G.S.; Lourenco, C.M.; Wright, A.F.; Auer-Grumbach, M.; Toscano, A.; Musumeci, O.; Valentino, M.L.; Caporali, L.; et al. Multi-system neurological disease is common in patients with OPA1 mutations. Brain 2010, 133, 771–786. [Google Scholar] [CrossRef] [PubMed]
- Kasai, H.; Hayami, H.; Yamaizumi, Z.; Saitô, H.; Nishimura, S. Detection and identification of mutagens and carcinogens as their adducts with guanosine derivatives. Nucleic Acids Res. 1984, 12, 2127–2136. [Google Scholar] [CrossRef] [Green Version]
- Maki, H. Origins of spontaneous mutations: Specificity and directionality of base-substitution, frameshift, and sequence-substitution mutageneses. Annu. Rev. Genet. 2002, 36, 279–303. [Google Scholar] [CrossRef]
- Nakabeppu, Y.; Kajitani, K.; Sakamoto, K.; Yamaguchi, H.; Tsuchimoto, D. MTH1, an oxidized purine nucleoside triphosphatase, prevents the cytotoxicity and neurotoxicity of oxidized purine nucleotides. DNA Repair 2006, 5, 761–772. [Google Scholar] [CrossRef]
- Chinchore, Y.; Begaj, T.; Wu, D.; Drokhlyansky, E.; Cepko, C.L. Glycolytic reliance promotes anabolism in photoreceptors. eLife 2017, 6, e25946. [Google Scholar] [CrossRef]
- Winkler, B.S. Glycolytic and oxidative metabolism in relation to retinal function. J. Gen. Physiol. 1981, 77, 667–692. [Google Scholar] [CrossRef] [Green Version]
- Bisbach, C.M.; Hass, D.T.; Robbings, B.M.; Rountree, A.M.; Sadilek, M.; Sweet, I.R.; Hurley, J.B. Succinate Can Shuttle Reducing Power from the Hypoxic Retina to the O2-Rich Pigment Epithelium. Cell Rep. 2020, 31, 107606. [Google Scholar] [CrossRef]
- Nishimura, D.Y.; Fath, M.; Mullins, R.F.; Searby, C.; Andrews, M.; Davis, R.; Andorf, J.L.; Mykytyn, K.; Swiderski, R.E.; Yang, B.; et al. Bbs2-null mice have neurosensory deficits, a defect in social dominance, and retinopathy associated with mislocalization of rhodopsin. Proc. Natl. Acad. Sci. USA 2004, 101, 16588–16593. [Google Scholar] [CrossRef] [Green Version]
- Abad, M.A.; Zou, J.; Medina-Pritchard, B.; Nigg, E.A.; Rappsilber, J.; Santamaria, A.; Jeyaprakash, A.A. Ska3 Ensures Timely Mitotic Progression by Interacting Directly with Microtubules and Ska1 Microtubule Binding Domain. Sci. Rep. 2016, 6, 34042. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Wang, M.Q.; Niu, W.B.; Wang, Y.J.; Liu, Y.Y.; Liu, L.Y.; Wang, M.; Zhong, J.; You, H.Y.; Wu, X.H.; et al. SKA3 promotes cell proliferation and migration in cervical cancer by activating the PI3K/Akt signaling pathway. Cancer Cell Int. 2018, 18, 183. [Google Scholar] [CrossRef] [PubMed]
- Kruman, I.I.; Wersto, R.P.; Cardozo-Pelaez, F.; Smilenov, L.; Chan, S.L.; Chrest, F.J.; Emokpae, R., Jr.; Gorospe, M.; Mattson, M.P. Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron 2004, 41, 549–561. [Google Scholar] [CrossRef] [Green Version]
| Gene Name | Description | Log2 Fold Change (Nrf1f/f;RhoiCre/Nrf1f/+) | Adjusted p Value |
|---|---|---|---|
| Ccdc87 | coiled-coil domain containing 87 | −1.6234 | 5.89 × 10−36 |
| Rnf207 | ring finger protein 207 | −1.3834 | 1.10 × 10−44 |
| AI429214 | expressed sequence AI429214 | −1.3369 | 6.84 × 10−24 |
| C030017K20Rik | RIKEN cDNA C030017K20 gene | −1.2807 | 1.19 × 10−31 |
| Zfp182 | zinc finger protein 182 | −1.2239 | 4.54 × 10−40 |
| Tbc1d24 | TBC1 domain family member 24 | −1.2234 | 9.22 × 10−120 |
| Cdc7 | cell division cycle 7 (S. cerevisiae) | −1.2132 | 4.45 × 10−43 |
| Samhd1 | SAM domain and HD domain 1 | −1.139 | 2.41 × 10−82 |
| Zc2hc1c | zinc finger C2HC-type containing 1C | −1.1341 | 4.26 × 10−23 |
| Wdr5b | WD repeat domain 5B | −1.1292 | 8.21 × 10−26 |
| Sprtn | SprT-like N-terminal domain | −1.1122 | 6.12 × 10−39 |
| H2-K2 | histocompatibility 2 K region locus 2 | −1.0906 | 1.58 × 10−22 |
| Shisa2 | shisa family member 2 | −1.0698 | 2.44 × 10−25 |
| Mlh3 | mutL homolog 3 (E. coli) | −1.0473 | 1.15 × 10−44 |
| Ankrd26 * | ankyrin repeat domain 26 | −1.0158 | 1.68 × 10−55 |
| Cog6 | component of oligomeric golgi complex 6 | −1.0071 | 4.78 × 10−49 |
| Zfp287 | zinc finger protein 287 | −0.99342 | 7.33 × 10−27 |
| Mtg2 * | mitochondrial ribosome associated GTPase 2 | −0.97742 | 5.34 × 10−33 |
| Katnal2 | katanin p60 subunit A-like 2 | −0.97416 | 3.88 × 10−34 |
| Mthfsd | methenyltetrahydrofolate synthetase domain containing | −0.97072 | 8.13 × 10−31 |
| Zfp791 | zinc finger protein 791 | −0.96542 | 3.25 × 10−19 |
| Zfp329 | zinc finger protein 329 | −0.9606 | 4.01 × 10−54 |
| Pot1b | protection of telomeres 1B | −0.95894 | 9.62 × 10−21 |
| Bbs2 | Bardet-Biedl syndrome 2 (human) | −0.95414 | 1.22 × 10−63 |
| Dynlrb2 | dynein light chain roadblock-type 2 | −0.94971 | 6.89 × 10−14 |
| Abhd5 | abhydrolase domain containing 5 | −0.9241 | 1.01 × 10−26 |
| Marveld3 | MARVEL (membrane-associating) domain containing 3 | −0.91459 | 6.08 × 10−15 |
| Ccdc166 | coiled-coil domain containing 166 | −0.90586 | 1.81 × 10−31 |
| Exoc8 | exocyst complex component 8 | −0.90494 | 6.33 × 10−36 |
| Stk38 | serine/threonine kinase 38 | −0.89765 | 2.81 × 10−60 |
| Tctex1d2 | Tctex1 domain containing 2 | −0.89357 | 9.03 × 10−14 |
| Gm26572 | predicted gene 26572 | −0.88514 | 1.06 × 10−16 |
| Rabif | RAB interacting factor | −0.87374 | 6.53 × 10−43 |
| Dynlt1c | dynein light chain Tctex-type 1C | −0.87284 | 3.64 × 10−11 |
| Spint2 | serine protease inhibitor Kunitz type 2 | −0.87063 | 2.51 × 10−43 |
| Fam98a | family with sequence similarity 98 member A | −0.86343 | 2.20 × 10−35 |
| Slx4 | SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) | −0.85413 | 9.39 × 10−34 |
| Mtrr | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase | −0.84843 | 1.96 × 10−18 |
| Setd4 | SET domain containing 4 | −0.83398 | 1.40 × 10−17 |
| Gcfc2 | GC-rich sequence DNA binding factor 2 | −0.83387 | 2.49 × 10−15 |
| Sox12 | SRY (sex determining region Y)-box 12 | −0.82933 | 7.00 × 10−21 |
| Sbf2 | SET binding factor 2 | −0.82821 | 2.80 × 10−50 |
| Nubp1 | nucleotide binding protein 1 | −0.82325 | 3.47 × 10−19 |
| Angptl6 | angiopoietin-like 6 | −0.82273 | 9.67 × 10−13 |
| Mrs2 * | MRS2 magnesium homeostasis factor homolog (S. cerevisiae) | −0.81398 | 6.57 × 10−35 |
| Pdrg1 | p53 and DNA damage regulated 1 | −0.79343 | 1.45 × 10−27 |
| Zfyve16 | zinc finger FYVE domain containing 16 | −0.78442 | 1.04 × 10−24 |
| Ints7 | integrator complex subunit 7 | −0.783 | 5.35 × 10−33 |
| Shmt2 * | serine hydroxymethyltransferase 2 (mitochondrial) | −0.78075 | 2.29 × 10−29 |
| Mettl22 | methyltransferase like 22 | −0.77958 | 2.18 × 10−16 |
| Ccdc93 | coiled-coil domain containing 93 | −0.77053 | 2.11 × 10−27 |
| Smg9 | smg-9 homolog nonsense mediated mRNA decay factor (C. elegans) | −0.76729 | 3.83 × 10−24 |
| Tmem143 * | transmembrane protein 143 | −0.75985 | 4.64 × 10−20 |
| Prkdc | protein kinase DNA activated catalytic polypeptide | −0.75349 | 1.45 × 10−18 |
| 2310061I04Rik * | RIKEN cDNA 2310061I04 gene | −0.75092 | 6.93 × 10−17 |
| Pls1 | plastin 1 (I-isoform) | −0.74708 | 2.22 × 10−10 |
| Nrtn | neurturin | −0.74655 | 2.40 × 10−12 |
| Nr2c1 | nuclear receptor subfamily 2 group C member 1 | −0.74308 | 2.67 × 10−19 |
| Hist1h1a | histone cluster 1 H1a | 0.74952 | 7.45 × 10−8 |
| Gm13421 | predicted gene 13421 | 0.77879 | 2.71 × 10−14 |
| Hist1h4c | histone cluster 1 H4c | 0.78665 | 1.79 × 10−8 |
| Bpifb6 | BPI fold containing family B member 6 | 0.79491 | 2.04 × 10−8 |
| Hist1h2be | histone cluster 1 H2be | 0.7962 | 2.31 × 10−14 |
| Nhej1 | nonhomologous end-joining factor 1 | 0.80335 | 1.01 × 10−8 |
| BC051142 | cDNA sequence BC051142 | 0.81214 | 1.83 ×10−10 |
| Hist1h2bj | histone cluster 1 H2bj | 0.83377 | 3.03 ×10−9 |
| Ska3 | spindle and kinetochore associated complex subunit 3 | 0.84721 | 1.54 ×10−9 |
| Gm11961 | predicted gene 11961 | 0.87529 | 2.79 × 10−15 |
| Hist1h1e | histone cluster 1 H1e | 0.93176 | 2.49 × 10−15 |
| Gm9959 | predicted gene 9959 | 0.93908 | 1.03 × 10−11 |
| Rbbp8 | retinoblastoma binding protein 8 | 0.97505 | 3.74 × 10−21 |
| Mlf1 | myeloid leukemia factor 1 | 0.98101 | 3.09 × 10−15 |
| Bcl2l12 | BCL2-like 12 (proline rich) | 0.98105 | 9.18 × 10−13 |
| Gm17101 | predicted gene 17101 | 0.98366 | 7.78 × 10−13 |
| Gm12480 | predicted gene 12480 | 0.98839 | 5.94 × 10−13 |
| Ino80c | INO80 complex subunit C | 0.99506 | 8.83 × 10−55 |
| Gm8251 | predicted gene 8251 | 1.1522 | 1.03 × 10−18 |
| Dsn1 | DSN1 MIND kinetochore complex component homolog (S. cerevisiae) | 1.1532 | 1.85 × 10−18 |
| Loxl4 | lysyl oxidase-like 4 | 1.2333 | 3.80 × 10−21 |
| Mutyh * | mutY homolog (E. coli) | 1.2731 | 3.28 × 10−38 |
| Zcwpw1 | zinc finger CW type with PWWP domain 1 | 1.4136 | 1.66 × 10−67 |
| Rad51ap1 | RAD51 associated protein 1 | 1.5557 | 1.07 × 10−41 |
| Cenpi | centromere protein I | 1.8589 | 4.28 × 10−52 |
| Fignl1 | fidgetin-like 1 | 2.0243 | 2.37 × 10−73 |
| Casc5 | cancer susceptibility candidate 5 | 2.7592 | 6.88 × 10−109 |
| Gene Name | Description | Log2 Fold change (Nrf1f/f;RhoiCre/Nrf1f/+) | Adjusted p Value |
|---|---|---|---|
| Ankrd26 | ankyrin repeat domain-containing protein 26 | −1.0158 | 1.68 × 10−55 |
| Mtg2 | mitochondrial ribosome associated GTPase 2 | −0.97742 | 5.34 × 10−33 |
| Mrs2 | MRS2 magnesium homeostasis factor homolog (S. cerevisiae) | −0.81398 | 6.57 × 10−35 |
| Shmt2 | serine hydroxymethyltransferase 2 (mitochondrial) | −0.78075 | 2.29 × 10−29 |
| Tmem143 | transmembrane protein 143 | −0.75985 | 4.64 × 10−20 |
| 2310061I04Rik | RIKEN cDNA 2310061I04 gene | −0.75092 | 4.12 × 10−19 |
| Fastkd3 | FAST kinase domains 3 | −0.73257 | 6.15 × 10−12 |
| Pstk | phosphoseryl tRNA kinase | −0.73031 | 1.28 × 10−16 |
| Alas1 | aminolevulinic acid synthase 1 | −0.70115 | 9.42 × 10−25 |
| Hsdl2 | hydroxysteroid dehydrogenase like 2 | −0.59599 | 1.75 × 10−20 |
| D2hgdh | D2 hydroxyglutarate dehydrogenase | −0.58842 | 1.56 × 10−11 |
| Sdr39u1 | short chain dehydrogenase/reductase family 39U member 1 | −0.55177 | 1.20 × 10−14 |
| Sdha | succinate dehydrogenase complex subunit A flavoprotein (Fp) | −0.54497 | 5.27 × 10−32 |
| Cecr5 | cat eye syndrome chromosome region candidate 5 | −0.53991 | 1.65 × 10−11 |
| Slc25a13 | solute carrier family 25 (mitochondrial carrier adenine nucleotide translocator) member 13 | −0.53462 | 4.91 × 10−4 |
| Acad10 | acyl Coenzyme A dehydrogenase family member 10 | −0.53414 | 2.61 × 10−6 |
| Pccb | propionyl Coenzyme A carboxylase beta polypeptide | −0.52005 | 2.09 ×10−15 |
| Exog | endo/exonuclease (5′-3′) endonuclease G-like | −0.51228 | 1.76 × 10−9 |
| Ide | insulin degrading enzyme | −0.50673 | 1.97 × 10−22 |
| Mcat | malonyl CoA:ACP acyltransferase (mitochondrial) | −0.48803 | 1.60 × 10−7 |
| Afg3l1 | AFG3-like AAA ATPase 1 | −0.48699 | 6.23 × 10−16 |
| 2810006K23Rik | RIKEN cDNA 2810006K23 gene | −0.47985 | 2.65 × 10−7 |
| Vars2 | valyl-tRNA synthetase 2 mitochondrial (putative) | −0.45605 | 9.55 × 10−7 |
| Mtpap | mitochondrial poly(A)_polymerase | −0.45337 | 1.37 × 10−5 |
| Dna2 | DNA replication helicase 2 homolog (yeast) | −0.44603 | 0.003128 |
| Mrpl49 | mitochondrial ribosomal protein L49 | −0.43346 | 1.64 × 10−8 |
| Nfu1 | NFU1 iron sulfur cluster scaffold homolog (S. cerevisiae) | −0.42766 | 7.19 × 10−9 |
| Rpusd3 | RNA pseudouridylate synthase domain containing 3 | −0.41583 | 1.73 × 10−4 |
| Abcb10 | ATP-binding cassette subfamily B (MDR/TAP) member 10 | −0.41548 | 3.37 × 10−12 |
| Ppa2 | pyrophosphatase (inorganic) 2 | −0.40886 | 9.23 × 10−6 |
| Bphl | biphenyl hydrolase like (serine hydrolase breast epithelial mucin associated antigen) | −0.39912 | 9.78 × 10−4 |
| Dhrs7b | dehydrogenase/reductase (SDR family) member 7B | −0.39831 | 3.88 × 10−6 |
| Cisd1 | CDGSH iron sulfur domain 1 | −0.3853 | 5.47 × 10−6 |
| Stx17 | syntaxin 17 | −0.38146 | 4.69 × 10−7 |
| Gpi1 | glucose phosphate isomerase 1 | −0.35742 | 2.27 × 10−6 |
| Ndufa8 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 8 | −0.34492 | 2.18 × 10−4 |
| Acaa1a | acetyl-Coenzyme A acyltransferase 1A | −0.33944 | 2.45 × 10−9 |
| Idh3a | isocitrate dehydrogenase 3 (NAD+) alpha | −0.32204 | 5.08 × 10−9 |
| Iba57 | IBA57 iron sulfur cluster assembly homolog (S. cerevisiae) | 0.33473 | 2.31 × 10−4 |
| Tha1 | threonine aldolase 1 | 0.34796 | 4.24 × 10−3 |
| Amacr | alpha methylacyl CoA racemase | 0.36987 | 1.82 × 10−3 |
| Aldh1l2 | aldehyde dehydrogenase 1 family member L2 | 0.37356 | 3.79 × 10−3 |
| Tfb1m | transcription factor B1 mitochondrial | 0.37658 | 4.31 × 10−3 |
| Mrpl54 | mitochondrial ribosomal protein L54 | 0.38391 | 2.29 × 10−4 |
| Mrpl36 | mitochondrial ribosomal protein L36 | 0.41616 | 9.30 × 10−5 |
| Nucb2 | nucleobindin 2 | 0.48404 | 4.16 × 10−16 |
| Sars2 | seryl-aminoacyl-tRNA synthetase 2 | 0.54314 | 2.87 × 10−10 |
| Ephx2 | epoxide hydrolase 2 cytoplasmic | 0.56004 | 1.71 × 10−14 |
| Ccdc51 | coiled-coil domain containing 51 | 0.57846 | 1.49 × 10−7 |
| Ccdc127 | coiled-coil domain containing 127 | 0.58535 | 1.02 × 10−22 |
| Rpl10a | ribosomal protein L10A | 0.64403 | 1.49 × 10−42 |
| Mutyh | mutY homolog (E. coli) | 1.2731 | 3.28 × 10−38 |
| GO Biological Process ID | Number of Focused Genes | Fold Enrichment | Raw p-Value | FDR | Gene Name |
|---|---|---|---|---|---|
| regulation of double-strand break repair (GO:2000779) | 6 | 18.07 | 1.35 × 10−6 | 3.05 × 10−3 | Pot1b, Rad51ap1, Fignl1, Rbbp8, Zcwpw1, Prkdc |
| double-strand break repair via homologous recombination (GO:0000724) | 5 | 16.42 | 1.69 × 10−5 | 1.91 × 10−2 | Cdc7, Slx4, Samhd1, Rad51ap1, Rbbp8 |
| regulation of DNA recombination (GO:0000018) | 6 | 15.04 | 3.75 × 10−6 | 6.57 × 10−3 | Pot1b, Rad51ap1, Fignl1, Rbbp8, Zcwpw1, Hist1h1a |
| positive regulation of DNA repair (GO:0045739) | 5 | 14.79 | 2.74 × 10−5 | 2.70 × 10−2 | Rad51ap1, Rbbp8, Zcwpw1, Ino80c, Prkdc |
| Gene Name | Description | Fold Change (Nrf1f/f;RhoiCre/Nrf1f/+) | Adjusted p Value |
|---|---|---|---|
| Hk1 | Hexokinase 1 | 1.24 | 5.35 × 10−10 |
| Gpi1 | Glucose-6-phsphate isomerase 1 | 0.78 | 2.27 × 10−6 |
| Pgam1 | Phosphoglycerate mutase 1 | 0.81 | 1.57 × 10−4 |
| Pgm2 | Phosphoglucomutase-2 | 0.80 | 1.34 × 10−3 |
| Eno1 | Enolase 1, alpha non-neuron | 0.82 | 5.76 × 10−3 |
| Pkm | Pyruvate kinase, muscle | 0.80 | 5.76 × 10−6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiyama, T.; Chen, C.-K.; Zhang, A.; Mao, C.-A. Differential Susceptibility of Retinal Neurons to the Loss of Mitochondrial Biogenesis Factor Nrf1. Cells 2022, 11, 2203. https://doi.org/10.3390/cells11142203
Kiyama T, Chen C-K, Zhang A, Mao C-A. Differential Susceptibility of Retinal Neurons to the Loss of Mitochondrial Biogenesis Factor Nrf1. Cells. 2022; 11(14):2203. https://doi.org/10.3390/cells11142203
Chicago/Turabian StyleKiyama, Takae, Ching-Kang Chen, Annie Zhang, and Chai-An Mao. 2022. "Differential Susceptibility of Retinal Neurons to the Loss of Mitochondrial Biogenesis Factor Nrf1" Cells 11, no. 14: 2203. https://doi.org/10.3390/cells11142203
APA StyleKiyama, T., Chen, C.-K., Zhang, A., & Mao, C.-A. (2022). Differential Susceptibility of Retinal Neurons to the Loss of Mitochondrial Biogenesis Factor Nrf1. Cells, 11(14), 2203. https://doi.org/10.3390/cells11142203






