Mechanosensitive Channel PIEZO1 Senses Shear Force to Induce KLF2/4 Expression via CaMKII/MEKK3/ERK5 Axis in Endothelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatment
2.2. Transfection and Infection
2.3. Shear Stress Application
2.4. Ca2+ Imaging
2.5. Immunofluorescence Staining
2.6. Western Blotting and Co-Immunoprecipitation
2.7. Reverse Transcription (RT) and Quantitative RT-PCR Analysis
2.8. Animals
2.9. Isolation of Mouse Aortas and Brain Endothelial Cells and Drug Treatment
2.10. Histology and Immunofluorescence
2.11. RNA Sequencing and Data Analysis
2.12. Statistical Analysis
3. Results
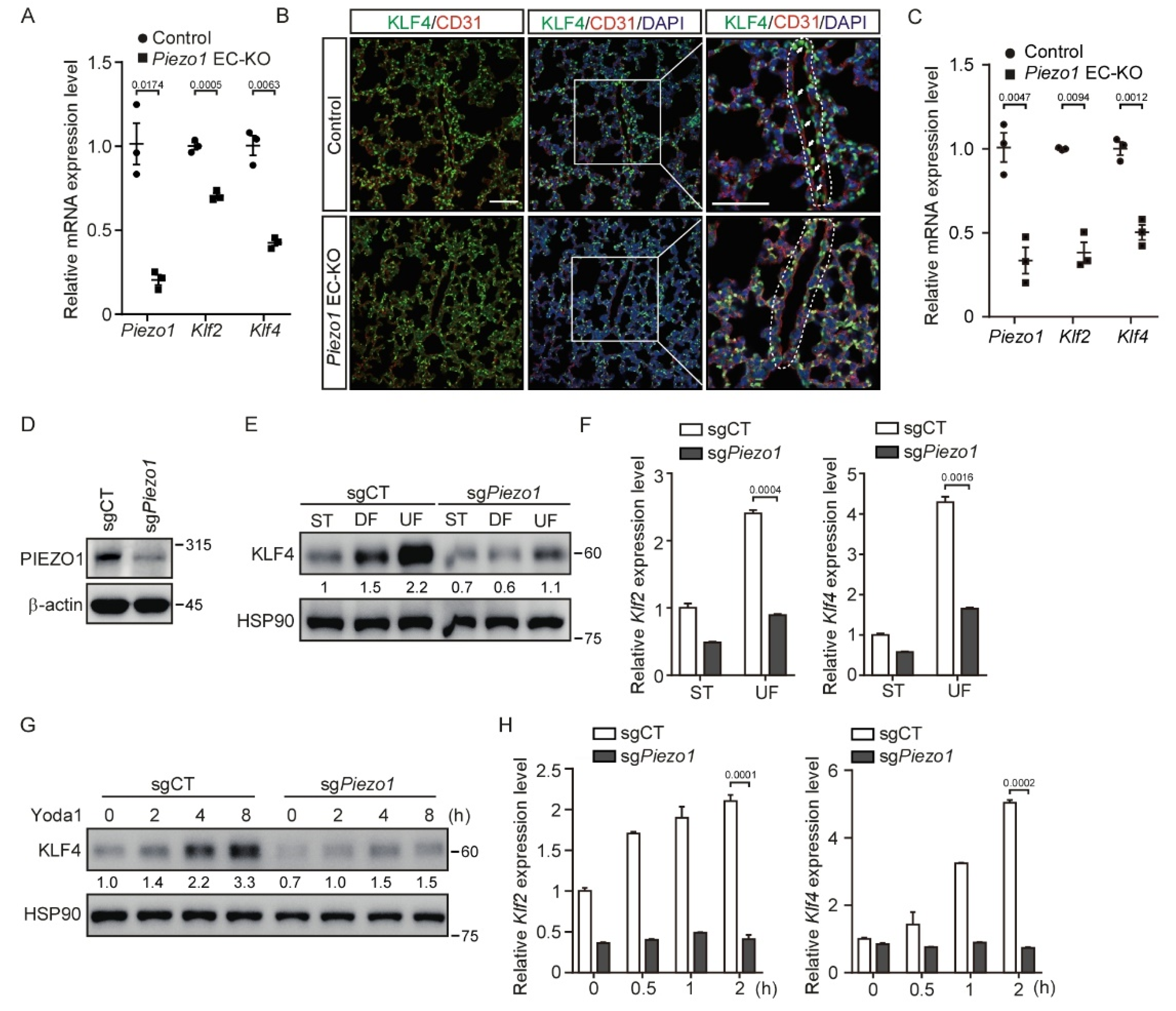
3.1. Shear Stress and Activation of PIEZO1 Enhance KLF2/4 Expression in Endothelial Cells
3.2. Deletion of Piezo1 in Endothelial Cells Suppresses Shear Stress-Mediated KLF2/4 Expression
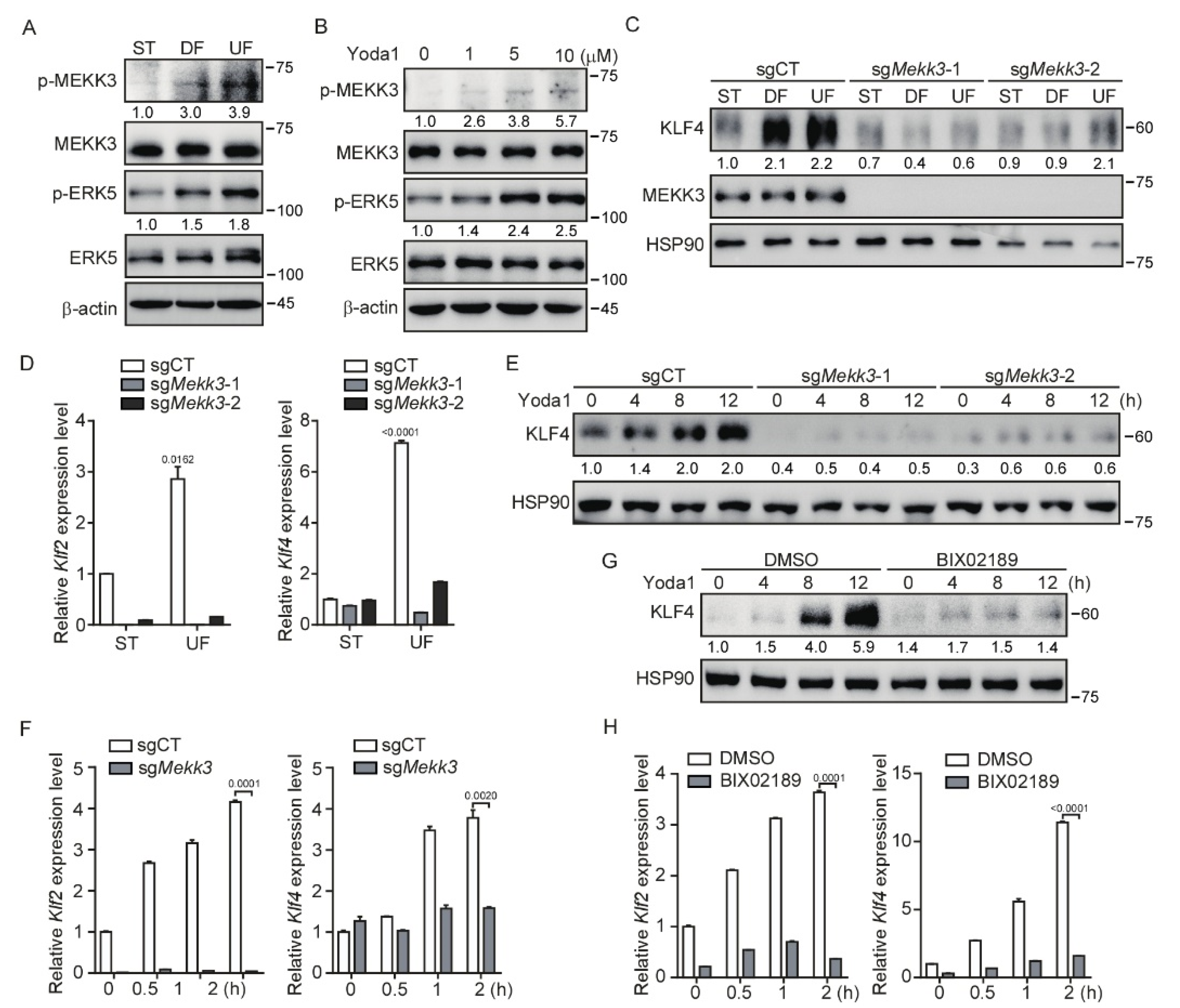
3.3. Mekk3 Deletion in Endothelial Cells Restrains PIEZO1-Mediated KLF2/4 Expression
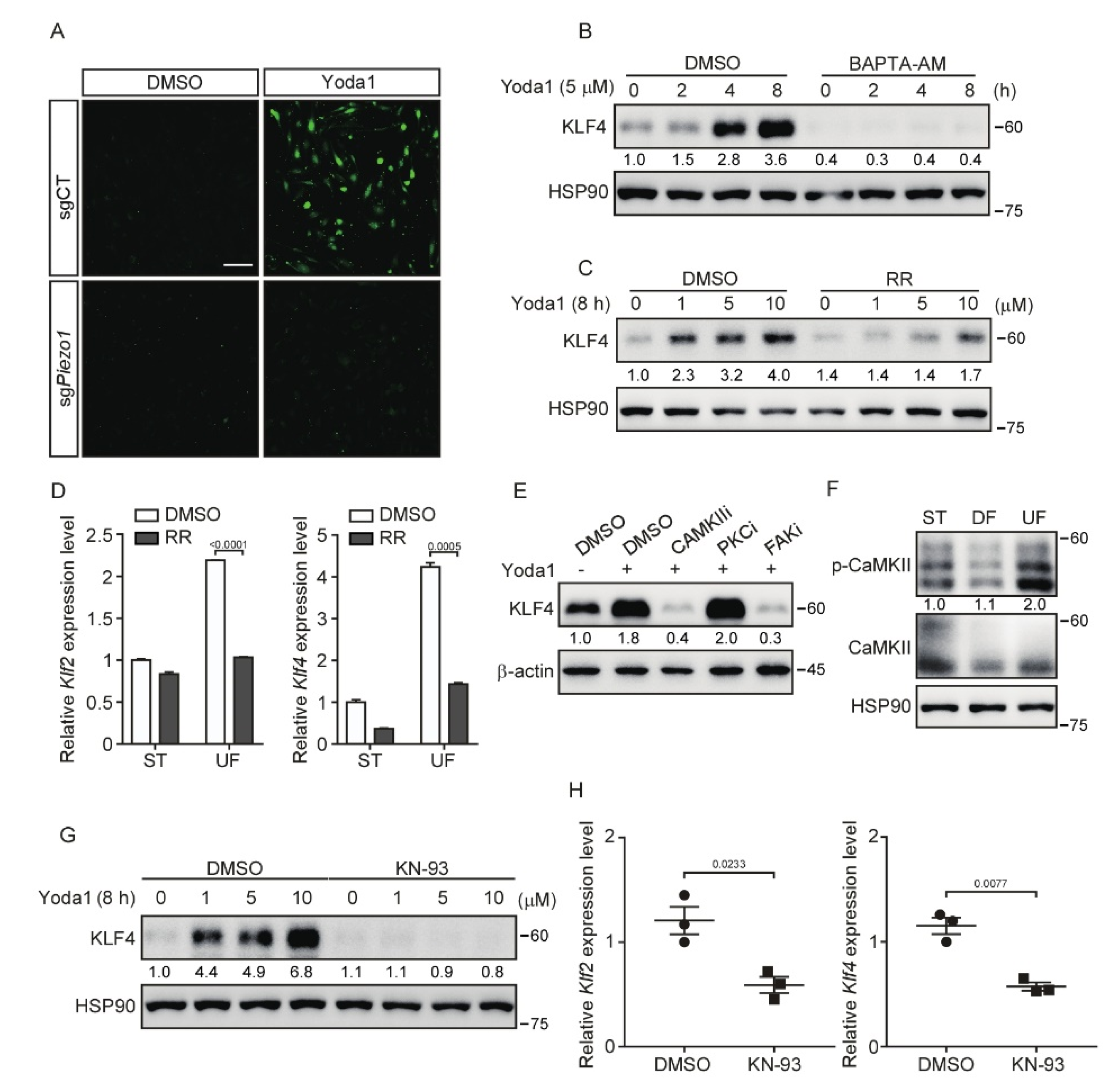
3.4. Inhibition of CaMKII Suppresses PIEZO1-Mediated KLF2/4 Expression
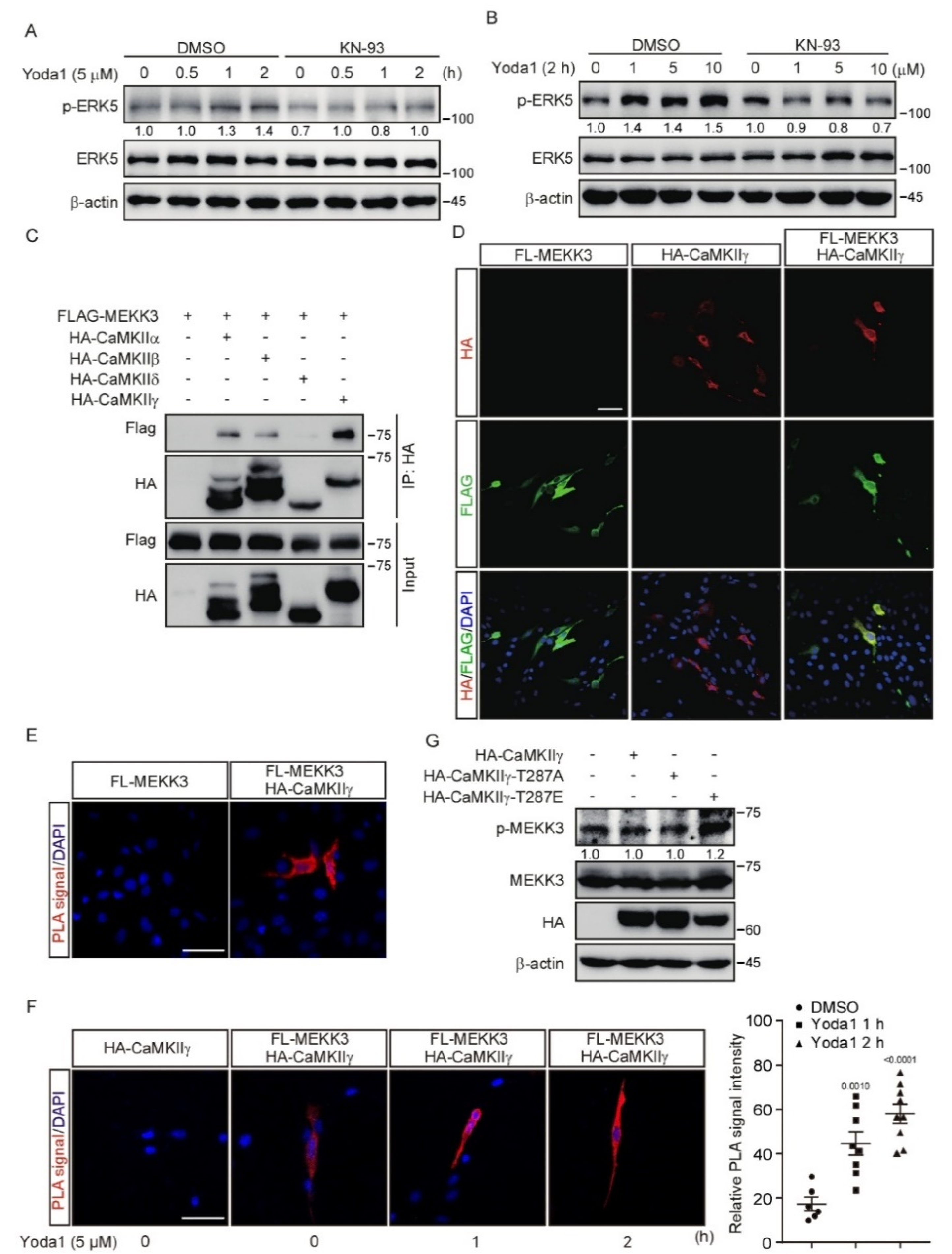
3.5. CaMKII Interacts with and Activates MEKK3
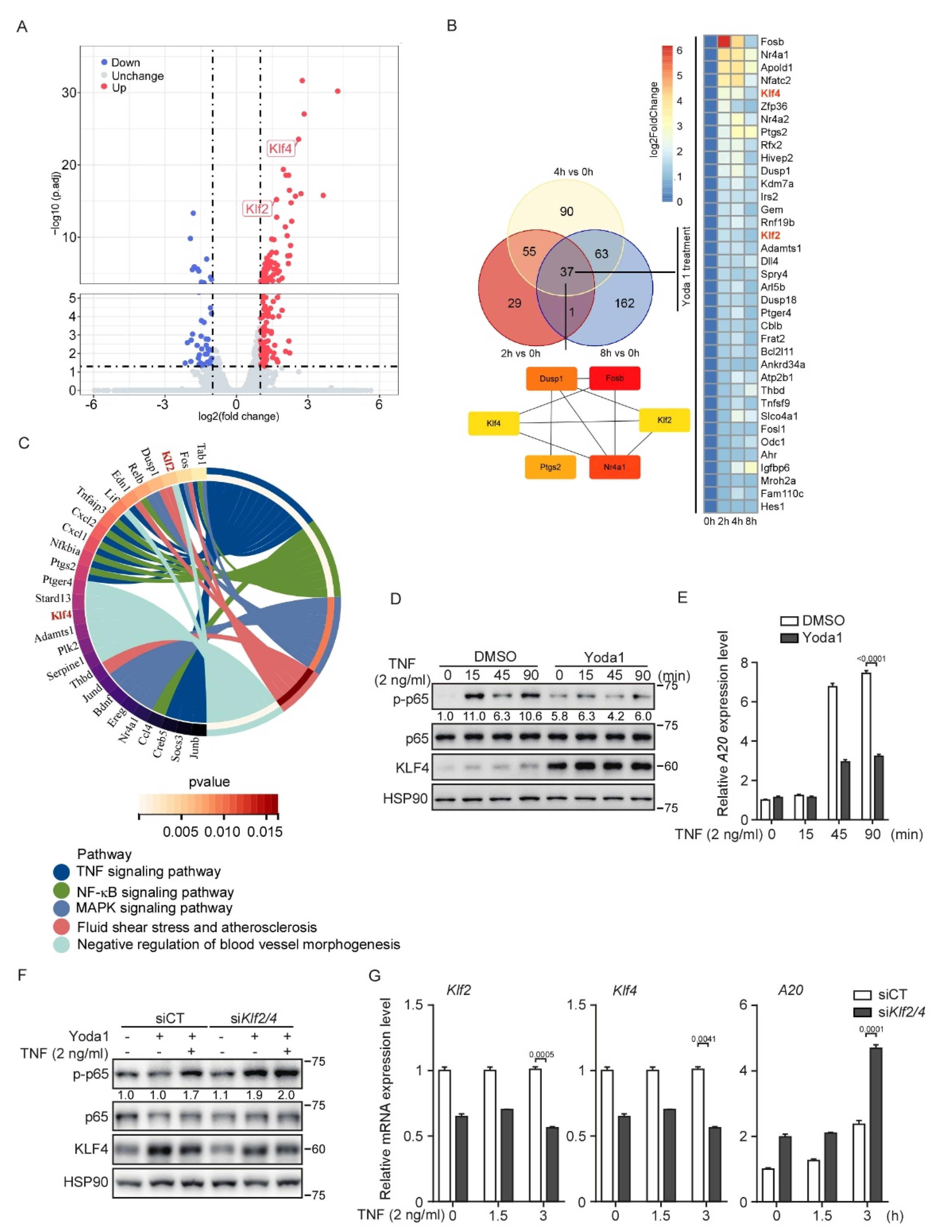
3.6. PIEZO1 Restrains the TNF-Induced NF-κB Activation in Endothelial Cells through KLF2/4
4. Discussion
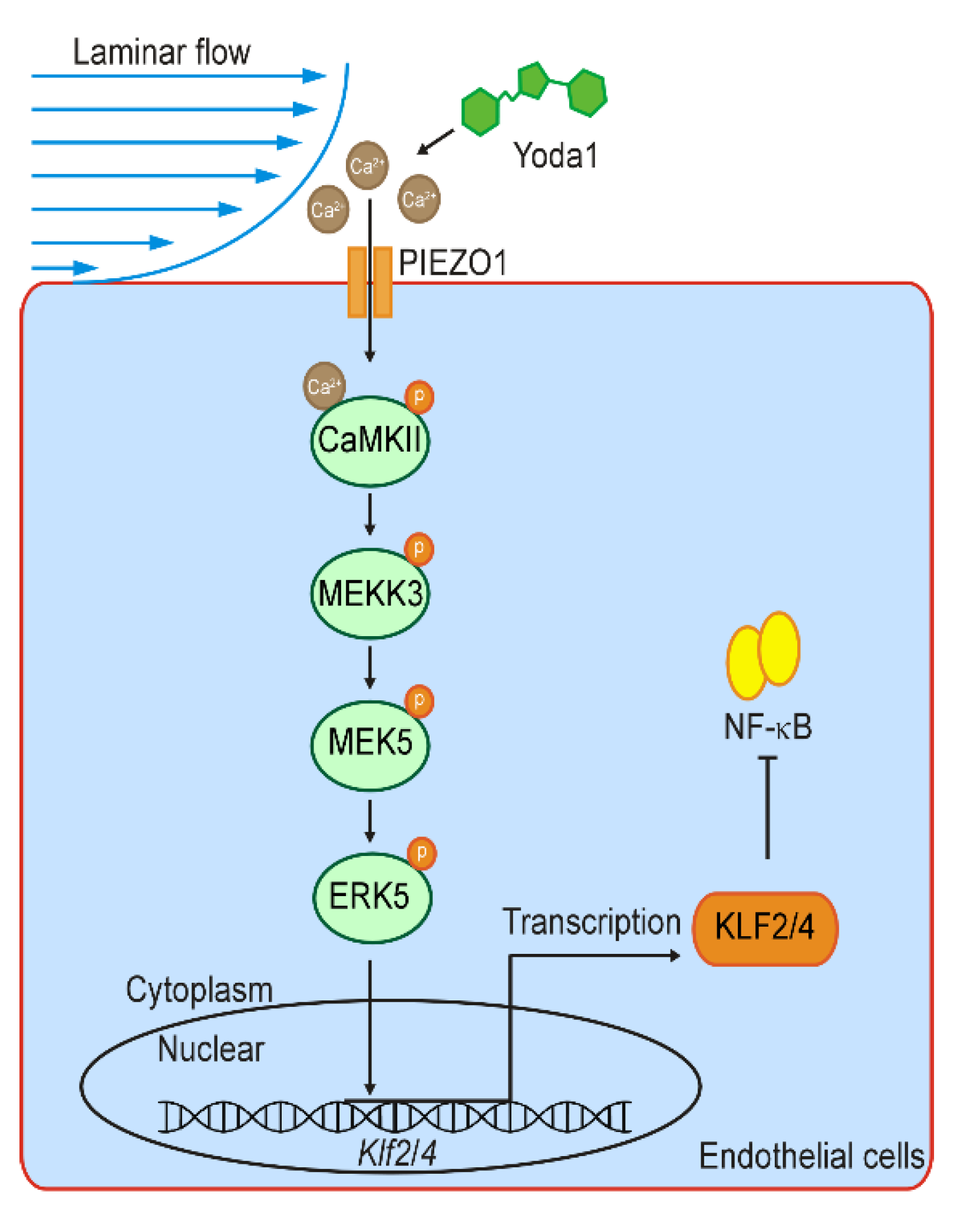
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barabutis, N. Growth Hormone Releasing Hormone in Endothelial Barrier Function. Trends Endocrinol. Metab. 2021, 32, 338–340. [Google Scholar] [CrossRef] [PubMed]
- Carlton, E.F.; McHugh, W.M.; McDonough, K.; Sturza, J.; Desch, K.; Cornell, T.T. Markers of Endothelial Dysfunction and Cytokines in High-Risk Pediatric Patients with Severe Sepsis. Am. J. Respir. Crit. Care Med. 2020, 201, 380–384. [Google Scholar] [CrossRef]
- Aalkjaer, C.; Nilsson, H.; De Mey, J.G.R. Sympathetic and Sensory-Motor Nerves in Peripheral Small Arteries. Physiol. Rev. 2021, 101, 495–544. [Google Scholar] [CrossRef] [PubMed]
- Souilhol, C.; Serbanovic-Canic, J.; Fragiadaki, M.; Chico, T.J.; Ridger, V.; Roddie, H.; Evans, P.C. Endothelial responses to shear stress in atherosclerosis: A novel role for developmental genes. Nat. Rev. Cardiol. 2020, 17, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Chien, S. Mechanotransduction and endothelial cell homeostasis: The wisdom of the cell. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H1209–H1224. [Google Scholar] [CrossRef] [Green Version]
- Davies, P.F.; Civelek, M.; Fang, Y.; Fleming, I. The atherosusceptible endothelium: Endothelial phenotypes in complex haemodynamic shear stress regions in vivo. Cardiovasc Res. 2013, 99, 315–327. [Google Scholar] [CrossRef] [Green Version]
- Davies, P.F. Flow-mediated endothelial mechanotransduction. Physiol. Rev. 1995, 75, 519–560. [Google Scholar] [CrossRef]
- Baeyens, N.; Bandyopadhyay, C.; Coon, B.G.; Yun, S.; Schwartz, M.A. Endothelial fluid shear stress sensing in vascular health and disease. J. Clin. Investig. 2016, 126, 821–828. [Google Scholar] [CrossRef]
- Xiao, L.; Harrison, D.G. Inflammation in Hypertension. Can. J. Cardiol. 2020, 36, 635–647. [Google Scholar] [CrossRef]
- Seta, F.; Cohen, R.A. The endothelium: Paracrine mediator of aortic dissection. Circulation 2014, 129, 2629–2632. [Google Scholar] [CrossRef] [Green Version]
- Kasikara, C.; Doran, A.C.; Cai, B.; Tabas, I. The role of non-resolving inflammation in atherosclerosis. J. Clin. Investig. 2018, 128, 2713–2723. [Google Scholar] [CrossRef] [Green Version]
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990-2019: Update From the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Dekker, R.J.; van Soest, S.; Fontijn, R.D.; Salamanca, S.; de Groot, P.G.; VanBavel, E.; Pannekoek, H.; Horrevoets, A.J. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2). Blood 2002, 100, 1689–1698. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; He, M.; Marin, T.; Shen, H.; Wang, W.T.; Lee, T.Y.; Hong, H.C.; Jiang, Z.L.; Garland, T., Jr.; Shyy, J.Y.; et al. Roles of KLF4 and AMPK in the inhibition of glycolysis by pulsatile shear stress in endothelial cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2103982118. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Hamik, A.; Nayak, L.; Tian, H.; Shi, H.; Lu, Y.; Sharma, N.; Liao, X.; Hale, A.; Boerboom, L.; et al. Endothelial Kruppel-like factor 4 protects against atherothrombosis in mice. J. Clin. Investig. 2012, 122, 4727–4731. [Google Scholar] [CrossRef] [Green Version]
- Simmons, R.D.; Kumar, S.; Jo, H. The role of endothelial mechanosensitive genes in atherosclerosis and omics approaches. Arch. Biochem. Biophys. 2016, 591, 111–131. [Google Scholar] [CrossRef] [Green Version]
- SenBanerjee, S.; Lin, Z.; Atkins, G.B.; Greif, D.M.; Rao, R.M.; Kumar, A.; Feinberg, M.W.; Chen, Z.; Simon, D.I.; Luscinskas, F.W.; et al. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J. Exp. Med. 2004, 199, 1305–1315. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Hou, B.; Tumova, S.; Muraki, K.; Bruns, A.; Ludlow, M.J.; Sedo, A.; Hyman, A.J.; McKeown, L.; Young, R.S.; et al. Piezo1 integration of vascular architecture with physiological force. Nature 2014, 515, 279–282. [Google Scholar] [CrossRef]
- Ranade, S.S.; Qiu, Z.; Woo, S.H.; Hur, S.S.; Murthy, S.E.; Cahalan, S.M.; Xu, J.; Mathur, J.; Bandell, M.; Coste, B.; et al. Piezo1, a mechanically activated ion channel, is required for vascular development in mice. Proc. Natl. Acad. Sci. USA 2014, 111, 10347–10352. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Chennupati, R.; Kaur, H.; Iring, A.; Wettschureck, N.; Offermanns, S. Endothelial cation channel PIEZO1 controls blood pressure by mediating flow-induced ATP release. J. Clin. Investig. 2016, 126, 4527–4536. [Google Scholar] [CrossRef] [Green Version]
- Albarran-Juarez, J.; Iring, A.; Wang, S.; Joseph, S.; Grimm, M.; Strilic, B.; Wettschureck, N.; Althoff, T.F.; Offermanns, S. Piezo1 and Gq/G11 promote endothelial inflammation depending on flow pattern and integrin activation. J. Exp. Med. 2018, 215, 2655–2672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedrich, E.E.; Hong, Z.; Xiong, S.; Zhong, M.; Di, A.; Rehman, J.; Komarova, Y.A.; Malik, A.B. Endothelial cell Piezo1 mediates pressure-induced lung vascular hyperpermeability via disruption of adherens junctions. Proc. Natl. Acad. Sci. USA 2019, 116, 12980–12985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iring, A.; Jin, Y.J.; Albarran-Juarez, J.; Siragusa, M.; Wang, S.; Dancs, P.T.; Nakayama, A.; Tonack, S.; Chen, M.; Kunne, C.; et al. Shear stress-induced endothelial adrenomedullin signaling regulates vascular tone and blood pressure. J. Clin. Investig. 2019, 129, 2775–2791. [Google Scholar] [CrossRef] [Green Version]
- Rode, B.; Shi, J.; Endesh, N.; Drinkhill, M.J.; Webster, P.J.; Lotteau, S.J.; Bailey, M.A.; Yuldasheva, N.Y.; Ludlow, M.J.; Cubbon, R.M.; et al. Piezo1 channels sense whole body physical activity to reset cardiovascular homeostasis and enhance performance. Nat. Commun. 2017, 8, 350. [Google Scholar] [CrossRef] [PubMed]
- Dardik, A.; Chen, L.; Frattini, J.; Asada, H.; Aziz, F.; Kudo, F.A.; Sumpio, B.E. Differential effects of orbital and laminar shear stress on endothelial cells. J. Vasc. Surg. 2005, 41, 869–880. [Google Scholar] [CrossRef] [Green Version]
- Regmi, S.; Fu, A.; Luo, K.Q. High Shear Stresses under Exercise Condition Destroy Circulating Tumor Cells in a Microfluidic System. Sci. Rep. 2017, 7, 39975. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; You, X.; Lotinun, S.; Zhang, L.; Wu, N.; Zou, W. Mechanical sensing protein PIEZO1 regulates bone homeostasis via osteoblast-osteoclast crosstalk. Nat. Commun. 2020, 11, 282. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.M.; Chen, A.F.; Zhang, K. Isolation and Primary Culture of Mouse Aortic Endothelial Cells. J. Vis. Exp. 2016, 52965. [Google Scholar] [CrossRef]
- Wu, W.; Xiao, H.; Laguna-Fernandez, A.; Villarreal, G., Jr.; Wang, K.C.; Geary, G.G.; Zhang, Y.; Wang, W.C.; Huang, H.D.; Zhou, J.; et al. Flow-Dependent Regulation of Kruppel-Like Factor 2 Is Mediated by MicroRNA-92a. Circulation 2011, 124, 633–641. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.; Meng, Q.; Qi, M.; Li, F.; Liu, B. Shear-Sensitive lncRNA AF131217.1 Inhibits Inflammation in HUVECs via Regulation of KLF4. Hypertension 2019, 73, e25–e34. [Google Scholar] [CrossRef]
- Wang, L.; Luo, J.Y.; Li, B.; Tian, X.Y.; Chen, L.J.; Huang, Y.; Liu, J.; Deng, D.; Lau, C.W.; Wan, S.; et al. Integrin-YAP/TAZ-JNK cascade mediates atheroprotective effect of unidirectional shear flow. Nature 2016, 540, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Yeh, Y.T.; Nguyen, P.; Limqueco, E.; Lopez, J.; Thorossian, S.; Guan, K.L.; Li, Y.J.; Chien, S. Flow-dependent YAP/TAZ activities regulate endothelial phenotypes and atherosclerosis. Proc. Natl. Acad. Sci. USA 2016, 113, 11525–11530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; He, J.; Lv, H.; Liu, Y.; Lv, X.; Zhang, C.; Zhu, Y.; Ai, D. c-Abl regulates YAPY357 phosphorylation to activate endothelial atherogenic responses to disturbed flow. J. Clin. Investig. 2019, 129, 1167–1179. [Google Scholar] [CrossRef] [PubMed]
- Dekker, R.J.; van Thienen, J.V.; Rohlena, J.; de Jager, S.C.; Elderkamp, Y.W.; Seppen, J.; de Vries, C.J.; Biessen, E.A.; van Berkel, T.J.; Pannekoek, H.; et al. Endothelial KLF2 links local arterial shear stress levels to the expression of vascular tone-regulating genes. Am. J. Pathol. 2005, 167, 609–618. [Google Scholar] [CrossRef] [Green Version]
- Bhagavathula, N.; Dame, M.K.; DaSilva, M.; Jenkins, W.; Aslam, M.N.; Perone, P.; Varani, J. Fibroblast response to gadolinium: Role for platelet-derived growth factor receptor. Investig. Radiol. 2010, 45, 769–777. [Google Scholar] [CrossRef] [Green Version]
- Payne, S.; De Val, S.; Neal, A. Endothelial-Specific Cre Mouse Models. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 2550–2561. [Google Scholar] [CrossRef] [Green Version]
- Kisanuki, Y.Y.; Hammer, R.E.; Miyazaki, J.; Williams, S.C.; Richardson, J.A.; Yanagisawa, M. Tie2-Cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev. Biol 2001, 230, 230–242. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Rawnsley, D.R.; Goddard, L.M.; Pan, W.; Cao, X.J.; Jakus, Z.; Zheng, H.; Yang, J.; Arthur, J.S.; Whitehead, K.J.; et al. The cerebral cavernous malformation pathway controls cardiac development via regulation of endocardial MEKK3 signaling and KLF expression. Dev. Cell 2015, 32, 168–180. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Tang, A.T.; Wong, W.Y.; Bamezai, S.; Goddard, L.M.; Shenkar, R.; Zhou, S.; Yang, J.; Wright, A.C.; Foley, M.; et al. Cerebral cavernous malformations arise from endothelial gain of MEKK3-KLF2/4 signalling. Nature 2016, 532, 122–126. [Google Scholar] [CrossRef] [Green Version]
- Murthy, S.E.; Dubin, A.E.; Patapoutian, A. Piezos thrive under pressure: Mechanically activated ion channels in health and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 771–783. [Google Scholar] [CrossRef]
- Cai, H.; Liu, D.; Garcia, J.G. CaM Kinase II-dependent pathophysiological signalling in endothelial cells. Cardiovasc. Res. 2008, 77, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Morita, T.; Kurihara, H.; Maemura, K.; Yoshizumi, M.; Nagai, R.; Yazaki, Y. Role of Ca2+ and protein kinase C in shear stress-induced actin depolymerization and endothelin 1 gene expression. Circ. Res. 1994, 75, 630–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Butler, P.; Wang, Y.; Hu, Y.; Han, D.C.; Usami, S.; Guan, J.L.; Chien, S. The role of the dynamics of focal adhesion kinase in the mechanotaxis of endothelial cells. Proc. Natl. Acad. Sci. USA 2002, 99, 3546–3551. [Google Scholar] [CrossRef] [Green Version]
- Geng, J.; Shi, Y.; Zhang, J.; Yang, B.; Wang, P.; Yuan, W.; Zhao, H.; Li, J.; Qin, F.; Hong, L.; et al. TLR4 signalling via Piezo1 engages and enhances the macrophage mediated host response during bacterial infection. Nat. Commun. 2021, 12, 3519. [Google Scholar] [CrossRef] [PubMed]
- Allen, K.L.; Hamik, A.; Jain, M.K.; McCrae, K.R. Endothelial cell activation by antiphospholipid antibodies is modulated by Kruppel-like transcription factors. Blood 2011, 117, 6383–6391. [Google Scholar] [CrossRef]
- Discher, D.E.; Janmey, P.; Wang, Y.L. Tissue cells feel and respond to the stiffness of their substrate. Science 2005, 310, 1139–1143. [Google Scholar] [CrossRef] [Green Version]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [Green Version]
- Niu, N.; Xu, S.; Xu, Y.; Little, P.J.; Jin, Z.G. Targeting Mechanosensitive Transcription Factors in Atherosclerosis. Trends Pharmacol. Sci. 2019, 40, 253–266. [Google Scholar] [CrossRef]
- Sweet, D.R.; Fan, L.; Hsieh, P.N.; Jain, M.K. Kruppel-Like Factors in Vascular Inflammation: Mechanistic Insights and Therapeutic Potential. Front. Cardiovasc. Med. 2018, 5, 6. [Google Scholar] [CrossRef] [Green Version]
- Chiu, J.J.; Chien, S. Effects of disturbed flow on vascular endothelium: Pathophysiological basis and clinical perspectives. Physiol. Rev. 2011, 91, 327–387. [Google Scholar] [CrossRef] [Green Version]
- Gimbrone, M.A., Jr.; Garcia-Cardena, G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ. Res. 2016, 118, 620–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkins, G.B.; Wang, Y.; Mahabeleshwar, G.H.; Shi, H.; Gao, H.; Kawanami, D.; Natesan, V.; Lin, Z.; Simon, D.I.; Jain, M.K. Hemizygous deficiency of Kruppel-like factor 2 augments experimental atherosclerosis. Circ. Res. 2008, 103, 690–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ando, J.; Yamamoto, K. Flow detection and calcium signalling in vascular endothelial cells. Cardiovasc. Res. 2013, 99, 260–268. [Google Scholar] [CrossRef] [Green Version]
- Aromolaran, A.A.; Blatter, L.A. Modulation of intracellular Ca2+ release and capacitative Ca2+ entry by CaMKII inhibitors in bovine vascular endothelial cells. Am. J. Physiol. Cell Physiol. 2005, 289, C1426–C1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, T.; Nemoto, S.; Ishida, K.; Taguchi, K.; Matsumoto, T.; Kamata, K. Involvement of CaM kinase II in the impairment of endothelial function and eNOS activity in aortas of Type 2 diabetic rats. Clin. Sci. 2012, 123, 375–386. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Ginnan, R.; Abdullaev, I.F.; Trebak, M.; Vincent, P.A.; Singer, H.A. Calcium/Calmodulin-dependent protein kinase II delta 6 (CaMKIIdelta6) and RhoA involvement in thrombin-induced endothelial barrier dysfunction. J. Biol. Chem. 2010, 285, 21303–21312. [Google Scholar] [CrossRef] [Green Version]
- Meoli, D.F.; White, R.J. Thrombin induces fibronectin-specific migration of pulmonary microvascular endothelial cells: Requirement of calcium/calmodulin-dependent protein kinase II. Am. J. Physiol. Lung Cell Mol. Physiol. 2009, 297, L706–L714. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Liu, X.; Gao, L.; Ji, Y.; Wang, L.; Zhang, C.; Dai, L.; Liu, J.; Ji, Z. Activation of Piezo1 by ultrasonic stimulation and its effect on the permeability of human umbilical vein endothelial cells. Biomed. Pharmacother. 2020, 131, 110796. [Google Scholar] [CrossRef]
- Lu, J.; Hu, Z.; Deng, Y.; Wu, Q.; Wu, M.; Song, H. MEKK2 and MEKK3 orchestrate multiple signals to regulate Hippo pathway. J. Biol. Chem. 2021, 296, 100400. [Google Scholar] [CrossRef]
- Bartoli, M.; Monneron, A.; Ladant, D. Interaction of calmodulin with striatin, a WD-repeat protein present in neuronal dendritic spines. J. Biol. Chem. 1998, 273, 22248–22253. [Google Scholar] [CrossRef] [Green Version]
- Castets, F.; Bartoli, M.; Barnier, J.V.; Baillat, G.; Salin, P.; Moqrich, A.; Bourgeois, J.P.; Denizot, F.; Rougon, G.; Calothy, G.; et al. A novel calmodulin-binding protein, belonging to the WD-repeat family, is localized in dendrites of a subset of CNS neurons. J. Cell Biol. 1996, 134, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.Z.; Marshall, K.L.; Min, S.; Daou, I.; Chapleau, M.W.; Abboud, F.M.; Liberles, S.D.; Patapoutian, A. PIEZOs mediate neuronal sensing of blood pressure and the baroreceptor reflex. Science 2018, 362, 464–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Chen, J.; Babicheva, A.; Jain, P.P.; Rodriguez, M.; Ayon, R.J.; Ravellette, K.S.; Wu, L.; Balistrieri, F.; Tang, H.; et al. Endothelial upregulation of mechanosensitive channel Piezo1 in pulmonary hypertension. Am. J. Physiol. Cell Physiol. 2021, 321, C1010–C1027. [Google Scholar] [CrossRef] [PubMed]
- Lhomme, A.; Gilbert, G.; Pele, T.; Deweirdt, J.; Henrion, D.; Baudrimont, I.; Campagnac, M.; Marthan, R.; Guibert, C.; Ducret, T.; et al. Stretch-activated Piezo1 Channel in Endothelial Cells Relaxes Mouse Intrapulmonary Arteries. Am. J. Respir Cell Mol. Biol 2019, 60, 650–658. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, D.; Zhang, C.; Yang, W.; Li, C.; Gao, Z.; Pei, K.; Li, Y. Piezo1 mediates endothelial atherogenic inflammatory responses via regulation of YAP/TAZ activation. Hum. Cell 2022, 35, 51–62. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Q.; Zou, Y.; Teng, P.; Chen, Z.; Wu, Y.; Dai, X.; Li, X.; Hu, Z.; Wu, S.; Xu, Y.; et al. Mechanosensitive Channel PIEZO1 Senses Shear Force to Induce KLF2/4 Expression via CaMKII/MEKK3/ERK5 Axis in Endothelial Cells. Cells 2022, 11, 2191. https://doi.org/10.3390/cells11142191
Zheng Q, Zou Y, Teng P, Chen Z, Wu Y, Dai X, Li X, Hu Z, Wu S, Xu Y, et al. Mechanosensitive Channel PIEZO1 Senses Shear Force to Induce KLF2/4 Expression via CaMKII/MEKK3/ERK5 Axis in Endothelial Cells. Cells. 2022; 11(14):2191. https://doi.org/10.3390/cells11142191
Chicago/Turabian StyleZheng, Qi, Yonggang Zou, Peng Teng, Zhenghua Chen, Yuefeng Wu, Xiaoyi Dai, Xiya Li, Zonghao Hu, Shengjun Wu, Yanhua Xu, and et al. 2022. "Mechanosensitive Channel PIEZO1 Senses Shear Force to Induce KLF2/4 Expression via CaMKII/MEKK3/ERK5 Axis in Endothelial Cells" Cells 11, no. 14: 2191. https://doi.org/10.3390/cells11142191
APA StyleZheng, Q., Zou, Y., Teng, P., Chen, Z., Wu, Y., Dai, X., Li, X., Hu, Z., Wu, S., Xu, Y., Zou, W., Song, H., & Ma, L. (2022). Mechanosensitive Channel PIEZO1 Senses Shear Force to Induce KLF2/4 Expression via CaMKII/MEKK3/ERK5 Axis in Endothelial Cells. Cells, 11(14), 2191. https://doi.org/10.3390/cells11142191





