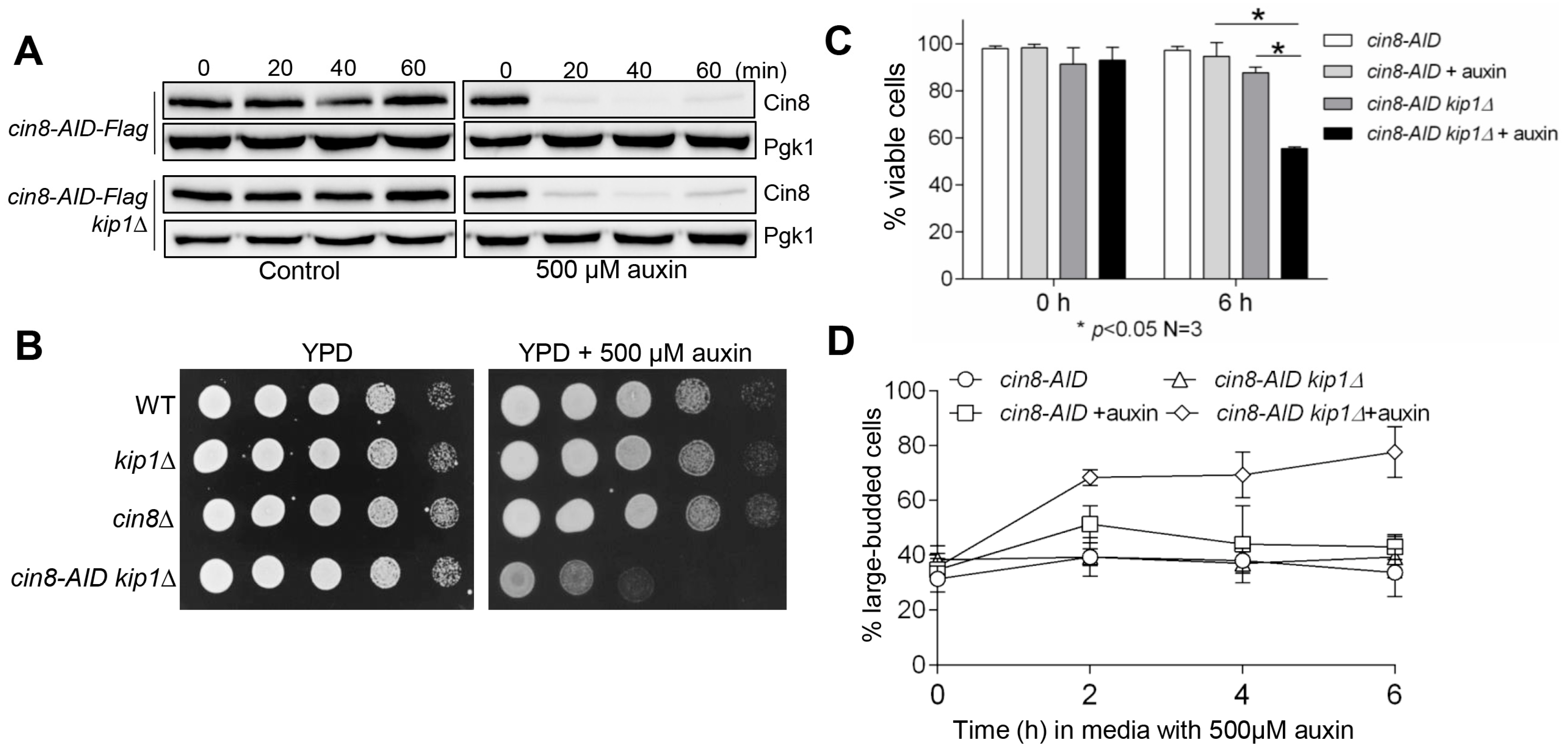
Figure 1.
The construction of cin8-AID strains. (A) Cin8-AID is degraded in the presence of auxin. The cin8-AID-3×Flag (3946-2-3, referred to as cin8-AID hereafter) and cin8-AID kip1∆ (3750-3-2) cells were grown in YPD medium to mid-log phase at 30 °C. Auxin (500 μM) was added into half of the cell cultures, and cells were harvested every 20 min. Cin8-AID protein levels were detected after Western blotting with anti-Flag antibody. Pgk1, loading control. (B) cin8-AID kip1∆ double mutants grow poorly in the presence of auxin. Saturated cells of wild-type (WT), kip1∆ (YBL063W), cin8∆ (YEL061C), and cin8-AID kip1∆ (3750-3-2) were 10-fold serially diluted onto YPD plates, both with and without 500 μM auxin. Growth was analyzed after a 2-day incubation at 30 °C. (C) The viability of cin8-AID and cin8-AID kip1∆ cells after growth in auxin. The cin8-AID (3946-2-3) and cin8-AID kip1∆ (3750-3-2) cells were grown in YPD medium at 30 °C to mid-log phase, and then 500 μM auxin was added into the cultures. Cells were collected at 0 and 6 h and spread onto YPD plates to examine plating efficiency. Cells that formed mini-colonies after overnight incubation at 30 °C were counted as viable. At least 300 cells were counted for each strain for the percentage of viable cells. The experiment was repeated three times, and statistical significance determined by * p < 0.05, using Kruskal–Wallis one-way ANOVA. (D) The accumulation of large-budded cells. Cells with the indicated genotypes were grown in the presence of auxin and collected at 2 h intervals to count budding index. A cell was counted as large-budded when the diameter of a daughter cell was greater than half of the diameter of the mother cell. Here shows the average percentage of large-budded cells from three independent experiments.
Figure 1.
The construction of cin8-AID strains. (A) Cin8-AID is degraded in the presence of auxin. The cin8-AID-3×Flag (3946-2-3, referred to as cin8-AID hereafter) and cin8-AID kip1∆ (3750-3-2) cells were grown in YPD medium to mid-log phase at 30 °C. Auxin (500 μM) was added into half of the cell cultures, and cells were harvested every 20 min. Cin8-AID protein levels were detected after Western blotting with anti-Flag antibody. Pgk1, loading control. (B) cin8-AID kip1∆ double mutants grow poorly in the presence of auxin. Saturated cells of wild-type (WT), kip1∆ (YBL063W), cin8∆ (YEL061C), and cin8-AID kip1∆ (3750-3-2) were 10-fold serially diluted onto YPD plates, both with and without 500 μM auxin. Growth was analyzed after a 2-day incubation at 30 °C. (C) The viability of cin8-AID and cin8-AID kip1∆ cells after growth in auxin. The cin8-AID (3946-2-3) and cin8-AID kip1∆ (3750-3-2) cells were grown in YPD medium at 30 °C to mid-log phase, and then 500 μM auxin was added into the cultures. Cells were collected at 0 and 6 h and spread onto YPD plates to examine plating efficiency. Cells that formed mini-colonies after overnight incubation at 30 °C were counted as viable. At least 300 cells were counted for each strain for the percentage of viable cells. The experiment was repeated three times, and statistical significance determined by * p < 0.05, using Kruskal–Wallis one-way ANOVA. (D) The accumulation of large-budded cells. Cells with the indicated genotypes were grown in the presence of auxin and collected at 2 h intervals to count budding index. A cell was counted as large-budded when the diameter of a daughter cell was greater than half of the diameter of the mother cell. Here shows the average percentage of large-budded cells from three independent experiments.
![Cells 11 02144 g001 Cells 11 02144 g001]()
Figure 2.
The cin8-AID kip1∆ mutants show defects in cell cycle and spindle formation. (A) Budding index. G1-arrested WT (JBY649), cin8-AID (3946-2-3), and cin8-AID kip1∆ (3889-3-1) cells with Pds1-18myc were released into 30 °C YPD medium containing 500 μM auxin. The α-factor was added back after 40 min release to block the following cell cycle. Samples were collected every 20 min to count the budding index. Here shows the average of large-budded cells after G1 release from three independent experiments. (B) Accumulation of anaphase inhibitor Pds1 in cin8-AID mutants treated with auxin. The cells in (A) were collected every 20 min to prepare protein samples. Western blotting was performed with anti-myc antibody. Pgk1, loading control. (C) Quantification of Pds1 levels. The relative Pds1 levels from the Western blotting results in (B) were plotted. Quantification of Pds1 levels is described in the Methods section. (D) cin8-AID and cin8-AID kip1∆ cells show mitotic defects. G1-arrested WT (4232-6-2), cin8-AID (4232-1-2), and cin8-AID kip1∆ (4232-7-2) cells with Mtw1-GFP and Spc110-mCherry were released into YPD, at 30 °C, containing 500 μM auxin. The α-factor was added back after 40 min to block the following cell cycle. Samples were collected every 20 min for the budding index (n = 3). (E) cin8-AID and cin8-AID kip1∆ cells display compromised kinetochore or SPB separation in the presence of auxin. The cells in (D) were collected and imaged at 100 and 120 min after G1 release to visualize the separation of kinetochore clusters (Mtw1-GFP) and SPBs (Spc110-mCherry). White arrows indicate unseparated Mtw1-GFP clusters and SPBs. Scale bar, 5 μm. The images are representative of three experimental repeats. (F) Quantification of kinetochore/SPB separation. Kinetochore/SPB separation phenotype was categorized as normal, separated SPB without Mtw1 cluster separation, or unseparated SPB and Mtw1 cluster. The experiment was repeated three times, and statistical significance was determined by * p < 0.05, using Kruskal–Wallis one-way ANOVA.
Figure 2.
The cin8-AID kip1∆ mutants show defects in cell cycle and spindle formation. (A) Budding index. G1-arrested WT (JBY649), cin8-AID (3946-2-3), and cin8-AID kip1∆ (3889-3-1) cells with Pds1-18myc were released into 30 °C YPD medium containing 500 μM auxin. The α-factor was added back after 40 min release to block the following cell cycle. Samples were collected every 20 min to count the budding index. Here shows the average of large-budded cells after G1 release from three independent experiments. (B) Accumulation of anaphase inhibitor Pds1 in cin8-AID mutants treated with auxin. The cells in (A) were collected every 20 min to prepare protein samples. Western blotting was performed with anti-myc antibody. Pgk1, loading control. (C) Quantification of Pds1 levels. The relative Pds1 levels from the Western blotting results in (B) were plotted. Quantification of Pds1 levels is described in the Methods section. (D) cin8-AID and cin8-AID kip1∆ cells show mitotic defects. G1-arrested WT (4232-6-2), cin8-AID (4232-1-2), and cin8-AID kip1∆ (4232-7-2) cells with Mtw1-GFP and Spc110-mCherry were released into YPD, at 30 °C, containing 500 μM auxin. The α-factor was added back after 40 min to block the following cell cycle. Samples were collected every 20 min for the budding index (n = 3). (E) cin8-AID and cin8-AID kip1∆ cells display compromised kinetochore or SPB separation in the presence of auxin. The cells in (D) were collected and imaged at 100 and 120 min after G1 release to visualize the separation of kinetochore clusters (Mtw1-GFP) and SPBs (Spc110-mCherry). White arrows indicate unseparated Mtw1-GFP clusters and SPBs. Scale bar, 5 μm. The images are representative of three experimental repeats. (F) Quantification of kinetochore/SPB separation. Kinetochore/SPB separation phenotype was categorized as normal, separated SPB without Mtw1 cluster separation, or unseparated SPB and Mtw1 cluster. The experiment was repeated three times, and statistical significance was determined by * p < 0.05, using Kruskal–Wallis one-way ANOVA.
![Cells 11 02144 g002 Cells 11 02144 g002]()
Figure 3.
In absence of the SAC, cin8 mutants lose viability. (A) Elimination of the SAC causes growth defect in cin8 mutant cells. WT, mad1∆ (4327-9-2), cin8-AID (3946-2-3), and cin8-AID mad1∆ (4236-1-3) cells were 10-fold serially diluted onto YPD plates, both with and without 500 μM auxin. Growth was analyzed after a 2-day incubation at 30 °C. (B) Elimination of the SAC results in chromosome missegregation in cin8 mutants. WT (4244-3-4), mad1∆ (4327-9-2), cin8-AID (4244-2-1), and cin8-AID mad1∆ (4236-1-3) cells containing CEN4-GFP (GFP-marked centromere of chromosome IV) and Spc110-mCherry (mCherry-marked SPB protein Spc110) were grown to mid-log phase in YPD medium at 30 °C. Auxin was added at 500 μM, and pictures were taken at 0 and 180 min. Images show chromosome missegregation in a cin8-AID mad1∆ cell after incubation in auxin media for 180 min. White arrows represent the SPB and CEN4-GFP in this cell. Scale bar, 5 μm. (C) Quantification of SPB and chromosome segregation defects. CEN4-GFP and SPB segregation at 180 min was counted for the cells in (B). CEN4-GFP and SPB segregation was categorized as normal, unseparated, or missegregated. Unseparated represents the absence of two clear CEN4-GFP and SPB dots. The experiment was repeated three times, and statistical significance was determined by p < 0.05, using Kruskal–Wallis one-way ANOVA. (D) Deficient SAC causes viability loss in cin8 cells. Cells in (B) were also collected at 180 min and spread onto YPD plates to count the plating efficiency after incubation at 30 °C overnight. The experiment was repeated three times, and statistical significance determined by a * p < 0.05, using the Wilcoxon rank sum test. (E) The kinetics of cell-cycle progression in synchronized cells lacking Cin8 and SAC. The same yeast strains listed in (B) were used in this experiment. G1-arrested cells were released into YPD, at 30 °C, containing 500 μM auxin. Samples were collected every 15 min to count the budding index. The α-factor was added back after a 40 min release to block the following cell cycle; n = 3. (F) Chromosome missegregation in synchronized cells lacking Cin8 and SAC. The same cells were also collected at 105 min after G1 release for imaging. White arrows represent SPB and CEN4-GFP in a cell showing chromosome missegregation. Scale bar, 5 μm. The pictures are representative of three experimental repeats. (G) Quantification of chromosome missegregation in synchronized cells lacking Cin8 and SAC. Segregation of CEN4-GFP and SPB at 105 min after G1 release was examined. The experiment was repeated three times and statistical significance determined by * p < 0.05, using Kruskal–Wallis one-way ANOVA.
Figure 3.
In absence of the SAC, cin8 mutants lose viability. (A) Elimination of the SAC causes growth defect in cin8 mutant cells. WT, mad1∆ (4327-9-2), cin8-AID (3946-2-3), and cin8-AID mad1∆ (4236-1-3) cells were 10-fold serially diluted onto YPD plates, both with and without 500 μM auxin. Growth was analyzed after a 2-day incubation at 30 °C. (B) Elimination of the SAC results in chromosome missegregation in cin8 mutants. WT (4244-3-4), mad1∆ (4327-9-2), cin8-AID (4244-2-1), and cin8-AID mad1∆ (4236-1-3) cells containing CEN4-GFP (GFP-marked centromere of chromosome IV) and Spc110-mCherry (mCherry-marked SPB protein Spc110) were grown to mid-log phase in YPD medium at 30 °C. Auxin was added at 500 μM, and pictures were taken at 0 and 180 min. Images show chromosome missegregation in a cin8-AID mad1∆ cell after incubation in auxin media for 180 min. White arrows represent the SPB and CEN4-GFP in this cell. Scale bar, 5 μm. (C) Quantification of SPB and chromosome segregation defects. CEN4-GFP and SPB segregation at 180 min was counted for the cells in (B). CEN4-GFP and SPB segregation was categorized as normal, unseparated, or missegregated. Unseparated represents the absence of two clear CEN4-GFP and SPB dots. The experiment was repeated three times, and statistical significance was determined by p < 0.05, using Kruskal–Wallis one-way ANOVA. (D) Deficient SAC causes viability loss in cin8 cells. Cells in (B) were also collected at 180 min and spread onto YPD plates to count the plating efficiency after incubation at 30 °C overnight. The experiment was repeated three times, and statistical significance determined by a * p < 0.05, using the Wilcoxon rank sum test. (E) The kinetics of cell-cycle progression in synchronized cells lacking Cin8 and SAC. The same yeast strains listed in (B) were used in this experiment. G1-arrested cells were released into YPD, at 30 °C, containing 500 μM auxin. Samples were collected every 15 min to count the budding index. The α-factor was added back after a 40 min release to block the following cell cycle; n = 3. (F) Chromosome missegregation in synchronized cells lacking Cin8 and SAC. The same cells were also collected at 105 min after G1 release for imaging. White arrows represent SPB and CEN4-GFP in a cell showing chromosome missegregation. Scale bar, 5 μm. The pictures are representative of three experimental repeats. (G) Quantification of chromosome missegregation in synchronized cells lacking Cin8 and SAC. Segregation of CEN4-GFP and SPB at 105 min after G1 release was examined. The experiment was repeated three times and statistical significance determined by * p < 0.05, using Kruskal–Wallis one-way ANOVA.
![Cells 11 02144 g003 Cells 11 02144 g003]()
Figure 4.
Elimination of the SAC suppresses metaphase arrest of cin8 kip1 cells. (A) mad1∆ partially suppresses the cell cycle delay in cin8 kip1 cells. G1-arrested WT (JBY649), mad1∆ (771-4-1), cin8-AID kip1∆ (3889-3-1), and cin8-AID kip1∆ mad1∆ (4003-1-3) cells containing Pds1-18myc were released into YPD, at 30 °C, containing 500 μM auxin. The α-factor was added back after 40 min release to block the following cell cycle. Samples were collected every 20 min to count budding index; n = 3. (B) mad1∆ alleviates the metaphase arrest in cin8 kip1 cells. The cells used in (A) were collected every 20 min to prepare protein samples. Western blotting was performed with anti-myc antibody to determine the level of Pds1-18myc. Pgk1, loading control. n = 3. (C) Quantification of Pds1 levels during cell cycle. The relative Pds1 levels of each strain during cell cycle were quantified by using the Western blot results in (B). (D) Elimination of the SAC results in chromosome missegregation in cin8 kip1 cells. WT (4244-3-4), cin8-AID kip1∆ (4260-2-4), and cin8-AID kip1∆ mad1∆ (4235-9-3) cells containing CEN4-GFP and Spc110-mCherry were grown to mid-log phase in YPD medium at 30 °C. Auxin was added at 500 μM. Samples were collected, and pictures were taken at 0 and 180 min. White arrows represent the SPB and CEN4-GFP in a cell showing chromosome missegregation. Scale bar, 5 μm. Pictures are representative from three experimental repeats. (E) Quantification of chromosome missegregation. Segregation of SPB and CEN4-GFP was counted at 180 min. Segregation of CEN4-GFP and SPB was categorized as normal, unseparated, or missegregated. Unseparated represents the absence of two clear CEN4-GFP and SPB dots. The experiment was repeated three times, and statistical significance was determined by * p < 0.05, using Kruskal–Wallis one-way ANOVA. (F) Deficient SAC causes viability loss in cin8 kip1 cells. Cells in (D) were also collected at 180 min and spread onto YPD to count the plating efficiency after overnight incubation at 30 °C. The experiment was repeated three times, and statistical significance was determined by * p < 0.05, using the Wilcoxon rank sum test.
Figure 4.
Elimination of the SAC suppresses metaphase arrest of cin8 kip1 cells. (A) mad1∆ partially suppresses the cell cycle delay in cin8 kip1 cells. G1-arrested WT (JBY649), mad1∆ (771-4-1), cin8-AID kip1∆ (3889-3-1), and cin8-AID kip1∆ mad1∆ (4003-1-3) cells containing Pds1-18myc were released into YPD, at 30 °C, containing 500 μM auxin. The α-factor was added back after 40 min release to block the following cell cycle. Samples were collected every 20 min to count budding index; n = 3. (B) mad1∆ alleviates the metaphase arrest in cin8 kip1 cells. The cells used in (A) were collected every 20 min to prepare protein samples. Western blotting was performed with anti-myc antibody to determine the level of Pds1-18myc. Pgk1, loading control. n = 3. (C) Quantification of Pds1 levels during cell cycle. The relative Pds1 levels of each strain during cell cycle were quantified by using the Western blot results in (B). (D) Elimination of the SAC results in chromosome missegregation in cin8 kip1 cells. WT (4244-3-4), cin8-AID kip1∆ (4260-2-4), and cin8-AID kip1∆ mad1∆ (4235-9-3) cells containing CEN4-GFP and Spc110-mCherry were grown to mid-log phase in YPD medium at 30 °C. Auxin was added at 500 μM. Samples were collected, and pictures were taken at 0 and 180 min. White arrows represent the SPB and CEN4-GFP in a cell showing chromosome missegregation. Scale bar, 5 μm. Pictures are representative from three experimental repeats. (E) Quantification of chromosome missegregation. Segregation of SPB and CEN4-GFP was counted at 180 min. Segregation of CEN4-GFP and SPB was categorized as normal, unseparated, or missegregated. Unseparated represents the absence of two clear CEN4-GFP and SPB dots. The experiment was repeated three times, and statistical significance was determined by * p < 0.05, using Kruskal–Wallis one-way ANOVA. (F) Deficient SAC causes viability loss in cin8 kip1 cells. Cells in (D) were also collected at 180 min and spread onto YPD to count the plating efficiency after overnight incubation at 30 °C. The experiment was repeated three times, and statistical significance was determined by * p < 0.05, using the Wilcoxon rank sum test.
![Cells 11 02144 g004 Cells 11 02144 g004]()
Figure 5.
The suppression of the cell-cycle delay in cin8 cells by tension checkpoint mutant dam1-3A. (A) dam1-3A is synthetically lethal with cin8-AID in the presence of auxin. WT (JBY649), dam1-3A (2425-7-2), cin8-AID (4332-5-4), and cin8-AID dam1-3A (4332-13-2) cells with Pds1-18myc were 10-fold serially diluted onto YPD plates, both with and without 500 μM auxin. Growth was analyzed after a 2-day incubation at 30 °C. (B) dam1-3A partially suppresses the cell-cycle delay in cin8-AID. The yeast strains listed in (A) were arrested in G1 and then released into 30 °C YPD containing 500 μM auxin. The α-factor was added back after 40 min release to block the following cell cycle. Cells were collected every 20 min to count the budding index; n = 3. (C) dam1-3A mutation suppresses the metaphase arrest in cin8 cells. The yeast cells in (B) were collected every 20 min to prepare protein samples. Western blotting was performed with anti-myc antibody to determine Pds1 protein levels. Pgk1, loading control. Pictures are representative of three experimental repeats. (D) Quantification of Pds1 levels. The relative Pds1 levels of each strain during cell cycle were quantified by using the Western blotting results from (C). Quantification of Pds1 levels is described in the Methods section.
Figure 5.
The suppression of the cell-cycle delay in cin8 cells by tension checkpoint mutant dam1-3A. (A) dam1-3A is synthetically lethal with cin8-AID in the presence of auxin. WT (JBY649), dam1-3A (2425-7-2), cin8-AID (4332-5-4), and cin8-AID dam1-3A (4332-13-2) cells with Pds1-18myc were 10-fold serially diluted onto YPD plates, both with and without 500 μM auxin. Growth was analyzed after a 2-day incubation at 30 °C. (B) dam1-3A partially suppresses the cell-cycle delay in cin8-AID. The yeast strains listed in (A) were arrested in G1 and then released into 30 °C YPD containing 500 μM auxin. The α-factor was added back after 40 min release to block the following cell cycle. Cells were collected every 20 min to count the budding index; n = 3. (C) dam1-3A mutation suppresses the metaphase arrest in cin8 cells. The yeast cells in (B) were collected every 20 min to prepare protein samples. Western blotting was performed with anti-myc antibody to determine Pds1 protein levels. Pgk1, loading control. Pictures are representative of three experimental repeats. (D) Quantification of Pds1 levels. The relative Pds1 levels of each strain during cell cycle were quantified by using the Western blotting results from (C). Quantification of Pds1 levels is described in the Methods section.
![Cells 11 02144 g005 Cells 11 02144 g005]()
Figure 6.
Tension checkpoint mutant dam1-3A causes chromosome missegregation in cells with depleted Cin8. (A) Images showing chromosome missegregation in cin8 dam1-3A mutants. WT (4244-3-4), cin8-AID (4244-2-1), dam1-3A (4330-7-4), and cin8-AID dam1-3A (4244-6-3) cells containing CEN4-GFP and Spc110-mCherry were grown to mid-log phase in YPD medium at 30 °C. Auxin was added to a final concentration of 500 μM. Pictures were taken at 0 and 180 min to visualize CEN4-GFP and Spc110-mCherry. White arrows represent SPB and CEN4-GFP in a cell showing chromosome missegregation. Scale bar, 5 μm. Pictures are representative from three experimental repeats. (B) The segregation of CEN4-GFP and SPBs. Segregation of CEN4-GFP and SPB was categorized as normal, unseparated, or missegregated. This experiment was repeated three times, and statistical significance was determined by * p < 0.05, using Kruskal–Wallis one-way ANOVA. (C) Viability loss in cin8-AID dam1-3A mutants after incubation in the presence of auxin. Cells in (B) were also collected at 180 min and spread onto YPD, and the plating efficiency was examined after overnight incubation at 30 °C. The experiment was repeated three times, and statistical significance was determined by * p < 0.05, using the Wilcoxon rank sum test.
Figure 6.
Tension checkpoint mutant dam1-3A causes chromosome missegregation in cells with depleted Cin8. (A) Images showing chromosome missegregation in cin8 dam1-3A mutants. WT (4244-3-4), cin8-AID (4244-2-1), dam1-3A (4330-7-4), and cin8-AID dam1-3A (4244-6-3) cells containing CEN4-GFP and Spc110-mCherry were grown to mid-log phase in YPD medium at 30 °C. Auxin was added to a final concentration of 500 μM. Pictures were taken at 0 and 180 min to visualize CEN4-GFP and Spc110-mCherry. White arrows represent SPB and CEN4-GFP in a cell showing chromosome missegregation. Scale bar, 5 μm. Pictures are representative from three experimental repeats. (B) The segregation of CEN4-GFP and SPBs. Segregation of CEN4-GFP and SPB was categorized as normal, unseparated, or missegregated. This experiment was repeated three times, and statistical significance was determined by * p < 0.05, using Kruskal–Wallis one-way ANOVA. (C) Viability loss in cin8-AID dam1-3A mutants after incubation in the presence of auxin. Cells in (B) were also collected at 180 min and spread onto YPD, and the plating efficiency was examined after overnight incubation at 30 °C. The experiment was repeated three times, and statistical significance was determined by * p < 0.05, using the Wilcoxon rank sum test.
![Cells 11 02144 g006 Cells 11 02144 g006]()
Figure 7.
Functional Ipl1 kinase is required for the cell-cycle delay in cin8 mutants. (A) cin8-AID ipl1-321 double mutants show growth defect on plates containing auxin. Saturated WT (JBY649), ipl1-321 (2715-6-4), cin8-AID (4332-5-4), and ipl1-321 cin8-AID (4338-8-1) cells containing Pds1-18myc were 10-fold serially diluted onto YPD plates, both with and without 500 μM auxin. Growth was analyzed after a 2-day incubation at 25 °C and 1-day incubation at 37 °C. (B) ipl1-321 suppresses the cell cycle delay in cin8 cells. G1-arrested WT (JBY649), ipl-321 (2715-6-4), cin8-AID (4332-5-4), and cin8-AID ipl1-321 (4333-8-1) cells containing Pds1-18myc were released into YPD at 25 °C containing 500 μM auxin. After 60 min release, α-factor was added back to block the following cell cycle. Samples were collected every 30 min to count budding index (n = 3). (C) ipl1-321 mutation suppresses the metaphase arrest in cin8 cells. The yeast cells in (B) were also collected every 30 min to prepare protein samples. Western blotting was performed with anti-myc antibody to determine Pds1 protein levels. Pgk1 was used as the loading control. (D) Quantification of Pds1 levels. The relative Pdsl levels were quantified by using the Western blotting results from (C). (E) A model showing the role of kinesin-5 motor Cin8 in chromosome segregation.
Figure 7.
Functional Ipl1 kinase is required for the cell-cycle delay in cin8 mutants. (A) cin8-AID ipl1-321 double mutants show growth defect on plates containing auxin. Saturated WT (JBY649), ipl1-321 (2715-6-4), cin8-AID (4332-5-4), and ipl1-321 cin8-AID (4338-8-1) cells containing Pds1-18myc were 10-fold serially diluted onto YPD plates, both with and without 500 μM auxin. Growth was analyzed after a 2-day incubation at 25 °C and 1-day incubation at 37 °C. (B) ipl1-321 suppresses the cell cycle delay in cin8 cells. G1-arrested WT (JBY649), ipl-321 (2715-6-4), cin8-AID (4332-5-4), and cin8-AID ipl1-321 (4333-8-1) cells containing Pds1-18myc were released into YPD at 25 °C containing 500 μM auxin. After 60 min release, α-factor was added back to block the following cell cycle. Samples were collected every 30 min to count budding index (n = 3). (C) ipl1-321 mutation suppresses the metaphase arrest in cin8 cells. The yeast cells in (B) were also collected every 30 min to prepare protein samples. Western blotting was performed with anti-myc antibody to determine Pds1 protein levels. Pgk1 was used as the loading control. (D) Quantification of Pds1 levels. The relative Pdsl levels were quantified by using the Western blotting results from (C). (E) A model showing the role of kinesin-5 motor Cin8 in chromosome segregation.
![Cells 11 02144 g007 Cells 11 02144 g007]()