‘Come together’—The Regulatory Interaction of Herpesviral Nuclear Egress Proteins Comprises Both Essential and Accessory Functions
Abstract
:1. Current Concept of Herpesviral Nuclear Egress Regulation
2. The Herpesviral Heterodimeric Core Nuclear Egress Complexes
2.1. Two Functionally Specialized Proteins Determine the Core of the Herpesviral NECs
2.2. The Hook-into-Groove Principle of Core NEC Heterodimerization
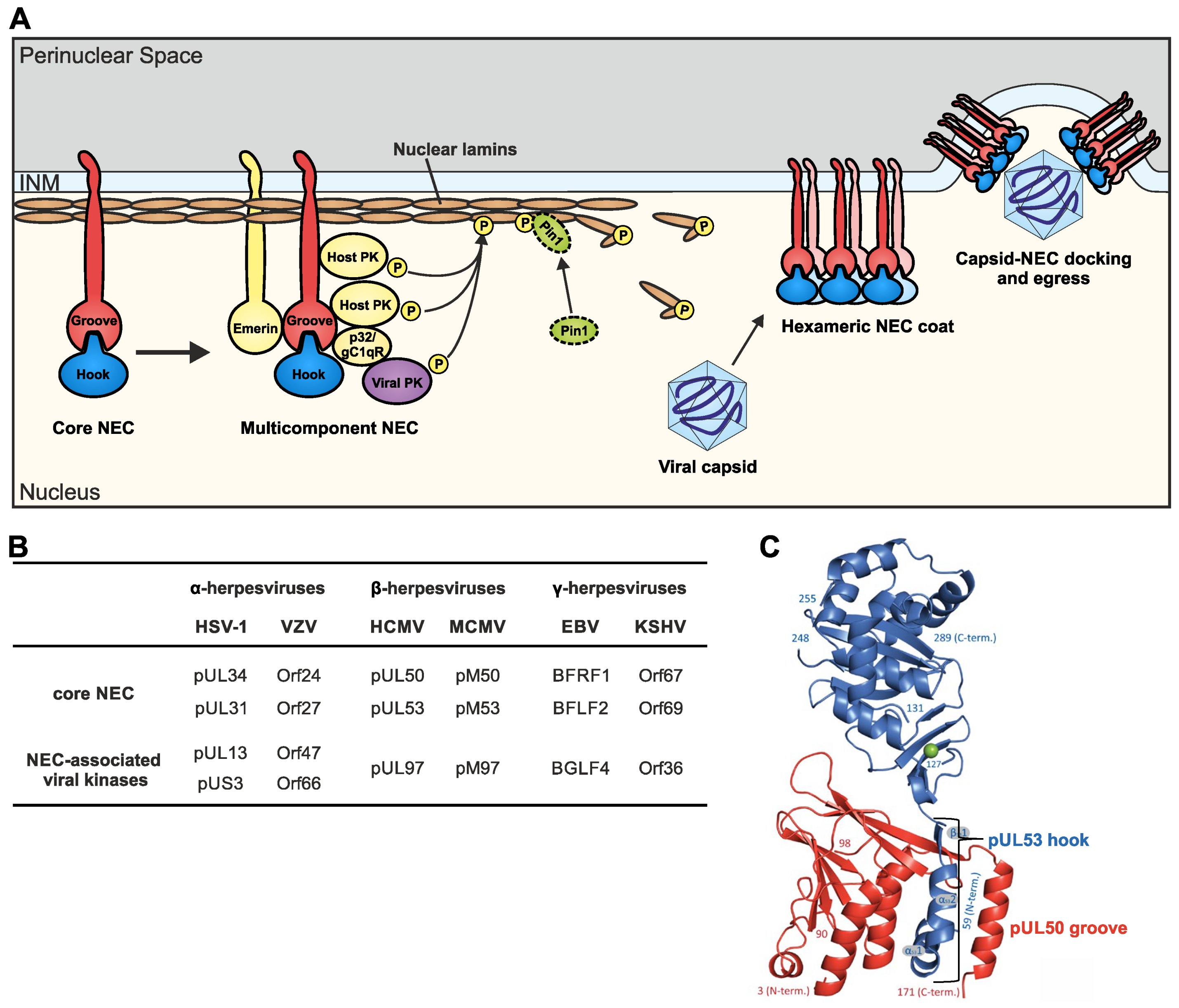
3. Multiple Viral and Host Components Can Be Associated with Nuclear Egress Complexes
3.1. Herpesviral Multicomponent NECs Represent Assembly Platforms That Recruit a Variety of Nuclear Proteins with Regulatory Activities
3.2. Similarities and Differences between the Constituents of Herpesviral Multicomponent NECs
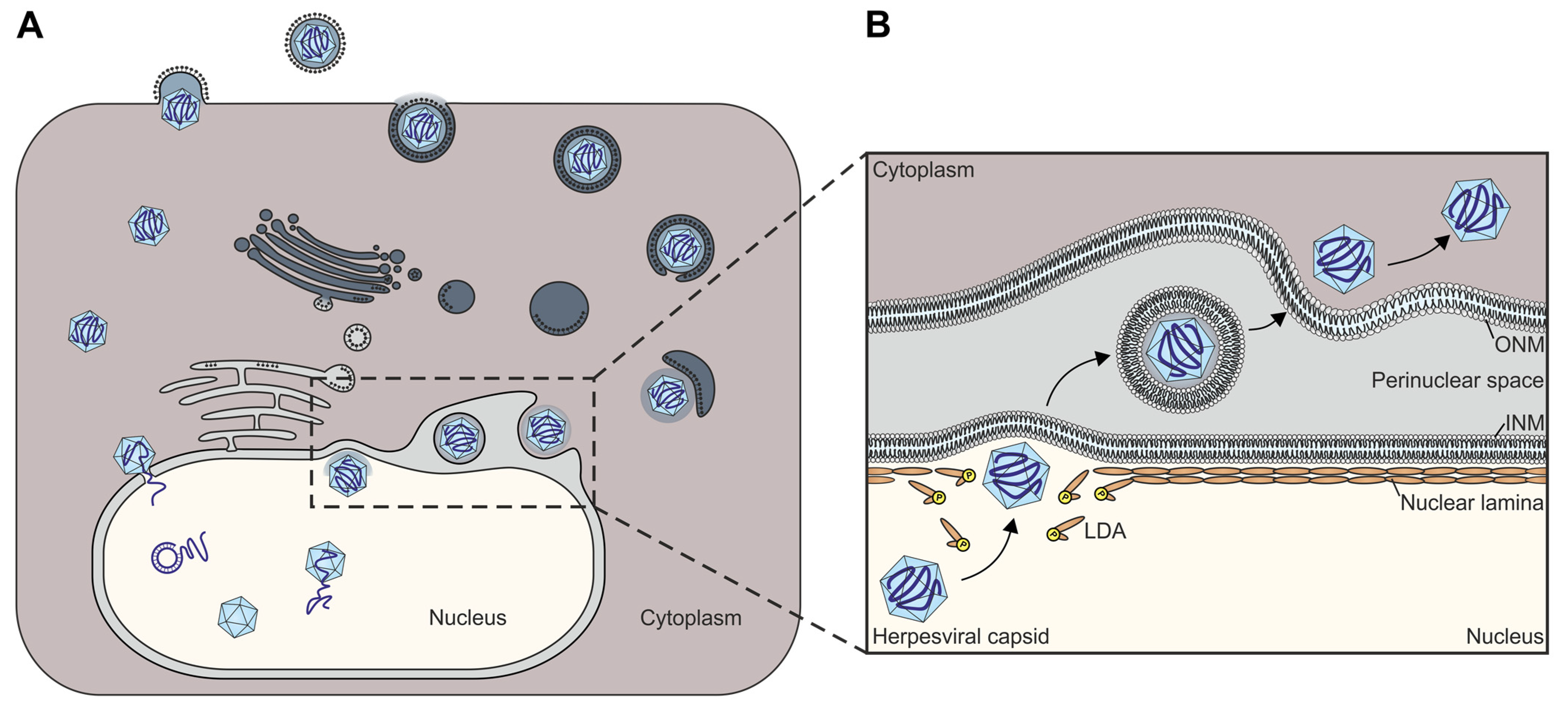
4. Main Functionality and Regulatory Roles Shared by Herpesviral NECs
4.1. The Key Role Played by Herpesviral NECs during the Viral Lytic Replication Cycle
4.2. Comparative Aspects of Herpesviral Nuclear Egress and Cellular NE-Specific Processes
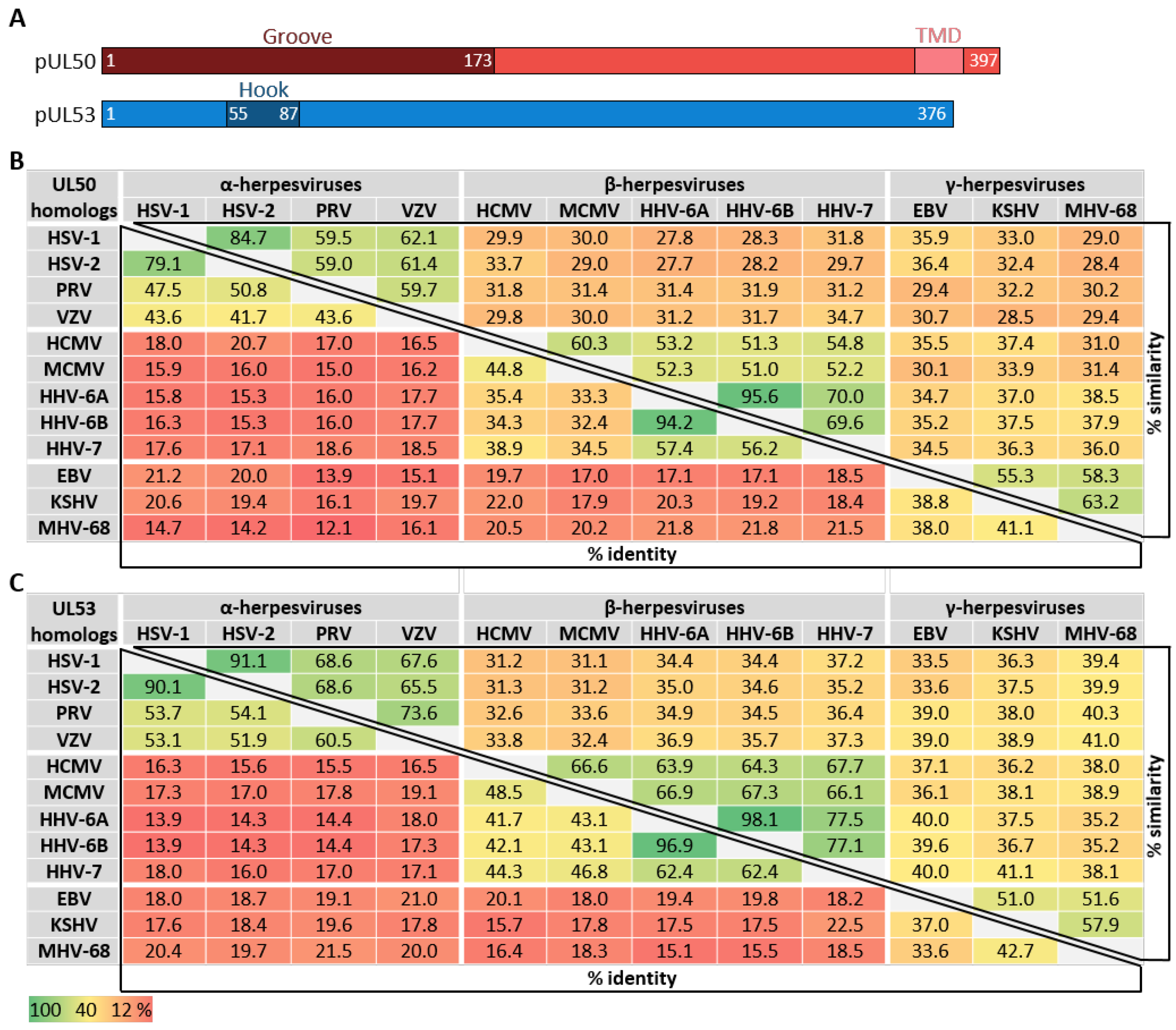
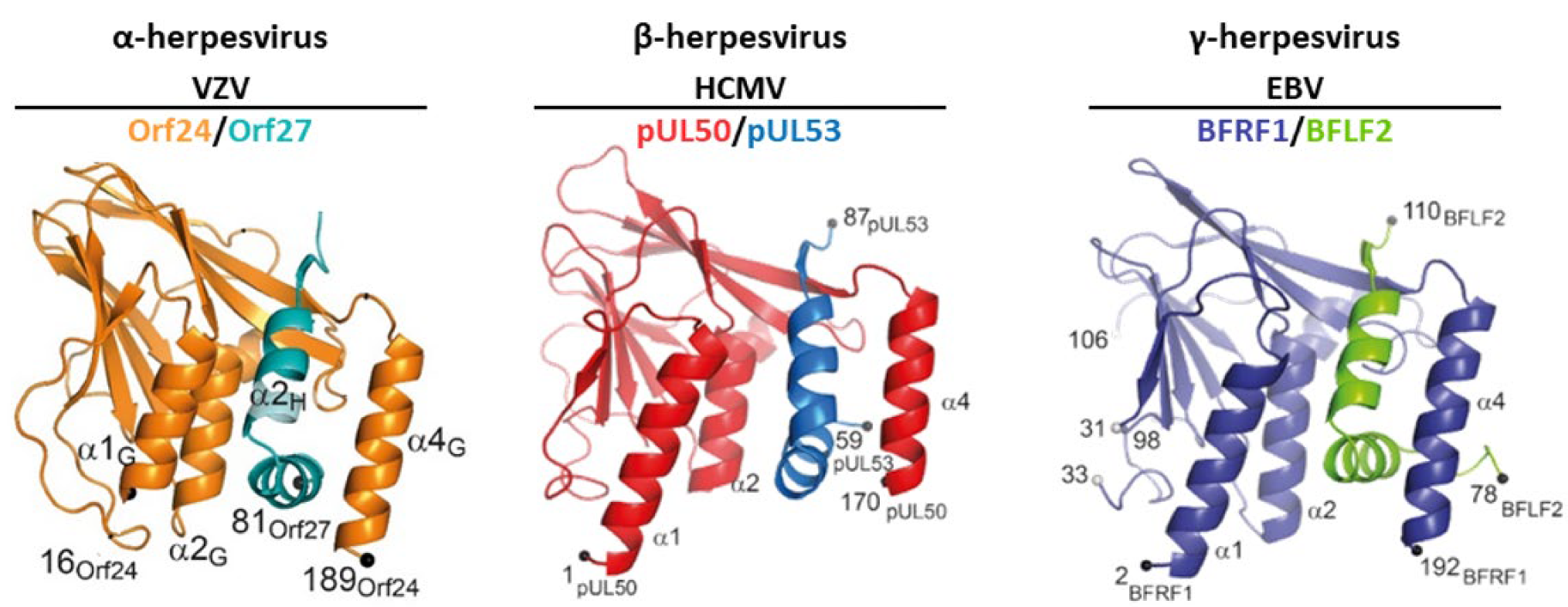
5. The Correlation between Herpesviral Core NEC Primary Sequences, Structural Conservation and Binding Properties
5.1. What Is the Degree of Conservation between the Primary NEC Amino Acid Sequences?
5.2. How Is the Correlation between NEC Sequences, Structures and Binding Properties?
6. Mutational Analysis Focusing on Specific Herpesviral NEC Functions
6.1. Phenotypical Features of Herpesviral Core NEC Proteins Determined by Viral Deletion Mutants
6.2. Regulated Viral Nuclear Egress Compared to Nuclear Envelope Breakdown (NEBD)
6.3. The Essential and Non-Essential Functional Properties of Herpesviral Core NECs
7. Validation of the NEC as a Unique Target of Anti-Herpesviral Therapy
7.1. Current Approaches and Proof-of-Concept That Support the Exploitation of the Herpesviral NEC as an Efficient Drug Target
7.2. The Mode of NEC-Directed Antiviral Drug Activity Achieved through Several Defined Mechanistic Principles
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Krug, L.T.; Pellett, P.E. The Family Herpesviridae: A Brief Introduction. In Fields Virology, 7th ed.; Howley, P.M., Knipe, D.M., Eds.; Wolter Kluwer: Warsaw, Poland, 2021; Volume 2, pp. 212–234. [Google Scholar]
- Griffiths, P.; Baraniak, I.; Reeves, M. The pathogenesis of human cytomegalovirus. J. Pathol. 2014, 235, 288–297. [Google Scholar] [CrossRef]
- Beck, M.; Hurt, E. The nuclear pore complex: Understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2016, 18, 73–89. [Google Scholar] [CrossRef]
- Panté, N.; Kann, M. Nuclear Pore Complex Is Able to Transport Macromolecules with Diameters of about ∼39 nm. Mol. Biol. Cell 2002, 13, 425–434. [Google Scholar] [CrossRef] [Green Version]
- Arii, J. Host and Viral Factors Involved in Nuclear Egress of Herpes Simplex Virus 1. Viruses 2021, 13, 754. [Google Scholar] [CrossRef]
- Bigalke, J.; Heldwein, E. Have NEC Coat, Will Travel: Structural Basis of Membrane Budding During Nuclear Egress in Herpesviruses. Adv. Virus Res. 2017, 97, 107–141. [Google Scholar] [CrossRef] [Green Version]
- Draganova, E.B.; Thorsen, M.K.; Heldwein, E.E. Nuclear Egress. Curr. Issues Mol. Biol. 2020, 41, 125–170. [Google Scholar] [CrossRef]
- Hellberg, T.; Paßvogel, L.; Schulz, K.S.; Klupp, B.G.; Mettenleiter, T.C. Nuclear Egress of Herpesviruses: The Prototypic Ve-sicular Nucleocytoplasmic Transport. Adv. Virus Res. 2016, 94, 81–140. [Google Scholar] [CrossRef]
- Lee, C.-P.; Chen, M.-R. Conquering the Nuclear Envelope Barriers by EBV Lytic Replication. Viruses 2021, 13, 702. [Google Scholar] [CrossRef]
- Lye, M.F.; Wilkie, A.R.; Filman, D.J.; Hogle, J.M.; Coen, D.M. Getting to and through the inner nuclear membrane during herpesvirus nuclear egress. Curr. Opin. Cell Biol. 2017, 46, 9–16. [Google Scholar] [CrossRef]
- Marschall, M.; Häge, S.; Conrad, M.; Alkhashrom, S.; Kicuntod, J.; Schweininger, J.; Kriegel, M.; Lösing, J.; Tillmanns, J.; Neipel, F.; et al. Nuclear Egress Complexes of HCMV and Other Herpesviruses: Solving the Puzzle of Sequence Coevolution, Conserved Structures and Subfamily-Spanning Binding Properties. Viruses 2020, 12, 683. [Google Scholar] [CrossRef]
- Marschall, M.; Muller, Y.A.; Diewald, B.; Sticht, H.; Milbradt, J. The human cytomegalovirus nuclear egress complex unites multiple functions: Recruitment of effectors, nuclear envelope rearrangement, and docking to nuclear capsids. Rev. Med. Virol. 2017, 27, e1934. [Google Scholar] [CrossRef] [PubMed]
- Mettenleiter, T.C.; Müller, F.; Granzow, H.; Klupp, B.G. The way out: What we know and do not know about herpesvirus nuclear egress. Cell. Microbiol. 2012, 15, 170–178. [Google Scholar] [CrossRef]
- Roller, R.J.; Baines, J.D. Herpesvirus Nuclear Egress. Advances in Anatomy, Embryology, and Cell Biology. In Cell Biology of Herpes Viruses; Osterrieder, K., Ed.; Springer International Publishing: Cham, Switzerland, 2017; Volume 223, pp. 143–169. [Google Scholar] [CrossRef]
- Johnson, D.C.; Baines, J.D. Herpesviruses remodel host membranes for virus egress. Nat. Rev. Genet. 2011, 9, 382–394. [Google Scholar] [CrossRef]
- Sanchez, V.; Britt, W. Human Cytomegalovirus Egress: Overcoming Barriers and Co-Opting Cellular Functions. Viruses 2021, 14, 15. [Google Scholar] [CrossRef]
- Crump, C. Virus Assembly and Egress of HSV. Adv. Exp. Med. Biol. 2018, 1045, 23–44. [Google Scholar] [CrossRef]
- Mettenleiter, T.C.; Klupp, B.G.; Granzow, H. Herpesvirus assembly: An update. Virus Res. 2009, 143, 222–234. [Google Scholar] [CrossRef]
- Roller, R.J.; Hassman, T.; Haugo-Crooks, A. Cell culture evolution of a HSV-1/VZV UL34/ORF24 chimeric virus reveals novel functions for HSV genes in capsid nuclear egress. J. Virol. 2021, 93. [Google Scholar] [CrossRef]
- Rose, A.; Schlieker, C. Alternative nuclear transport for cellular protein quality control. Trends Cell Biol. 2012, 22, 509–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speese, S.D.; Ashley, J.; Jokhi, V.; Nunnari, J.; Barria, R.; Li, Y.; Ataman, B.; Koon, A.; Chang, Y.-T.; Li, Q.; et al. Nuclear Envelope Budding Enables Large Ribonucleoprotein Particle Export during Synaptic Wnt Signaling. Cell 2012, 149, 832–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeev-Ben-Mordehai, T.; Weberruß, M.; Lorenz, M.; Cheleski, J.; Hellberg, T.; Whittle, C.; El Omari, K.; Vasishtan, D.; Dent, K.C.; Harlos, K.; et al. Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights into Inner Nuclear Membrane Remodeling. Cell Rep. 2015, 13, 2645–2652. [Google Scholar] [CrossRef] [Green Version]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, M.; Bender, B.J.; Kamil, J.P.; Lye, M.F.; Pesola, J.M.; Reim, N.I.; Hogle, J.M.; Coen, D.M. Human Cytomegalovirus UL97 Phosphorylates the Viral Nuclear Egress Complex. J. Virol. 2015, 89, 523–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonntag, E.; Milbradt, J.; Svrlanska, A.; Strojan, H.; Häge, S.; Kraut, A.; Hesse, A.-M.; Amin, B.; Sonnewald, U.; Couté, Y.; et al. Protein kinases responsible for the phosphorylation of the nuclear egress core complex of human cytomegalovirus. J. Gen. Virol. 2017, 98, 2569–2581. [Google Scholar] [CrossRef] [PubMed]
- Milbradt, J.; Hutterer, C.; Bahsi, H.; Wagner, S.; Sonntag, E.; Horn, A.H.C.; Kaufer, B.B.; Mori, Y.; Sticht, H.; Fossen, T.; et al. The Prolyl Isomerase Pin1 Promotes the Herpesvirus-Induced Phosphorylation-Dependent Disassembly of the Nuclear Lamina Required for Nucleocytoplasmic Egress. PLoS Pathog. 2016, 12, e1005825. [Google Scholar] [CrossRef] [Green Version]
- Milbradt, J.; Kraut, A.; Hutterer, C.; Sonntag, E.; Schmeiser, C.; Ferro, M.; Wagner, S.; Lenac, T.; Claus, C.; Pinkert, S.; et al. Proteomic Analysis of the Multimeric Nuclear Egress Complex of Human Cytomegalovirus. Mol. Cell. Proteom. 2014, 13, 2132–2146. [Google Scholar] [CrossRef] [Green Version]
- Häge, S.; Sonntag, E.; Svrlanska, A.; Borst, E.M.; Stilp, A.-C.; Horsch, D.; Müller, R.; Kropff, B.; Milbradt, J.; Stamminger, T.; et al. Phenotypical Characterization of the Nuclear Egress of Recombinant Cytomegaloviruses Reveals Defective Replication upon ORF-UL50 Deletion but Not pUL50 Phosphosite Mutation. Viruses 2021, 13, 165. [Google Scholar] [CrossRef]
- Lemnitzer, F.; Raschbichler, V.; Kolodziejczak, D.; Israel, L.; Imhof, A.; Bailer, S.M.; Koszinowski, U.; Ruzsics, Z. Mouse cytomegalovirus egress protein pM50 interacts with cellular endophilin-A2. Cell. Microbiol. 2012, 15, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Kato, A.; Oyama, M.; Kozuka-Hata, H.; Arii, J.; Kawaguchi, Y. Role of Host Cell p32 in Herpes Simplex Virus 1 De-Envelopment during Viral Nuclear Egress. J. Virol. 2015, 89, 8982–8998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lye, M.F.; Sharma, M.; El Omari, K.; Filman, D.; Schuermann, J.P.; Hogle, J.M.; Coen, D.M. Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex. EMBO J. 2015, 34, 2937–2952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, Y.A.; Hage, S.; Alkhashrom, S.; Hollriegl, T.; Weigert, S.; Dolles, S.; Hof, K.; Walzer, S.A.; Egerer-Sieber, C.; Conrad, M.; et al. High-resolution crystal structures of two prototypical beta- and gamma-herpesviral nuclear egress complexes unravel the determinants of subfamily specificity. J. Biol. Chem. 2020, 295. [Google Scholar] [CrossRef]
- Walzer, S.A.; Egerer-Sieber, C.; Sticht, H.; Sevvana, M.; Hohl, K.; Milbradt, J.; Muller, Y.A.; Marschall, M. Crystal Structure of the Human Cytomegalovirus pUL50-pUL53 Core Nuclear Egress Complex Provides Insight into a Unique Assembly Scaffold for Virus-Host Protein Interactions. J. Biol. Chem. 2015, 290, 27452–27458. [Google Scholar] [CrossRef] [Green Version]
- Diewald, B.; Socher, E.; Söldner, C.A.; Sticht, H. Conformational Dynamics of Herpesviral NEC Proteins in Different Oligomerization States. Int. J. Mol. Sci. 2018, 19, 2908. [Google Scholar] [CrossRef] [Green Version]
- Häge, S.; Sonntag, E.; Borst, E.M.; Tannig, P.; Seyler, L.; Bäuerle, T.; Bailer, S.M.; Lee, C.-P.; Müller, R.; Wangen, C.; et al. Patterns of Autologous and Nonautologous Interactions between Core Nuclear Egress Complex (NEC) Proteins of alpha, beta and gamma Herpesviruses. Viruses 2020, 12, 303. [Google Scholar] [CrossRef] [Green Version]
- Milbradt, J.; Sonntag, E.; Wagner, S.; Strojan, H.; Wangen, C.; Rovis, T.L.; Lisnic, B.; Jonjic, S.; Sticht, H.; Britt, W.J.; et al. Human Cytomegalovirus Nuclear Capsids Associate with the Core Nuclear Egress Complex and the Viral Protein Kinase pUL97. Viruses 2018, 10, 35. [Google Scholar] [CrossRef] [Green Version]
- Wilkie, A.R.; Sharma, M.; Coughlin, M.; Pesola, J.M.; Ericsson, M.; Lawler, J.L.; Fernandez, R.; Coen, D.M. Human Cytomegalovirus Nuclear Egress Complex Subunit, UL53, Associates with Capsids and Myosin Va, but Is Not Important for Capsid Localization towards the Nuclear Periphery. Viruses 2022, 14, 479. [Google Scholar] [CrossRef]
- Marschall, M.; Marzi, A.; aus dem Siepen, P.; Jochmann, R.; Kalmer, M.; Auerochs, S.; Lischka, P.; Leis, M.; Stamminger, T. Cellular p32 Recruits Cytomegalovirus Kinase pUL97 to Redistribute the Nuclear Lamina. J. Biol. Chem. 2005, 280, 33357–33367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milbradt, J.; Auerochs, S.; Marschall, M. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J. Gen. Virol. 2007, 88, 2642–2650. [Google Scholar] [CrossRef]
- Milbradt, J.; Auerochs, S.; Sticht, H.; Marschall, M. Cytomegaloviral proteins that associate with the nuclear lamina: Components of a postulated nuclear egress complex. J. Gen. Virol. 2009, 90, 579–590. [Google Scholar] [CrossRef]
- Milbradt, J.; Webel, R.; Auerochs, S.; Sticht, H.; Marschall, M. Novel Mode of Phosphorylation-triggered Reorganization of the Nuclear Lamina during Nuclear Egress of Human Cytomegalovirus. J. Biol. Chem. 2010, 285, 13979–13989. [Google Scholar] [CrossRef] [Green Version]
- Sonntag, E.; Hamilton, S.T.; Bahsi, H.; Wagner, S.; Jonjic, S.; Rawlinson, W.D.; Marschall, M.; Milbradt, J. Cytomegalovirus pUL50 is the multi-interacting determinant of the core nuclear egress complex (NEC) that recruits cellular accessory NEC components. J. Gen. Virol. 2016, 97, 1676–1685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Changotra, H.; Turk, S.M.; Artigues, A.; Thakur, N.; Gore, M.; Muggeridge, M.I.; Hutt-Fletcher, L.M. Epstein–Barr virus glycoprotein gM can interact with the cellular protein p32 and knockdown of p32 impairs virus. Virology 2016, 489, 223–232. [Google Scholar] [CrossRef] [Green Version]
- Goodrum, F.; Britt, W.; Mocarski, E.S. Cytomegalovirus in Fields Virology, 7th ed.; Howley, P.M., Knipe, D.M., Damania, B.A., Cohen, J.I., Eds.; Wolters Kluwer: Warsaw, Poland, 2021; Volume 2, pp. 389–444. [Google Scholar]
- Oberstein, A.; Perlman, D.H.; Shenk, T.; Terry, L.J. Human cytomegalovirus pUL97 kinase induces global changes in the infected cell phosphoproteome. Proteomics 2015, 15, 2006–2022. [Google Scholar] [CrossRef] [Green Version]
- Stahl, J.A.; Chavan, S.S.; Sifford, J.M.; MacLeod, V.; Voth, D.E.; Edmondson, R.D.; Forrest, J.C. Phosphoproteomic Analyses Reveal Signaling Pathways That Facilitate Lytic Gammaherpesvirus Replication. PLoS Pathog. 2013, 9, e1003583. [Google Scholar] [CrossRef] [Green Version]
- Weekes, M.P.; Tomasec, P.; Huttlin, E.L.; Fielding, C.A.; Nusinow, D.; Stanton, R.J.; Wang, E.C.Y.; Aicheler, R.; Murrell, I.; Wilkinson, G.W.G.; et al. Quantitative Temporal Viromics: An Approach to Investigate Host-Pathogen Interaction. Cell 2014, 157, 1460–1472. [Google Scholar] [CrossRef] [Green Version]
- Beltran, P.M.J.; Cristea, I.M. The life cycle and pathogenesis of human cytomegalovirus infection: Lessons from proteomics. Expert Rev. Proteom. 2014, 11, 697–711. [Google Scholar] [CrossRef]
- Maeda, F.; Arii, J.; Hirohata, Y.; Maruzuru, Y.; Koyanagi, N.; Kato, A.; Kawaguchi, Y. Herpes Simplex Virus 1 UL34 Protein Regulates the Global Architecture of the Endoplasmic Reticulum in Infected Cells. J. Virol. 2017, 91, e00271-17. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.K.; Kim, Y.J.; Han, T.-H.; Milbradt, J.; Marschall, M.; Ahn, J.-H. Transmembrane Protein pUL50 of Human Cytomegalovirus Inhibits ISGylation by Downregulating UBE1L. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.K.; Hyeon, S.; Ahn, J.-H. The Human Cytomegalovirus Transmembrane Protein pUL50 Induces Loss of VCP/p97 and Is Regulated by a Small Isoform of pUL50. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Gonnella, R.; Dimarco, M.; Farina, G.A.; Santarelli, R.; Valia, S.; Faggioni, A.; Angeloni, A.; Cirone, M.; Farina, A. BFRF1 protein is involved in EBV-mediated autophagy manipulation. Microbes Infect. 2020, 22, 585–591. [Google Scholar] [CrossRef]
- Wang, P.; Deng, Y.; Guo, Y.; Xu, Z.; Li, Y.; Ou, X.; Xie, L.; Lu, M.; Zhong, J.; Li, B.; et al. Epstein-Barr Virus Early Protein BFRF1 Suppresses IFN-β Activity by Inhibiting the Activation of IRF3. Front. Immunol. 2020, 11, 513383. [Google Scholar] [CrossRef]
- Liu, G.; Kung, H.; Chen, C.; Huang, C.; Wang, Y.; Yu, C.; Lee, C. Improving nuclear envelope dynamics by EBV BFRF1 facilitates intranuclear component clearance through autophagy. FASEB J. 2018, 32, 3968–3983. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.-P.; Chen, M.-R. Escape of herpesviruses from the nucleus. Rev. Med. Virol. 2010, 20, 214–230. [Google Scholar] [CrossRef] [PubMed]
- Bigalke, J.M.; Heldwein, E.E. Nuclear Exodus: Herpesviruses Lead the Way. Annu. Rev. Virol. 2016, 3, 387–409. [Google Scholar] [CrossRef] [Green Version]
- Bailer, S.M. Venture from the Interior—Herpesvirus pUL31 Escorts Capsids from Nucleoplasmic Replication Compartments to Sites of Primary Envelopment at the Inner Nuclear Membrane. Cells 2017, 6, 46. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.Y.; Ikegami, K. Nuclear lamin phosphorylation: An emerging role in gene regulation and pathogenesis of laminopathies. Nucleus 2020, 11, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Gershburg, E.; Pagano, J.S. Conserved herpesvirus protein kinases. Biochim. Biophys. Acta Proteins Proteom. 2008, 1784, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Fradkin, L.G.; Budnik, V. This bud’s for you: Mechanisms of cellular nucleocytoplasmic trafficking via nuclear envelope budding. Curr. Opin. Cell Biol. 2016, 41, 125–131. [Google Scholar] [CrossRef] [Green Version]
- Morrison, L.A.; DeLassus, G.S. Breach of the nuclear lamina during assembly of herpes simplex viruses. Nucleus 2011, 2, 271–276. [Google Scholar] [CrossRef] [Green Version]
- Cibulka, J.; Fraiberk, M.; Forstova, J. Nuclear Actin and Lamins in Viral Infections. Viruses 2012, 4, 325–347. [Google Scholar] [CrossRef] [Green Version]
- Arii, J.; Shindo, K.; Koyanagi, N.; Kato, A.; Kawaguchi, Y. Multiple Roles of the Cytoplasmic Domain of Herpes Simplex Virus 1 Envelope Glycoprotein D in Infected Cells. J. Virol. 2016, 90, 10170–10181. [Google Scholar] [CrossRef] [Green Version]
- Bigalke, J.M.; Heuser, T.; Nicastro, D.; Heldwein, E.E. Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat. Commun. 2014, 5, 4131. [Google Scholar] [CrossRef] [PubMed]
- Marschall, M.; Stamminger, T. Molecular targets for antiviral therapy of cytomegalovirus infections. Future Microbiol. 2009, 4, 731–742. [Google Scholar] [CrossRef]
- Chang, Y.E.; Van Sant, C.; Krug, P.W.; Sears, A.E.; Roizman, B. The null mutant of the U(L)31 gene of herpes simplex virus 1: Construction and phenotype in infected cells. J. Virol. 1997, 71, 8307–8315. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, W.; Klupp, B.G.; Granzow, H.; Osterrieder, N.; Mettenleiter, T.C. The Interacting UL31 and UL34 Gene Products of Pseudorabies Virus Are Involved in Egress from the Host-Cell Nucleus and Represent Components of Primary Enveloped but Not Mature Virions. J. Virol. 2002, 76, 364–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, L.; Tanaka, M.; Kawaguchi, Y.; Baines, J.D. Cell lines that support replication of a novel herpes simplex virus 1 UL31 deletion mutant can properly target UL34 protein to the nuclear rim in the absence of UL31. Virology 2004, 329, 68–76. [Google Scholar] [CrossRef] [Green Version]
- Roller, R.J.; Zhou, Y.; Schnetzer, R.; Ferguson, J.; DeSalvo, D. Herpes Simplex Virus Type 1 U L 34 Gene Product Is Required for Viral Envelopment. J. Virol. 2000, 74, 117–129. [Google Scholar] [CrossRef] [Green Version]
- Schweininger, J.; Kriegel, M.; Häge, S.; Conrad, M.; Alkhashrom, S.; Lösing, J.; Weiler, S.; Tillmanns, J.; Egerer-Sieber, C.; Decker, A.; et al. The crystal structure of the varicella-zoster Orf24-Orf27 nuclear egress complex spotlights multiple determinants of herpesvirus subfamily specificity. J. Biol. Chem. 2022, 298, 101625. [Google Scholar] [CrossRef]
- Ye, G.-J.; Roizman, B. The essential protein encoded by the UL 31 gene of herpes simplex virus 1 depends for its stability on the presence of U L 34 protein. Proc. Natl. Acad. Sci. USA 2000, 97, 11002–11007. [Google Scholar] [CrossRef] [Green Version]
- Bigalke, J.M.; Heldwein, E.E. Structural basis of membrane budding by the nuclear egress complex of herpesviruses. EMBO J. 2015, 34, 2921–2936. [Google Scholar] [CrossRef]
- Sudol, M.; Sliwa, K.; Russo, T. Functions of WW domains in the nucleus. FEBS Lett. 2001, 490, 190–195. [Google Scholar] [CrossRef] [Green Version]
- Zarrinpar, A.; Lim, W.A. Converging on proline: The mechanism of WW domain peptide recognition. Nat. Genet. 2000, 7, 611–613. [Google Scholar] [CrossRef]
- De Magistris, P.; Antonin, W. The Dynamic Nature of the Nuclear Envelope. Curr. Biol. 2018, 28, R487–R497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ungricht, R.; Kutay, U. Mechanisms and functions of nuclear envelope remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 229–245. [Google Scholar] [CrossRef]
- Heald, R.; McKeon, F. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell 1990, 61, 579–589. [Google Scholar] [CrossRef]
- Peter, M.; Heitlinger, E.; Häner, M.; Aebi, U.; Nigg, E. Disassembly of in vitro formed lamin head-to-tail polymers by CDC2 kinase. EMBO J. 1991, 10, 1535–1544. [Google Scholar] [CrossRef]
- Thompson, L.J.; Fields, A.P. betaII protein kinase C is required for the G2/M phase transition of cell cycle. J. Biol. Chem. 1996, 271, 15045–15053. [Google Scholar] [CrossRef] [Green Version]
- Hamirally, S.; Kamil, J.P.; Ndassa-Colday, Y.M.; Lin, A.J.; Jahng, W.J.; Baek, M.-C.; Noton, S.; Silva, L.A.; Simpson-Holley, M.; Knipe, D.M.; et al. Viral Mimicry of Cdc2/Cyclin-Dependent Kinase 1 Mediates Disruption of Nuclear Lamina during Human Cytomegalovirus Nuclear Egress. PLoS Pathog. 2009, 5, e1000275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muranyi, W.; Haas, J.; Wagner, M.; Krohne, G.; Koszinowski, U.H. Cytomegalovirus Recruitment of Cellular Kinases to Dissolve the Nuclear Lamina. Science 2002, 297, 854–857. [Google Scholar] [CrossRef]
- Sharma, M.; Kamil, J.P.; Coughlin, M.; Reim, N.I.; Coen, D.M. Human Cytomegalovirus UL50 and UL53 Recruit Viral Protein Kinase UL97, Not Protein Kinase C, for Disruption of Nuclear Lamina and Nuclear Egress in Infected Cells. J. Virol. 2014, 88, 249–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, Y.; Zhou, S.; Gao, S.; Deng, H. Remodeling of host membranes during herpesvirus assembly and egress. Protein Cell 2018, 10, 315–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hume, A.J.; Finkel, J.S.; Kamil, J.P.; Coen, D.M.; Culbertson, M.R.; Kalejta, R.F. Phosphorylation of Retinoblastoma Protein by Viral Protein with Cyclin-Dependent Kinase Function. Science 2008, 320, 797–799. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Kato, K.; Tanaka, M.; Kanamori, M.; Nishiyama, Y.; Yamanashi, Y. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1delta. J. Virol. 2003, 77, 2359–2368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuny, C.V.; Chinchilla, K.; Culbertson, M.R.; Kalejta, R.F. Cyclin-Dependent Kinase-Like Function Is Shared by the Beta- and Gamma- Subset of the Conserved Herpesvirus Protein Kinases. PLoS Pathog. 2010, 6, e1001092. [Google Scholar] [CrossRef] [PubMed]
- Mou, F.; Forest, T.; Baines, J.D. U S 3 of Herpes Simplex Virus Type 1 Encodes a Promiscuous Protein Kinase That Phosphorylates and Alters Localization of Lamin A/C in Infected Cells. J. Virol. 2007, 81, 6459–6470. [Google Scholar] [CrossRef] [Green Version]
- Hertel, L. Herpesviruses and Intermediate Filaments: Close Encounters with the Third Type. Viruses 2011, 3, 1015–1040. [Google Scholar] [CrossRef] [Green Version]
- Ng, T.; Talarico, C.; Burnette, T.C.; Biron, K.; Roizman, B. Partial Substitution of the Functions of the Herpes Simplex Virus 1 UL13 Gene by the Human Cytomegalovirus UL97 Gene. Virology 1996, 225, 347–358. [Google Scholar] [CrossRef] [Green Version]
- Romaker, D.; Schregel, V.; Maurer, K.; Auerochs, S.; Marzi, A.; Sticht, H.; Marschall, M. Analysis of the Structure-Activity Relationship of Four Herpesviral UL97 Subfamily Protein Kinases Reveals Partial but not Full Functional Conservation†. J. Med. Chem. 2006, 49, 7044–7053. [Google Scholar] [CrossRef]
- Wild, M.; Kicuntod, J.; Seyler, L.; Wangen, C.; Bertzbach, L.D.; Conradie, A.M.; Kaufer, B.B.; Wagner, S.; Michel, D.; Eickhoff, J.; et al. Combinatorial Drug Treatments Reveal Promising Anticytomegaloviral Profiles for Clinically Relevant Pharmaceutical Kinase Inhibitors (PKIs). Int. J. Mol. Sci. 2021, 22, 575. [Google Scholar] [CrossRef]
- Shindo, K.; Kato, A.; Koyanagi, N.; Sagara, H.; Arii, J.; Kawaguchi, Y. Characterization of a Herpes Simplex Virus 1 (HSV-1) Chimera in Which the Us3 Protein Kinase Gene Is Replaced with the HSV-2 Us3 Gene. J. Virol. 2016, 90, 457–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schütz, M.; Thomas, M.; Wangen, C.; Wagner, S.; Rauschert, L.; Errerd, T.; Kießling, M.; Sticht, H.; Milbradt, J.; Marschall, M. The peptidyl-prolyl cis/trans isomerase Pin1 interacts with three early regulatory proteins of human cytomegalovirus. Virus Res. 2020, 285, 198023. [Google Scholar] [CrossRef] [PubMed]
- Draganova, E.B.; Valentin, J.; Heldwein, E.E. The Ins and Outs of Herpesviral Capsids: Divergent Structures and Assembly Mechanisms across the Three Subfamilies. Viruses 2021, 13, 1913. [Google Scholar] [CrossRef]
- Hagen, C.; Dent, K.C.; Zeev-Ben-Mordehai, T.; Grange, M.; Bosse, J.B.; Whittle, C.; Klupp, B.G.; Siebert, C.A.; Vasishtan, D.; Bäuerlein, F.J.; et al. Structural Basis of Vesicle Formation at the Inner Nuclear Membrane. Cell 2015, 163, 1692–1701. [Google Scholar] [CrossRef] [Green Version]
- Thorsen, M.K.; Lai, A.; Lee, M.W.; Hoogerheide, D.P.; Wong, G.C.L.; Freed, J.H.; Heldwein, E.E. Highly Basic Clusters in the Herpes Simplex Virus 1 Nuclear Egress Complex Drive Membrane Budding by Inducing Lipid Ordering. mBio 2021, 12, e0154821. [Google Scholar] [CrossRef]
- Luitweiler, E.M.; Henson, B.W.; Pryce, E.N.; Patel, V.; Coombs, G.; McCaffery, J.M.; Desai, P.J. Interactions of the Kaposi’s Sarcoma-Associated Herpesvirus Nuclear Egress Complex: ORF69 Is a Potent Factor for Remodeling Cellular Membranes. J. Virol. 2013, 87, 3915–3929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santarelli, R.; Farina, A.; Granato, M.; Gonnella, R.; Raffa, S.; Leone, L.; Bei, R.; Modesti, A.; Frati, L.; Torrisi, M.R.; et al. Identification and Characterization of the Product Encoded by ORF69 of Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2008, 82, 4562–4572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnee, M.; Ruzsics, Z.; Bubeck, A.; Koszinowski, U.H. Common and Specific Properties of Herpesvirus UL34/UL31 Protein Family Members Revealed by Protein ComplementationAssay. J. Virol. 2006, 80, 11658–11666. [Google Scholar] [CrossRef] [Green Version]
- Farina, A.; Feederle, R.; Raffa, S.; Gonnella, R.; Santarelli, R.; Frati, L.; Angeloni, A.; Torrisi, M.R.; Faggioni, A.; Delecluse, H.-J. BFRF1 of Epstein-Barr Virus Is Essential for Efficient Primary Viral Envelopment and Egress. J. Virol. 2005, 79, 3703–3712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granato, M.; Feederle, R.; Farina, A.; Gonnella, R.; Santarelli, R.; Hub, B.; Faggioni, A.; Delecluse, H.-J. Deletion of Epstein-Barr Virus BFLF2 Leads to Impaired Viral DNA Packaging and Primary Egress as Well as to the Production of Defective Viral Particles. J. Virol. 2008, 82, 4042–4051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klupp, B.G.; Granzow, H.; Mettenleiter, T.C. Primary Envelopment of Pseudorabies Virus at the Nuclear Membrane Requires the UL34 Gene Product. J. Virol. 2000, 74, 10063–10073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Häge, S.; Büscher, N.; Pakulska, V.; Hahn, F.; Adrait, A.; Krauter, S.; Borst, E.M.; Schlötzer-Schrehardt, U.; Couté, Y.; Plachter, B.; et al. The Complex Regulatory Role of Cytomegalovirus Nuclear Egress Protein pUL50 in the Production of Infectious Virus. Cells 2021, 10, 3119. [Google Scholar] [CrossRef] [PubMed]
- Couté, Y.; Kraut, A.; Zimmermann, C.; Büscher, N.; Hesse, A.-M.; Bruley, C.; De Andrea, M.; Wangen, C.; Hahn, F.; Marschall, M.; et al. Mass Spectrometry-Based Characterization of the Virion Proteome, Phosphoproteome, and Associated Kinase Activity of Human Cytomegalovirus. Microorganisms 2020, 8, 820. [Google Scholar] [CrossRef]
- Cross, T.; Griffiths, G.; Deacon, E.; Sallis, R.; Gough, M.; Watters, D.; Lord, J.M. PKC-δ is an apoptotic lamin kinase. Oncogene 2000, 19, 2331–2337. [Google Scholar] [CrossRef] [Green Version]
- Marschall, M.; Feichtinger, S.; Milbradt, J. Regulatory Roles of Protein Kinases in Cytomegalovirus Replication. Adv. Virus Res. 2011, 80, 69–101. [Google Scholar] [CrossRef]
- Park, R.; Baines, J.D. Herpes Simplex Virus Type 1 Infection Induces Activation and Recruitment of Protein Kinase C to the Nuclear Membrane and Increased Phosphorylation of Lamin B. J. Virol. 2006, 80, 494–504. [Google Scholar] [CrossRef] [Green Version]
- Barnes, J.; Wilson, D.W. Seeking Closure: How Do Herpesviruses Recruit the Cellular ESCRT Apparatus? J. Virol. 2019, 93. [Google Scholar] [CrossRef] [Green Version]
- Calistri, A.; Reale, A.; Palù, G.; Parolin, C. Why Cells and Viruses Cannot Survive without an ESCRT. Cells 2021, 10, 483. [Google Scholar] [CrossRef]
- Olmos, Y.; Hodgson, L.; Mantell, J.; Verkade, P.; Carlton, J.G. ESCRT-III controls nuclear envelope reformation. Nature 2015, 522, 236–239. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.; Etingov, I.; Panté, N. Effect of Viral Infection on the Nuclear Envelope and Nuclear Pore Complex. Int. Rev. Cell Mol. Biol. 2012, 299, 117–159. [Google Scholar] [CrossRef]
- Mettenleiter, T.C. Breaching the Barrier—The Nuclear Envelope in Virus Infection. J. Mol. Biol. 2016, 428, 1949–1961. [Google Scholar] [CrossRef]
- Molenberghs, F.; Bogers, J.J.; De Vos, W.H. Confined no more: Viral mechanisms of nuclear entry and egress. Int. J. Biochem. Cell Biol. 2020, 129, 105875. [Google Scholar] [CrossRef]
- Close, W.L.; Anderson, A.N.; Pellett, P.E. Betaherpesvirus Virion Assembly and Egress. In Human Herpesviruses; Advances in Experimental Medicine and Biology Series; Springer: Singapore, 2018; Volume 1045, pp. 167–207. [Google Scholar] [CrossRef]
- Heming, J.D.; Conway, J.F.; Homa, F.L. Herpesvirus Capsid Assembly and DNA Packaging. Cell Biol. Herpes Viruses 2017, 223, 119–142. [Google Scholar] [CrossRef] [Green Version]
- Kornfeind, E.M.; Visalli, R.J. Human herpesvirus portal proteins: Structure, function, and antiviral prospects. Rev. Med. Virol. 2018, 28, e1972. [Google Scholar] [CrossRef] [PubMed]
- Wofford, A.S.; McCusker, I.; Green, J.C.; Vensko, T.A.; Pellett, P.E. Betaherpesvirus assembly and egress: Recent advances illuminate the path. Adv. Virus Res. 2020, 108, 337–392. [Google Scholar] [CrossRef] [PubMed]
- Klupp, B.G.; Granzow, H.; Mettenleiter, T.C. Nuclear Envelope Breakdown Can Substitute for Primary Envelopment-Mediated Nuclear Egress of Herpesviruses. J. Virol. 2011, 85, 8285–8292. [Google Scholar] [CrossRef] [Green Version]
- Schulz, K.S.; Klupp, B.G.; Granzow, H.; Paßvogel, L.; Mettenleiter, T.C. Herpesvirus nuclear egress: Pseudorabies Virus can simultaneously induce nuclear envelope breakdown and exit the nucleus via the envelopment–deenvelopment-pathway. Virus Res. 2015, 209, 76–86. [Google Scholar] [CrossRef]
- Grimm, K.S.; Klupp, B.G.; Granzow, H.; Müller, F.M.; Fuchs, W.; Mettenleiter, T.C. Analysis of Viral and Cellular Factors Influencing Herpesvirus-Induced Nuclear Envelope Breakdown. J. Virol. 2012, 86, 6512–6521. [Google Scholar] [CrossRef] [Green Version]
- Bubeck, A.; Wagner, M.; Ruzsics, Z.; Lötzerich, M.; Iglesias, M.; Singh, I.R.; Koszinowski, U.H. Comprehensive Mutational Analysis of a Herpesvirus Gene in the Viral Genome Context Reveals a Region Essential for Virus Replication. J. Virol. 2004, 78, 8026–8035. [Google Scholar] [CrossRef] [Green Version]
- Lötzerich, M.; Ruzsics, Z.; Koszinowski, U.H. Functional Domains of Murine Cytomegalovirus Nuclear Egress Protein M53/p38. J. Virol. 2006, 80, 73–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alkhashrom, S.; Kicuntod, J.; Häge, S.; Schweininger, J.; Muller, Y.; Lischka, P.; Marschall, M.; Eichler, J. Exploring the Human Cytomegalovirus Core Nuclear Egress Complex as a Novel Antiviral Target: A New Type of Small Molecule Inhibitors. Viruses 2021, 13, 471. [Google Scholar] [CrossRef] [PubMed]
- Kicuntod, J.; Alkhashrom, S.; Häge, S.; Diewald, B.; Müller, R.; Hahn, F.; Lischka, P.; Sticht, H.; Eichler, J.; Marschall, M. Properties of Oligomeric Interaction of the Cytomegalovirus Core Nuclear Egress Complex (NEC) and Its Sensitivity to an NEC Inhibitory Small Molecule. Viruses 2021, 13, 462. [Google Scholar] [CrossRef]
- Häge, S.; Horsch, D.; Stilp, A.-C.; Kicuntod, J.; Müller, R.; Hamilton, S.T.; Egilmezer, E.; Rawlinson, W.D.; Stamminger, T.; Sonntag, E.; et al. A quantitative nuclear egress assay to investigate the nucleocytoplasmic capsid release of human cytomegalovirus. J. Virol. Methods 2020, 283, 113909. [Google Scholar] [CrossRef]
- Steingruber, M.; Marschall, M. The Cytomegalovirus Protein Kinase pUL97: Host Interactions, Regulatory Mechanisms and Antiviral Drug Targeting. Microorganisms 2020, 8, 515. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Coen, D.M. Comparison of Effects of Inhibitors of Viral and Cellular Protein Kinases on Human Cytomegalovirus Disruption of Nuclear Lamina and Nuclear Egress. J. Virol. 2014, 88, 10982–10985. [Google Scholar] [CrossRef] [Green Version]
- Kicuntod, J.; Häge, S.; Hahn, F.; Sticht, H.; Marschall, M. The Oligomeric Assemblies of Cytomegalovirus Core Nuclear Egress Proteins Are Associated with Host Kinases and Show Sensitivity to Antiviral Kinase Inhibitors. Viruses 2022, 14, 1021. [Google Scholar] [CrossRef]


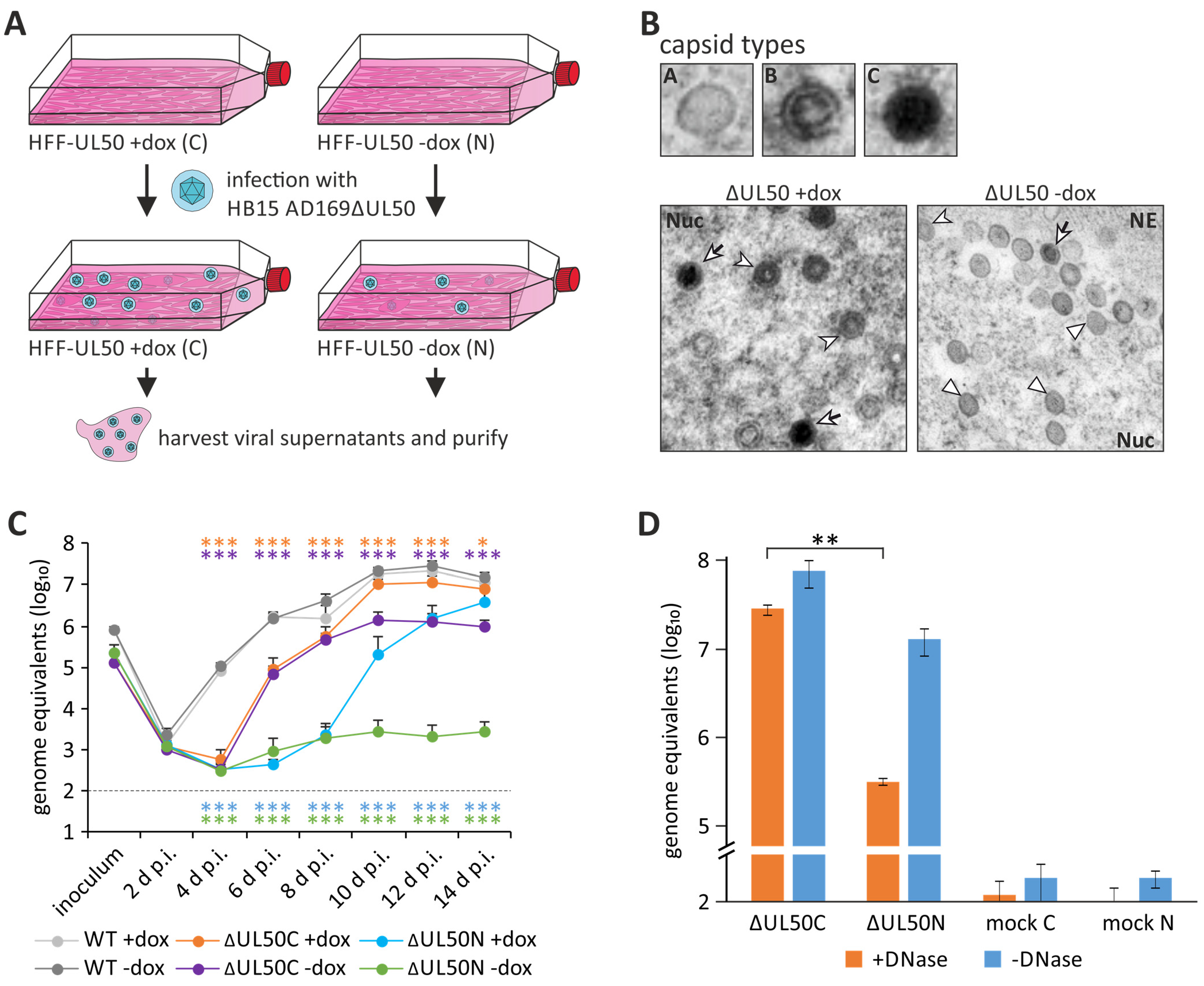
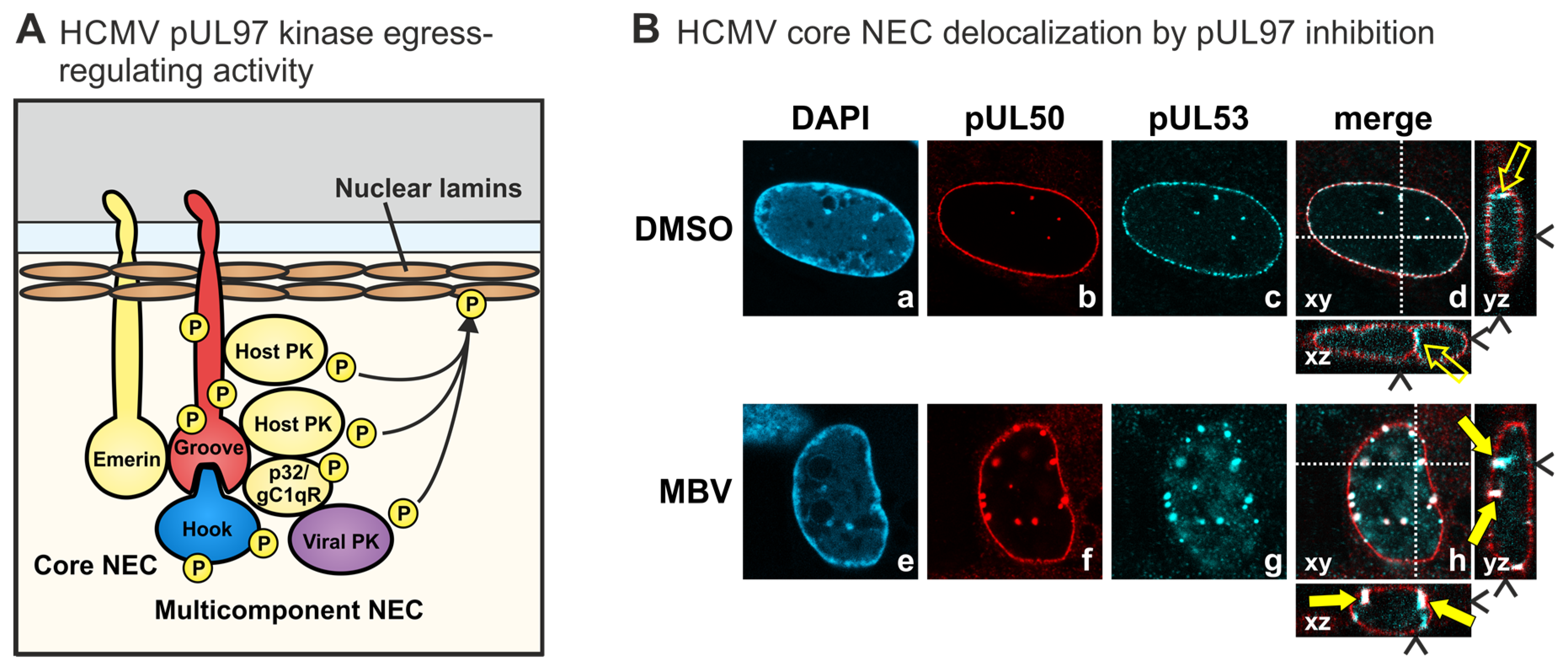
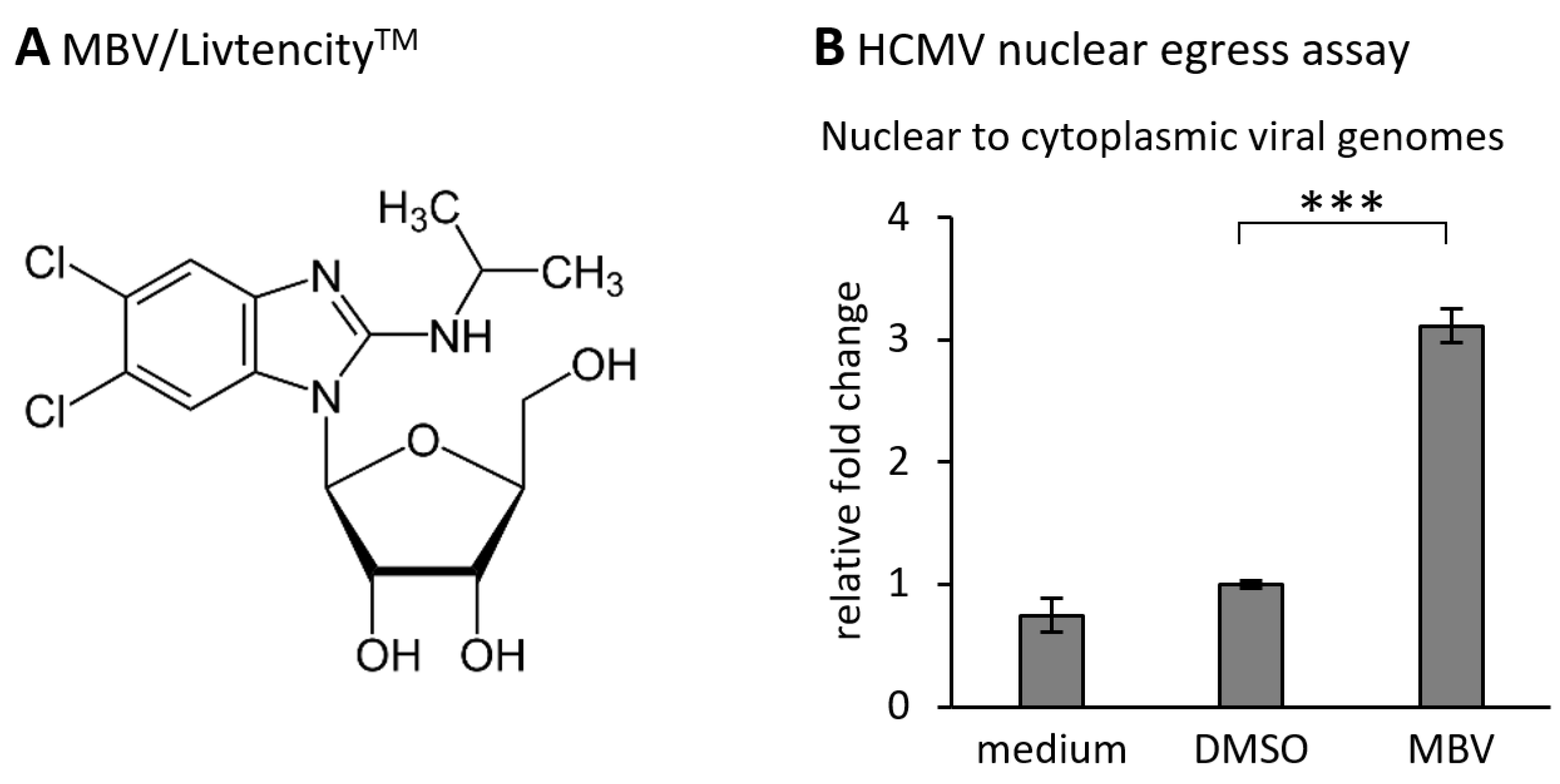
| Amino Acid Sequences | Poorly Conserved |
|---|---|
| 3D structure of core NEC | Almost fully conserved |
| Principle of hook-into-groove interaction | Identical |
| Multicomponent interaction properties | In part conserved |
| Overall nuclear egress functionality | Conserved in final outcome |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Häge, S.; Marschall, M. ‘Come together’—The Regulatory Interaction of Herpesviral Nuclear Egress Proteins Comprises Both Essential and Accessory Functions. Cells 2022, 11, 1837. https://doi.org/10.3390/cells11111837
Häge S, Marschall M. ‘Come together’—The Regulatory Interaction of Herpesviral Nuclear Egress Proteins Comprises Both Essential and Accessory Functions. Cells. 2022; 11(11):1837. https://doi.org/10.3390/cells11111837
Chicago/Turabian StyleHäge, Sigrun, and Manfred Marschall. 2022. "‘Come together’—The Regulatory Interaction of Herpesviral Nuclear Egress Proteins Comprises Both Essential and Accessory Functions" Cells 11, no. 11: 1837. https://doi.org/10.3390/cells11111837
APA StyleHäge, S., & Marschall, M. (2022). ‘Come together’—The Regulatory Interaction of Herpesviral Nuclear Egress Proteins Comprises Both Essential and Accessory Functions. Cells, 11(11), 1837. https://doi.org/10.3390/cells11111837





