Systematic Characterization of the Disruption of Intestine during Liver Tumor Progression in the xmrk Oncogene Transgenic Zebrafish Model
Abstract
:1. Introduction
2. Material and Methods
2.1. Zebrafish Maintenance and Doxycycline Treatment
2.2. Sample Collection and Histological, Immunohistochemistry and TUNEL Assays
2.3. Imaging and Analysis of Histological, Immunohistochemistry and TUNEL Samples
2.4. RNA Extraction, Library Preparation and Sequencing
2.5. Bioinformatics Analysis of Sequencing Data
3. Results
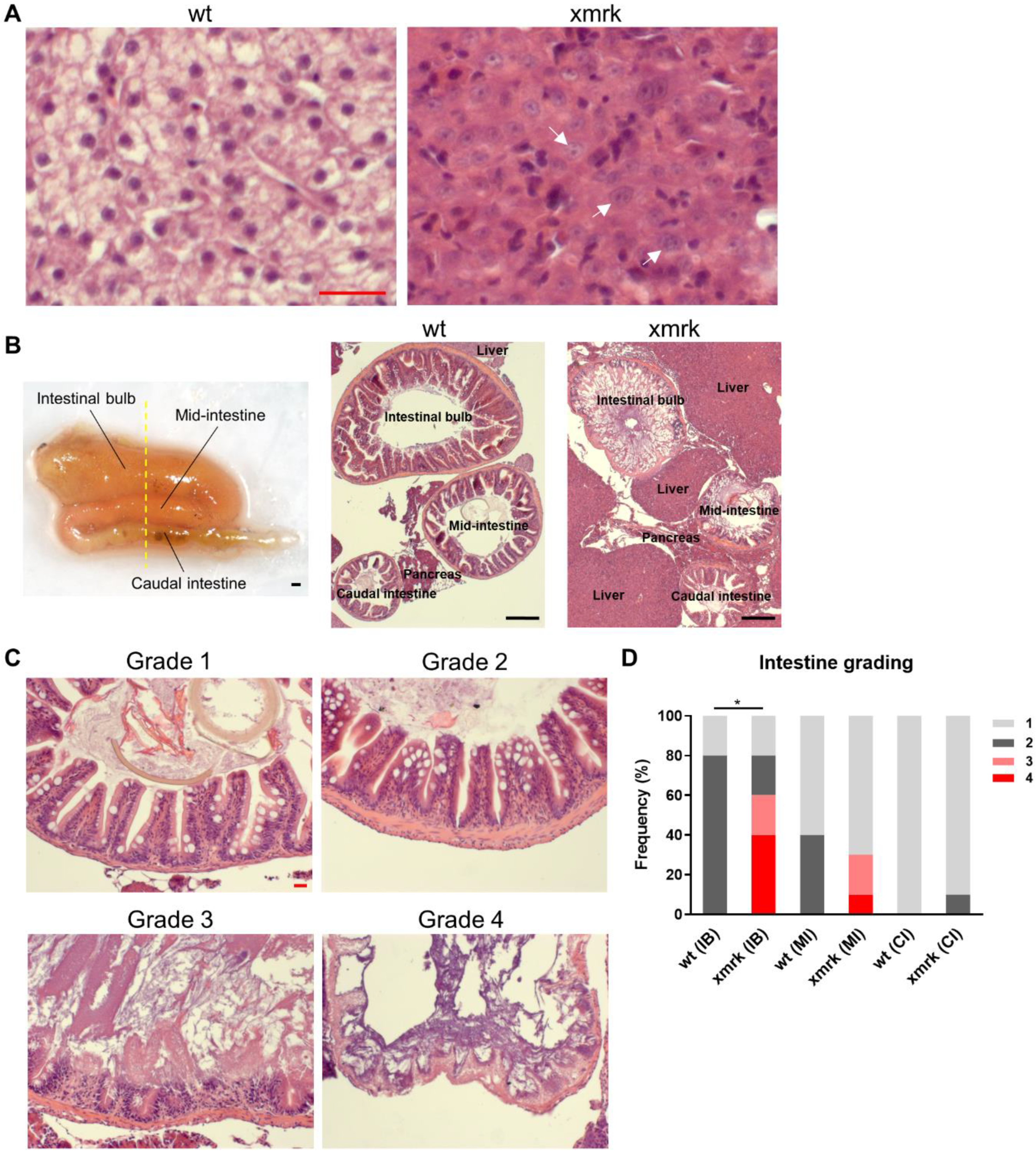
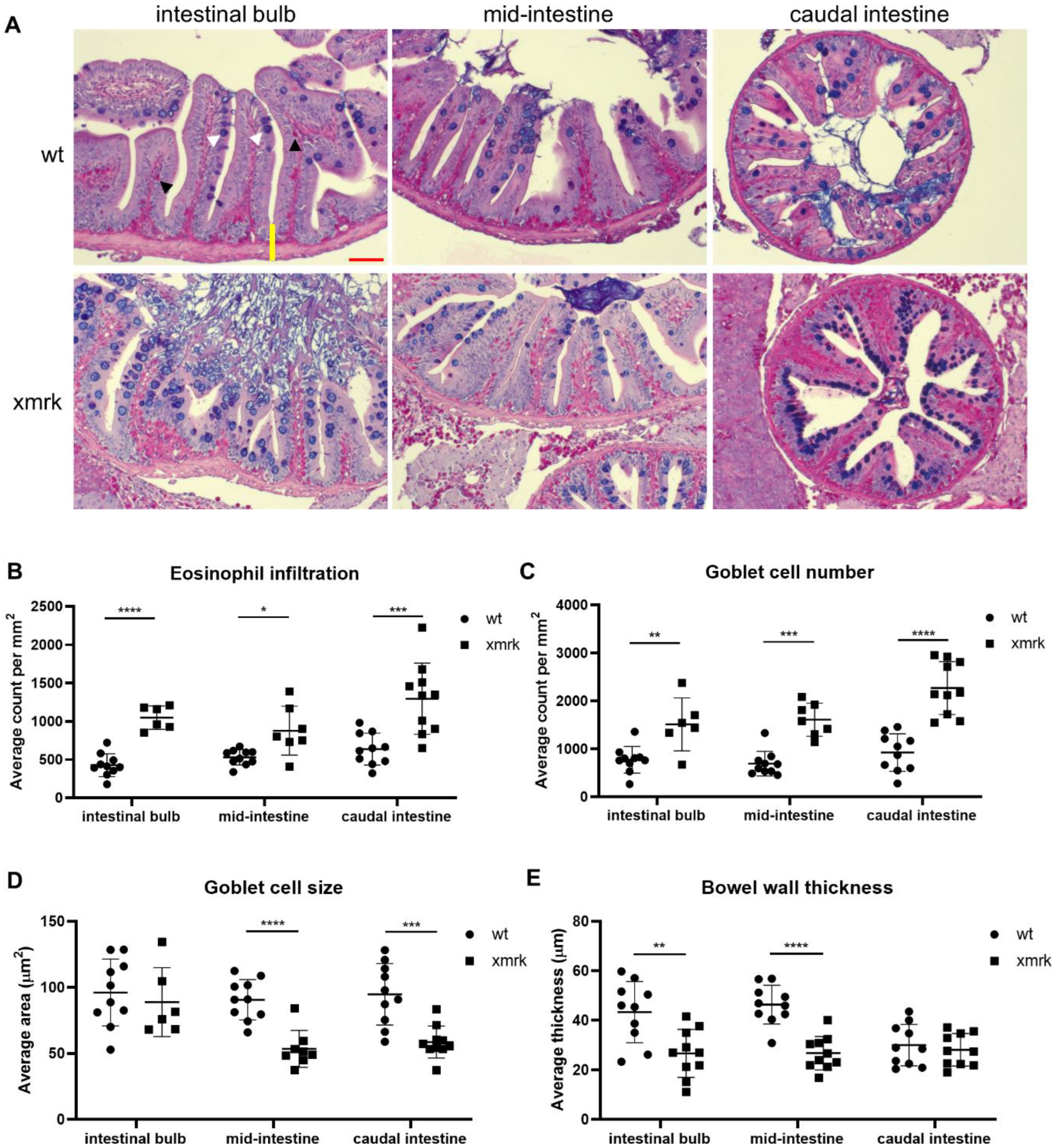
3.1. Morphological and Cellular Disruption of Intestine in HCC Fish
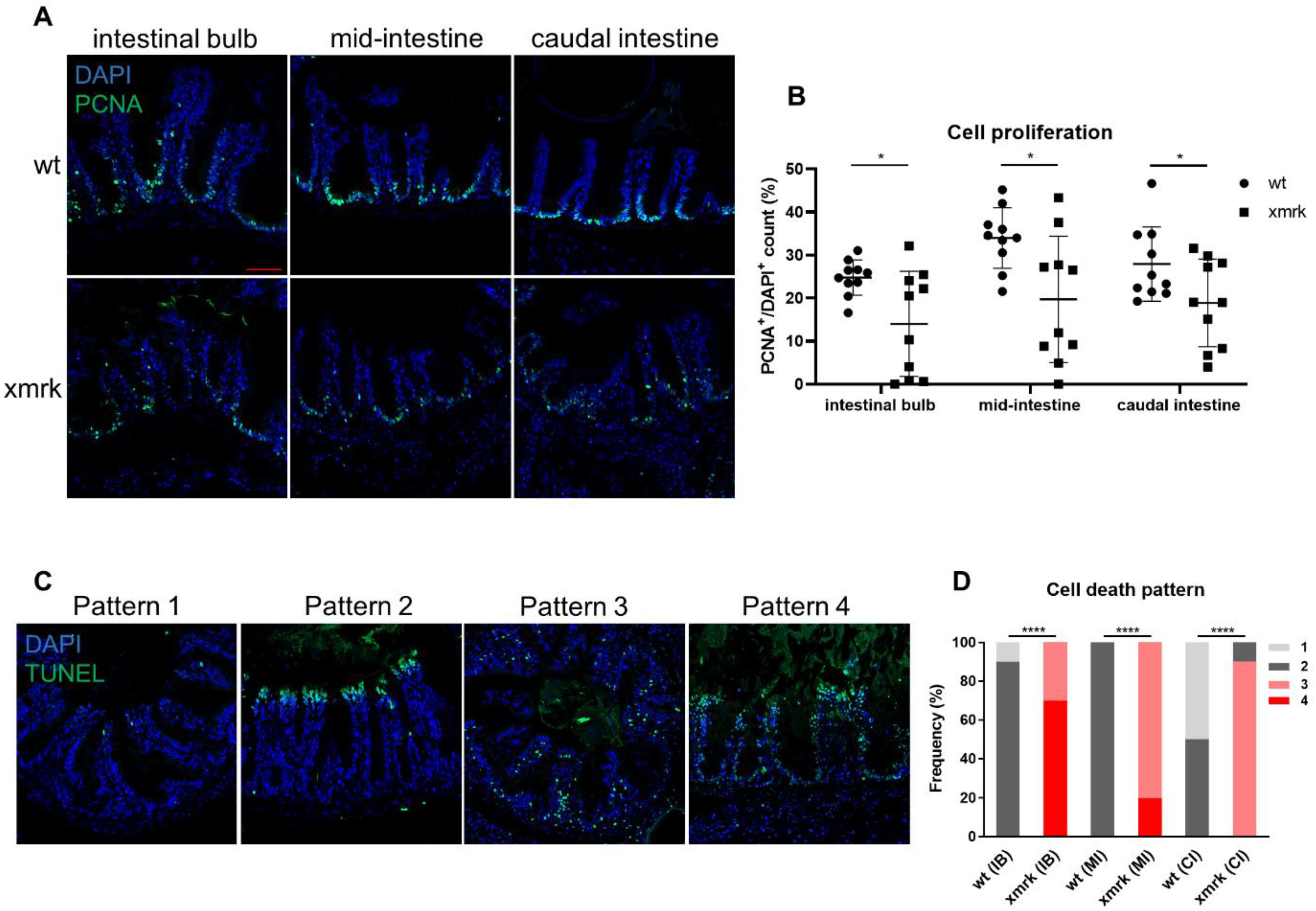
3.2. Disruption of Intestinal Epithelial Cell Renewal in HCC Fish
3.3. Progressive Disruption of Intestine during Liver Tumor Progression
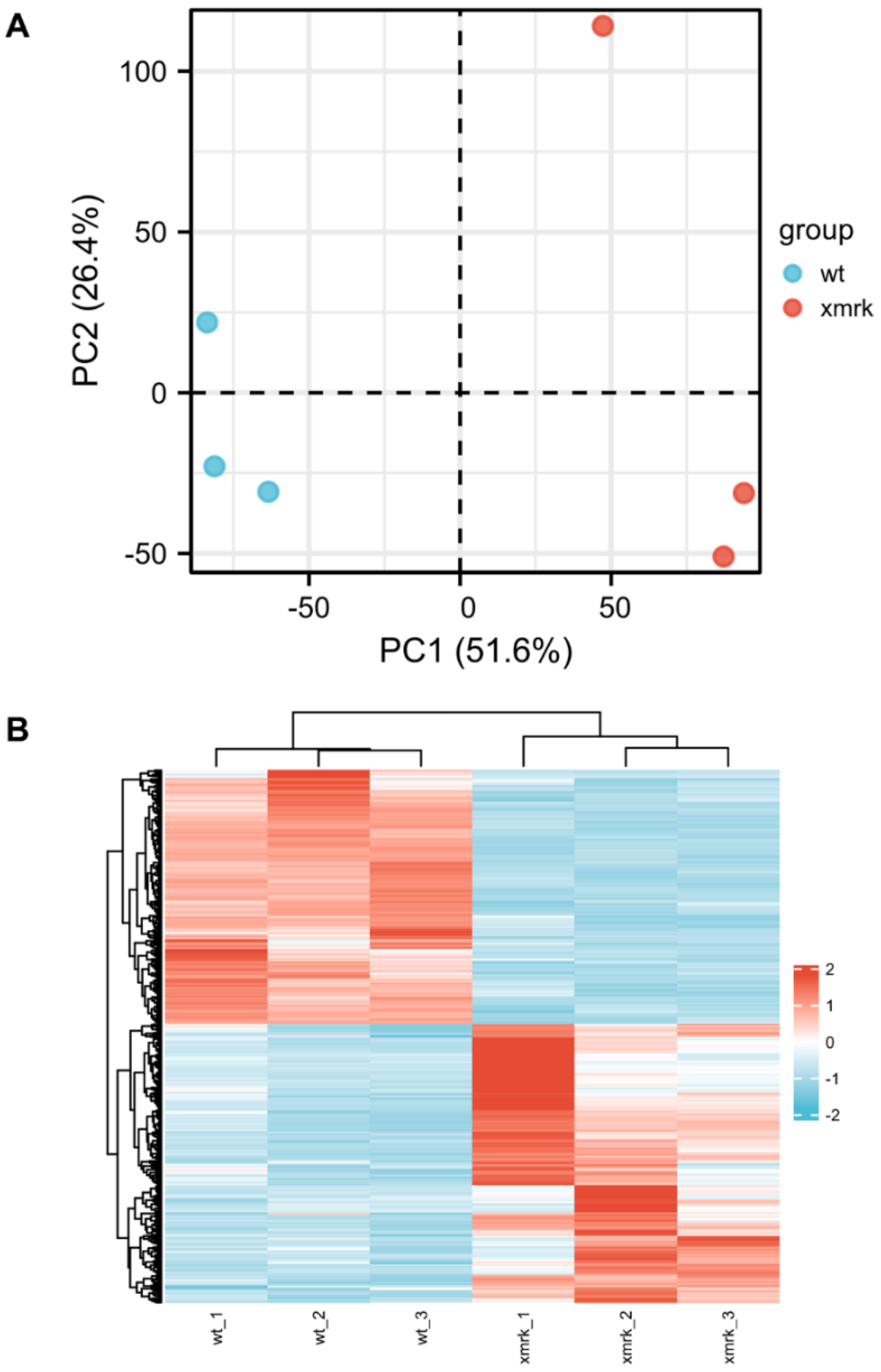
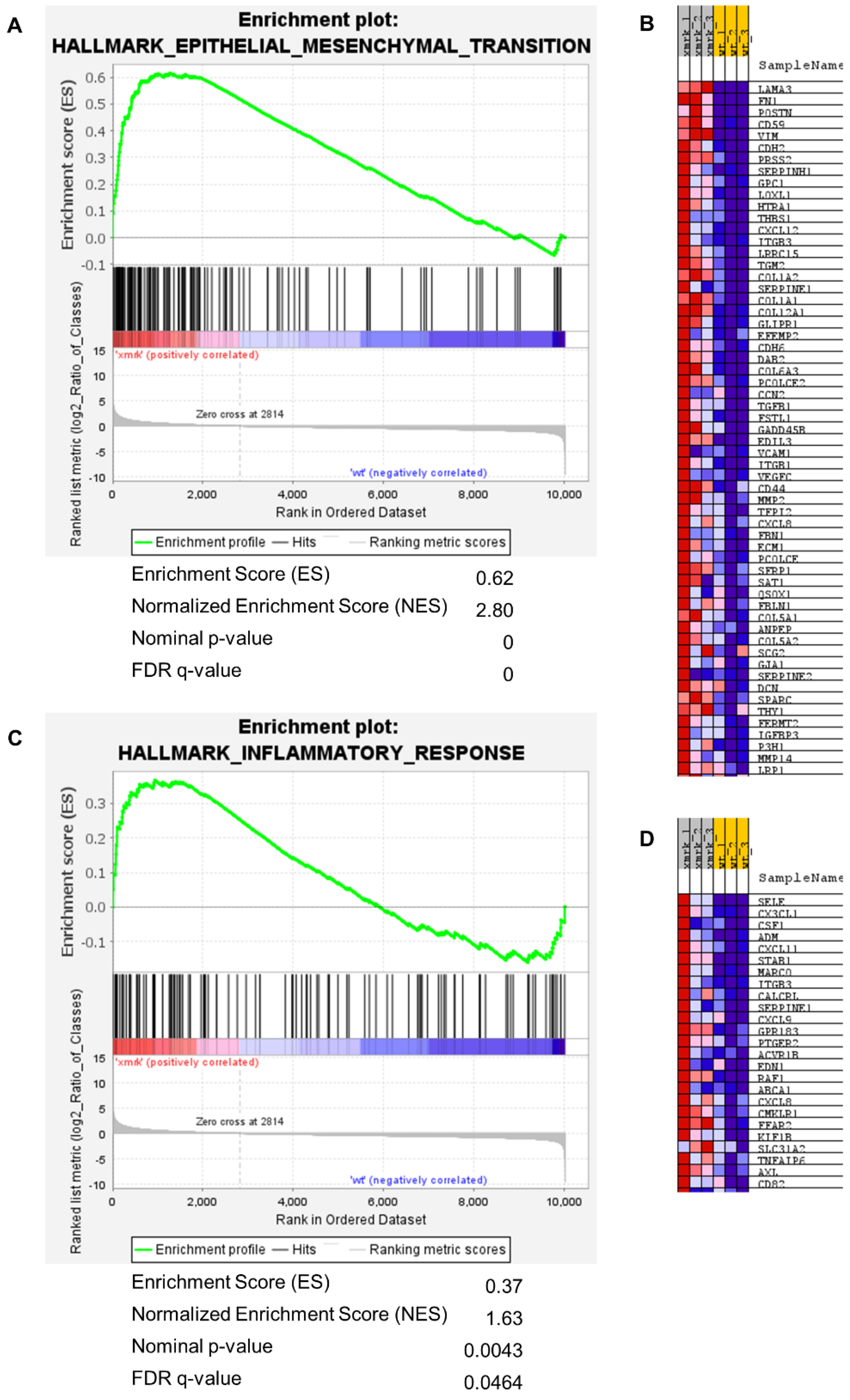
3.4. Transcriptomic Change of Intestine in HCC Fish
3.5. Comparison of xmrk Intestine with Human Intestinal Disease Conditions
3.6. Deregulation of Neutrophil-Related Genes and Intestinal-Function-Related Genes during HCC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr. Biol. 2020, 30, R921–R925. [Google Scholar] [CrossRef] [PubMed]
- McAllister, S.S.; Weinberg, R.A. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat. Cell Biol. 2014, 16, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, M.R.; Svoronos, N.; Perales-Puchalt, A.; Conejo-Garcia, J.R. The Tumor Macroenvironment: Cancer-Promoting Networks Beyond Tumor Beds. Adv. Cancer Res. 2015, 128, 235–262. [Google Scholar] [CrossRef] [Green Version]
- Martin, O.A.; Redon, C.E.; Nakamura, A.J.; Dickey, J.S.; Georgakilas, A.G.; Bonner, W.M. Systemic DNA damage related to cancer. Cancer Res. 2011, 71, 3437–3441. [Google Scholar] [CrossRef] [Green Version]
- Redon, C.E.; Dickey, J.S.; Nakamura, A.J.; Kareva, I.G.; Naf, D.; Nowsheen, S.; Kryston, T.B.; Bonner, W.M.; Georgakilas, A.G.; Sedelnikova, O.A. Tumors induce complex DNA damage in distant proliferative tissues in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 17992–17997. [Google Scholar] [CrossRef] [Green Version]
- Argiles, J.M.; Stemmler, B.; Lopez-Soriano, F.J.; Busquets, S. Inter-tissue communication in cancer cachexia. Nat. Rev. Endocrinol. 2018, 15, 9–20. [Google Scholar] [CrossRef]
- Tripathi, A.; Debelius, J.; Brenner, D.A.; Karin, M.; Loomba, R.; Schnabl, B.; Knight, R. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 397–411. [Google Scholar] [CrossRef]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef] [Green Version]
- Schwabe, R.F.; Greten, T.F. Gut microbiome in HCC—Mechanisms, diagnosis and therapy. J. Hepatol. 2020, 72, 230–238. [Google Scholar] [CrossRef] [Green Version]
- Ponziani, F.R.; Bhoori, S.; Castelli, C.; Putignani, L.; Rivoltini, L.; Del Chierico, F.; Sanguinetti, M.; Morelli, D.; Paroni Sterbini, F.; Petito, V.; et al. Hepatocellular Carcinoma Is Associated With Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 2019, 69, 107–120. [Google Scholar] [CrossRef] [PubMed]
- White, R.; Rose, K.; Zon, L. Zebrafish cancer: The state of the art and the path forward. Nat. Rev. Cancer 2013, 13, 624–636. [Google Scholar] [CrossRef] [PubMed]
- McConnell, A.M.; Noonan, H.R.; Zon, L.I. Reeling in the Zebrafish Cancer Models. Annu. Rev. Cancer Biol. 2021, 5, 331–350. [Google Scholar] [CrossRef]
- Wrighton, P.J.; Oderberg, I.M.; Goessling, W. There Is Something Fishy About Liver Cancer: Zebrafish Models of Hepatocellular Carcinoma. Cell Mol. Gastroenterol. Hepatol. 2019, 8, 347–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Huang, X.; Zhan, H.; Zeng, Z.; Li, C.; Spitsbergen, J.M.; Meierjohann, S.; Schartl, M.; Gong, Z. Inducible and repressable oncogene-addicted hepatocellular carcinoma in Tet-on xmrk transgenic zebrafish. J. Hepatol. 2012, 56, 419–425. [Google Scholar] [CrossRef]
- Chew, T.W.; Liu, X.J.; Liu, L.; Spitsbergen, J.M.; Gong, Z.; Low, B.C. Crosstalk of Ras and Rho: Activation of RhoA abates Kras-induced liver tumorigenesis in transgenic zebrafish models. Oncogene 2014, 33, 2717–2727. [Google Scholar] [CrossRef] [Green Version]
- Zheng, W.; Li, Z.; Nguyen, A.T.; Li, C.; Emelyanov, A.; Gong, Z. Xmrk, kras and myc transgenic zebrafish liver cancer models share molecular signatures with subsets of human hepatocellular carcinoma. PLoS ONE 2014, 9, e91179. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, A.T.; Emelyanov, A.; Koh, C.H.; Spitsbergen, J.M.; Parinov, S.; Gong, Z. An inducible kras(V12) transgenic zebrafish model for liver tumorigenesis and chemical drug screening. Dis. Models Mech. 2012, 5, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Nguyen, A.T.; Spitsbergen, J.M.; Gong, Z. Myc-induced liver tumors in transgenic zebrafish can regress in tp53 null mutation. PLoS ONE 2015, 10, e0117249. [Google Scholar] [CrossRef]
- Lee, A.Q.; Li, Y.; Gong, Z. Inducible Liver Cancer Models in Transgenic Zebrafish to Investigate Cancer Biology. Cancers 2021, 13, 5148. [Google Scholar] [CrossRef]
- Li, Y.; Li, H.; Spitsbergen, J.M.; Gong, Z. Males develop faster and more severe hepatocellular carcinoma than females in kras(V12) transgenic zebrafish. Sci. Rep. 2017, 7, 41280. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Agrawal, I.; Gong, Z. Reversion of tumor hepatocytes to normal hepatocytes during liver tumor regression in an oncogene-expressing transgenic zebrafish model. Dis. Models Mech. 2019, 12, dmm039578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Yan, C.; Wang, X.; Gong, Z. Leptin induces muscle wasting in a zebrafish kras-driven hepatocellular carcinoma (HCC) model. Dis. Models Mech. 2019, 12, dmm038240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.W.; Sun, Y.; Lin, L.I.; Liu, D.; Gong, Z. Exacerbation of Liver Tumor Metastasis in twist1a+/xmrk+ Double Transgenic Zebrafish following Lipopolysaccharide or Dextran Sulphate Sodium Exposure. Pharmaceuticals 2021, 14, 867. [Google Scholar] [CrossRef]
- Wallace, K.N.; Akhter, S.; Smith, E.M.; Lorent, K.; Pack, M. Intestinal growth and differentiation in zebrafish. Mech. Dev. 2005, 122, 157–173. [Google Scholar] [CrossRef]
- Lickwar, C.R.; Camp, J.G.; Weiser, M.; Cocchiaro, J.L.; Kingsley, D.M.; Furey, T.S.; Sheikh, S.Z.; Rawls, J.F. Genomic dissection of conserved transcriptional regulation in intestinal epithelial cells. PLoS Biol. 2017, 15, e2002054. [Google Scholar] [CrossRef]
- Wang, Z.; Du, J.; Lam, S.H.; Mathavan, S.; Matsudaira, P.; Gong, Z. Morphological and molecular evidence for functional organization along the rostrocaudal axis of the adult zebrafish intestine. BMC Genom. 2010, 11, 392. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.W.; Ho, Y.J.; Ciou, S.C.; Gong, Z. Innovative Disease Model: Zebrafish as an in vivo Platform for Intestinal Disorder and Tumors. Biomedicines 2017, 5, 58. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Cock, P.J.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Brugman, S.; Liu, K.Y.; Lindenbergh-Kortleve, D.; Samsom, J.N.; Furuta, G.T.; Renshaw, S.A.; Willemsen, R.; Nieuwenhuis, E.E. Oxazolone-induced enterocolitis in zebrafish depends on the composition of the intestinal microbiota. Gastroenterology 2009, 137, 1757–1767.e1. [Google Scholar] [CrossRef]
- Li, C.; Barton, C.; Henke, K.; Daane, J.; Treaster, S.; Caetano-Lopes, J.; Tanguay, R.L.; Harris, M.P. celsr1a is essential for tissue homeostasis and onset of aging phenotypes in the zebrafish. eLife 2020, 9, e50523. [Google Scholar] [CrossRef]
- Witte, M.; Huitema, L.F.; Nieuwenhuis, E.E.; Brugman, S. Deficiency in macrophage-stimulating protein results in spontaneous intestinal inflammation and increased susceptibility toward epithelial damage in zebrafish. Zebrafish 2014, 11, 542–550. [Google Scholar] [CrossRef]
- Gurtner, A.; Gonzalez-Perez, I.; Arnold, I.C. Intestinal eosinophils, homeostasis and response to bacterial intrusion. In Seminars in Immunopathology; Springer: Berlin/Heidelberg, Germany, 2021; pp. 295–306. [Google Scholar]
- Yang, S.; Yu, M. Role of goblet cells in intestinal barrier and mucosal immunity. J. Inflamm. Res. 2021, 14, 3171. [Google Scholar] [CrossRef]
- Grondin, J.A.; Kwon, Y.H.; Far, P.M.; Haq, S.; Khan, W.I. Mucins in intestinal mucosal defense and inflammation: Learning from clinical and experimental studies. Front. Immunol. 2020, 11, 2054. [Google Scholar] [CrossRef]
- Jain, U.; Otley, A.R.; Van Limbergen, J.; Stadnyk, A.W. The complement system in inflammatory bowel disease. Inflamm. Bowel Dis. 2014, 20, 1628–1637. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Huang, B.; Jin, T.; Ocansey, D.K.W.; Jiang, J.; Mao, F. Intestinal Fibrosis in Inflammatory Bowel Disease and the Prospects of Mesenchymal Stem Cell Therapy. Front. Immunol. 2022, 13, 835005. [Google Scholar] [CrossRef] [PubMed]
- Speca, S.; Giusti, I.; Rieder, F.; Latella, G. Cellular and molecular mechanisms of intestinal fibrosis. World J. Gastroenterol. 2012, 18, 3635–3661. [Google Scholar] [CrossRef]
- Stadnicki, A. Involvement of coagulation and hemostasis in inflammatory bowel diseases. Curr. Vasc. Pharmacol. 2012, 10, 659–669. [Google Scholar] [CrossRef]
- Rieder, F.; Fiocchi, C. Intestinal fibrosis in IBD—A dynamic, multifactorial process. Nat. Rev. Gastroenterol. Hepatol. 2009, 6, 228–235. [Google Scholar] [CrossRef]
- Barbara, G.; Barbaro, M.R.; Fuschi, D.; Palombo, M.; Falangone, F.; Cremon, C.; Marasco, G.; Stanghellini, V. Inflammatory and microbiota-related regulation of the intestinal epithelial barrier. Front. Nutr. 2021, 8, 718356. [Google Scholar] [CrossRef]
- Odenwald, M.A.; Turner, J.R. The intestinal epithelial barrier: A therapeutic target? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 9–21. [Google Scholar] [CrossRef]
- Berritto, D.; Iacobellis, F.; Mazzei, M.A.; Volterrani, L.; Guglielmi, G.; Brunese, L.; Grassi, R. MDCT in ischaemic colitis: How to define the aetiology and acute, subacute and chronic phase of damage in the emergency setting. Br. J. Radiol. 2016, 89, 20150821. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, T.; Oliveira, M.I.; Castro, R.; Araújo, B.; Viamonte, B.; Cunha, R. Bowel wall thickening at CT: Simplifying the diagnosis. Insights Imaging 2014, 5, 195–208. [Google Scholar] [CrossRef] [Green Version]
- Erben, U.; Loddenkemper, C.; Doerfel, K.; Spieckermann, S.; Haller, D.; Heimesaat, M.M.; Zeitz, M.; Siegmund, B.; Kuhl, A.A. A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int. J. Clin. Exp. Pathol. 2014, 7, 4557–4576. [Google Scholar] [PubMed]
- Feakins, R.M. Inflammatory bowel disease biopsies: Updated British Society of Gastroenterology reporting guidelines. J. Clin. Pathol. 2013, 66, 1005–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geiger, B.M.; Gras-Miralles, B.; Ziogas, D.C.; Karagiannis, A.K.; Zhen, A.; Fraenkel, P.; Kokkotou, E. Intestinal upregulation of melanin-concentrating hormone in TNBS-induced enterocolitis in adult zebrafish. PLoS ONE 2013, 8, e83194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldsmith, J.R.; Cocchiaro, J.L.; Rawls, J.F.; Jobin, C. Glafenine-induced intestinal injury in zebrafish is ameliorated by mu-opioid signaling via enhancement of Atf6-dependent cellular stress responses. Dis. Models Mech. 2013, 6, 146–159. [Google Scholar] [CrossRef] [Green Version]
- Patankar, J.V.; Becker, C. Cell death in the gut epithelium and implications for chronic inflammation. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 543–556. [Google Scholar] [CrossRef]
- Birchenough, G.M.; Johansson, M.E.; Gustafsson, J.K.; Bergstrom, J.H.; Hansson, G.C. New developments in goblet cell mucus secretion and function. Mucosal Immunol. 2015, 8, 712–719. [Google Scholar] [CrossRef] [Green Version]
- Vancamelbeke, M.; Vermeire, S. The intestinal barrier: A fundamental role in health and disease. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 821–834. [Google Scholar] [CrossRef]
- Kim, J.J.; Khan, W.I. Goblet cells and mucins: Role in innate defense in enteric infections. Pathogens 2013, 2, 55–70. [Google Scholar] [CrossRef] [Green Version]
- Oehlers, S.H.; Flores, M.V.; Hall, C.J.; Crosier, K.E.; Crosier, P.S. Retinoic acid suppresses intestinal mucus production and exacerbates experimental enterocolitis. Dis. Models Mech. 2012, 5, 457–467. [Google Scholar] [CrossRef] [Green Version]
- Oehlers, S.H.; Flores, M.V.; Okuda, K.S.; Hall, C.J.; Crosier, K.E.; Crosier, P.S. A chemical enterocolitis model in zebrafish larvae that is dependent on microbiota and responsive to pharmacological agents. Dev. Dyn. 2011, 240, 288–298. [Google Scholar] [CrossRef]
- Ara, T.; Hashimoto, D.; Hayase, E.; Noizat, C.; Kikuchi, R.; Hasegawa, Y.; Matsuda, K.; Ono, S.; Matsuno, Y.; Ebata, K.; et al. Intestinal goblet cells protect against GVHD after allogeneic stem cell transplantation via Lypd8. Sci. Transl. Med. 2020, 12, eaaw0720. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Lu, Y.F.; Li, I.C.; Hwang, S.P. Zebrafish Agr2 is required for terminal differentiation of intestinal goblet cells. PLoS ONE 2012, 7, e34408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegyi, P.; Maleth, J.; Walters, J.R.; Hofmann, A.F.; Keely, S.J. Guts and Gall: Bile Acids in Regulation of Intestinal Epithelial Function in Health and Disease. Physiol. Rev. 2018, 98, 1983–2023. [Google Scholar] [CrossRef] [Green Version]
- National Institute of Diabetes and Digestive and Kidney Diseases. Bile Acids. Available online: https://www.ncbi.nlm.nih.gov/books/NBK548626/ (accessed on 27 April 2022).
- Sauerbruch, T.; Hennenberg, M.; Trebicka, J.; Beuers, U. Bile Acids, Liver Cirrhosis, and Extrahepatic Vascular Dysfunction. Front. Physiol. 2021, 12, 718783. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fu, X.; Van Ness, C.; Meng, Z.; Ma, X.; Huang, W. Bile Acid Receptors and Liver Cancer. Curr. Pathobiol. Rep. 2013, 1, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Feng, J.; Li, J.; Yu, Q.; Ji, J.; Wu, J.; Dai, W.; Guo, C. The gut microbiome-bile acid axis in hepatocarcinogenesis. Biomed. Pharmacother. 2021, 133, 111036. [Google Scholar] [CrossRef]
- Shi, N.; Li, N.; Duan, X.; Niu, H. Interaction between the gut microbiome and mucosal immune system. Mil. Med. Res. 2017, 4, 14. [Google Scholar] [CrossRef]
- Fournier, B.; Parkos, C. The role of neutrophils during intestinal inflammation. Mucosal Immunol. 2012, 5, 354–366. [Google Scholar] [CrossRef]
- Jacobs, I.; Ceulemans, M.; Wauters, L.; Breynaert, C.; Vermeire, S.; Verstockt, B.; Vanuytsel, T. Role of Eosinophils in Intestinal Inflammation and Fibrosis in Inflammatory Bowel Disease: An Overlooked Villain? Front. Immunol. 2021, 12, 754413. [Google Scholar] [CrossRef]
- Loktionov, A. Eosinophils in the gastrointestinal tract and their role in the pathogenesis of major colorectal disorders. World J. Gastroenterol. 2019, 25, 3503–3526. [Google Scholar] [CrossRef]
- Wéra, O.; Lancellotti, P.; Oury, C. The Dual Role of Neutrophils in Inflammatory Bowel Diseases. J. Clin. Med. 2016, 5, 118. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Rothenberg, M.E. Roles and Regulation of Gastrointestinal Eosinophils in Immunity and Disease. J. Immunol. 2014, 193, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, S.; Hug, S.; Stratmann, A.E.P.; Erber, M.; Vidoni, L.; Knapp, C.L.; Thomaß, B.D.; Fauler, M.; Nilsson, B.; Nilsson Ekdahl, K.; et al. Interleukin 8 Elicits Rapid Physiological Changes in Neutrophils That Are Altered by Inflammatory Conditions. J. Innate Immun. 2021, 13, 225–241. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, S.; Reyes-Aldasoro, C.C.; Candel, S.; Renshaw, S.A.; Mulero, V.; Calado, Â. Cxcl8 (IL-8) mediates neutrophil recruitment and behavior in the zebrafish inflammatory response. J. Immunol. 2013, 190, 4349–4359. [Google Scholar] [CrossRef]
- Erger, R.A.; Casale, T.B. Interleukin-8 is a potent mediator of eosinophil chemotaxis through endothelium and epithelium. Am. J. Physiol. 1995, 268, L117–L122. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, N.; Koizumi, H.; Murata, M.; Mano, K.; Ohta, K. Nuclear factor kappa B mediates interleukin-8 production in eosinophils. Int. Arch. Allergy Immunol. 1999, 120, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Shimshoni, E.; Yablecovitch, D.; Baram, L.; Dotan, I.; Sagi, I. ECM remodelling in IBD: Innocent bystander or partner in crime? The emerging role of extracellular molecular events in sustaining intestinal inflammation. Gut 2015, 64, 367–372. [Google Scholar] [CrossRef] [Green Version]
- Dohan, A.; Faraoun, S.A.; Barral, M.; Guerrache, Y.; Boudiaf, M.; Dray, X.; Hoeffel, C.; Allez, M.; Farges, O.; Beaugerie, L.; et al. Extra-intestinal malignancies in inflammatory bowel diseases: An update with emphasis on MDCT and MR imaging features. Diagn. Interv. Imaging 2015, 96, 871–883. [Google Scholar] [CrossRef] [Green Version]
- Erichsen, R.; Olen, O.; Sachs, M.C.; Pedersen, L.; Halfvarson, J.; Askling, J.; Ekbom, A.; Ludvigsson, J.F.; Sorensen, H.T. Hepatobiliary Cancer Risk in Patients with Inflammatory Bowel Disease: A Scandinavian Population-Based Cohort Study. Cancer Epidemiol. Biomark. Prev. 2021, 30, 886–894. [Google Scholar] [CrossRef]
- Nguyen, T.B.; Do, D.N.; Nguyen, T.T.P.; Nguyen, T.L.; Nguyen-Thanh, T.; Nguyen, H.T. Immune-related biomarkers shared by inflammatory bowel disease and liver cancer. PLoS ONE 2022, 17, e0267358. [Google Scholar] [CrossRef]
- Jiang, J.W.; Chen, X.H.; Ren, Z.; Zheng, S.S. Gut microbial dysbiosis associates hepatocellular carcinoma via the gut-liver axis. Hepatobiliary Pancreat. Dis. Int. 2019, 18, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.X.; Schwabe, R.F. The gut microbiome and liver cancer: Mechanisms and clinical translation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 527–539. [Google Scholar] [CrossRef] [PubMed]


| Category | Pathway Name | NES | FDR q-Value |
|---|---|---|---|
| Extracellular Matrix Organization | EXTRACELLULAR_MATRIX_ORGANIZATION | 2.78 | 0 |
| ECM_PROTEOGLYCANS | 2.62 | 0 | |
| ELASTIC_FIBRE_FORMATION | 2.54 | 1.94 × 10–4 | |
| MOLECULES_ASSOCIATED_WITH_ELASTIC_FIBRES | 2.39 | 4.60 × 10–4 | |
| COLLAGEN_FORMATION | 1.92 | 3.33 × 10–2 | |
| ASSEMBLY_OF_COLLAGEN_FIBRILS_AND_OTHER_MULTIMERIC_STRUCTURES | 2.02 | 2.12 × 10–2 | |
| NON_INTEGRIN_MEMBRANE_ECM_INTERACTIONS | 2.46 | 2.33 × 10–4 | |
| SYNDECAN_INTERACTIONS | 2.01 | 2.21 × 10–2 | |
| ACTIVATION_OF_MATRIX_METALLOPROTEINASES | 2.43 | 2.04 × 10–4 | |
| DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | 2.37 | 5.83 × 10–4 | |
| LAMININ_INTERACTIONS | 2.29 | 1.59 × 10–3 | |
| INTEGRIN_CELL_SURFACE_INTERACTIONS | 2.28 | 1.78 × 10–3 | |
| Hemostasis | PLATELET_AGGREGATION_PLUG_FORMATION | 2.53 | 1.55 × 10–4 |
| RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2 | 1.98 | 2.53 × 10–2 | |
| CELL_SURFACE_INTERACTIONS_AT_THE_VASCULAR_WALL | 1.96 | 2.91 × 10–2 | |
| FORMATION_OF_FIBRIN_CLOT_CLOTTING_CASCADE | 1.92 | 3.41 × 10–2 | |
| Cell Cycle | CELL_CYCLE_CHECKPOINTS | –1.98 | 1.11 × 10–3 |
| STABILIZATION_OF_P53 | –2.42 | 0 | |
| G1_S_DNA_DAMAGE_CHECKPOINTS | –2.38 | 0 | |
| G2_M_CHECKPOINTS | –2.30 | 4.83 × 10–5 | |
| CELL_CYCLE_MITOTIC | –1.90 | 3.43 × 10–3 | |
| MITOTIC_G1_PHASE_AND_G1_S_TRANSITION | –2.31 | 5.31 × 10–5 | |
| S_PHASE | –2.28 | 4.25 × 10–5 | |
| MITOTIC_G2_G2_M_PHASES | –1.83 | 6.75 × 10–3 | |
| M_PHASE | –1.73 | 1.83 × 10–2 | |
| MITOTIC_METAPHASE_AND_ANAPHASE | –1.76 | 1.42 × 10–2 | |
| CHROMOSOME_MAINTENANCE | –1.96 | 1.32 × 10–3 | |
| TELOMERE_MAINTENANCE | –1.91 | 2.70 × 10–3 | |
| DNA Replication | DNA_REPLICATION | –2.33 | 5.90 × 10–5 |
| DNA_REPLICATION_PRE_INITIATION | –2.39 | 0 | |
| SYNTHESIS_OF_DNA | –2.37 | 0 | |
| DNA_STRAND_ELONGATION | –2.20 | 3.03 × 10–5 | |
| SWITCHING_OF_ORIGINS_TO_A_POST_REPLICATIVE_STATE | –2.11 | 1.94 × 10–4 | |
| DNA Repair | DNA_REPAIR | –1.69 | 2.78 × 10–2 |
| TRANSCRIPTION_COUPLED_NUCLEOTIDE_EXCISION_REPAIR_TC_NER | –1.67 | 3.13 × 10–2 | |
| BASE_EXCISION_REPAIR | –1.80 | 9.68 × 10–3 | |
| DNA_DOUBLE_STRAND_BREAK_REPAIR | –1.77 | 1.30 × 10–2 | |
| HOMOLOGY_DIRECTED_REPAIR | –1.74 | 1.63 × 10–2 | |
| DNA_DAMAGE_BYPASS | –1.80 | 9.04 × 10–3 | |
| Transcription | RNA_POLYMERASE_II_TRANSCRIPTION_TERMINATION | –1.74 | 1.64 × 10–2 |
| REGULATION_OF_RUNX2_EXPRESSION_AND_ACTIVITY | –2.18 | 2.79 × 10–5 | |
| REGULATION_OF_RUNX3_EXPRESSION_AND_ACTIVITY | –2.30 | 5.06 × 10–5 | |
| RUNX1_REGULATES_TRANSCRIPTION_OF_GENES_INVOLVED_IN_DIFFERENTIATION_OF_HSCS | –2.04 | 5.20 × 10–4 | |
| TP53_REGULATES_TRANSCRIPTION_OF_CELL_DEATH_GENES | –1.76 | 1.48 × 10–2 | |
| GENE_SILENCING_BY_RNA | –1.66 | 3.18 × 10–2 | |
| Metabolism of RNA | REGULATION_OF_MRNA_STABILITY_BY_PROTEINS_THAT_BIND_AU_RICH_ELEMENTS | –2.10 | 2.31 × 10–4 |
| PROCESSING_OF_CAPPED_INTRON_CONTAINING_PRE_MRNA | –1.97 | 1.27 × 10–3 | |
| MRNA_SPLICING | –1.93 | 2.04 × 10–3 | |
| TRANSPORT_OF_MATURE_TRANSCRIPT_TO_CYTOPLASM | –1.83 | 6.69 × 10–3 | |
| SNRNP_ASSEMBLY | –1.67 | 3.01 × 10–2 | |
| Metabolism of Proteins | TRANSLATION | –1.84 | 6.77 × 10–3 |
| MITOCHONDRIAL_TRANSLATION | –2.41 | 0 | |
| DEUBIQUITINATION | –1.83 | 6.67 × 10–3 | |
| NEDDYLATION | –1.75 | 1.64 × 10–2 | |
| ASPARAGINE_N_LINKED_GLYCOSYLATION | –1.61 | 4.66 × 10–2 | |
| Immune System | COMPLEMENT_CASCADE | 1.96 | 2.87 × 10–2 |
| INITIAL_TRIGGERING_OF_COMPLEMENT | 2.02 | 2.15 × 10–2 | |
| TNFR2_NON_CANONICAL_NF_KB_PATHWAY | –2.29 | 4.42 × 10–5 | |
| INTERLEUKIN_1_SIGNALING | –2.22 | 3.32 × 10–5 | |
| INTERLEUKIN_12_FAMILY_SIGNALING | –1.72 | 2.10 × 10–2 | |
| C_TYPE_LECTIN_RECEPTORS_CLRS | –2.08 | 2.55 × 10–4 | |
| FC_EPSILON_RECEPTOR_FCERI_SIGNALING | –2.12 | 1.98 × 10–4 | |
| DDX58_IFIH1_MEDIATED_INDUCTION_OF_INTERFERON_ALPHA_BETA | –1.67 | 3.15 × 10–2 | |
| ROS_AND_RNS_PRODUCTION_IN_PHAGOCYTES | –1.66 | 3.30 × 10–2 | |
| SIGNALING_BY_THE_B_CELL_RECEPTOR_BCR | –2.23 | 3.43 × 10–5 | |
| CLASS_I_MHC_MEDIATED_ANTIGEN_PROCESSING_PRESENTATION | –1.66 | 3.31 × 10–2 | |
| Metabolism | CHOLESTEROL_BIOSYNTHESIS | 2.56 | 2.59 × 10–4 |
| BIOLOGICAL_OXIDATIONS | 1.90 | 3.71 × 10–2 | |
| METABOLISM_OF_POLYAMINES | –2.28 | 4.08 × 10–5 | |
| METABOLISM_OF_COFACTORS | –1.84 | 6.70 × 10–3 | |
| THE_CITRIC_ACID_TCA_CYCLE_AND_RESPIRATORY_ELECTRON_TRANSPORT | –1.72 | 2.12 × 10–2 | |
| RESPIRATORY_ELECTRON_TRANSPORT | –1.95 | 1.75 × 10–3 | |
| COMPLEX_I_BIOGENESIS | –1.78 | 1.15 × 10–2 | |
| CITRIC_ACID_CYCLE_TCA_CYCLE | –1.62 | 4.30 × 10–2 | |
| Protein Localization, Transport of Small Molecules | PROTEIN_LOCALIZATION | –1.81 | 8.57 × 10–3 |
| MITOCHONDRIAL_PROTEIN_IMPORT | –2.08 | 2.64 × 10–4 | |
| ABC_FAMILY_PROTEINS_MEDIATED_TRANSPORT | –2.12 | 2.02 × 10–4 | |
| PLASMA_LIPOPROTEIN_CLEARANCE | –1.63 | 4.09 × 10–2 | |
| Cellular Responses to Stimuli | CELLULAR_RESPONSE_TO_HYPOXIA | –2.48 | 0 |
| HSP90_CHAPERONE_CYCLE_FOR_STEROID_HORMONE_RECEPTORS_SHR_IN_THE_PRESENCE_OF_LIGAND | –1.86 | 4.85 × 10–3 | |
| CELLULAR_RESPONSE_TO_CHEMICAL_STRESS | –1.83 | 7.10 × 10–3 | |
| HSF1_ACTIVATION | –1.68 | 2.93 × 10–2 | |
| ATTENUATION_PHASE | –1.60 | 4.95 × 10–2 | |
| Signal Transduction | GPCR_LIGAND_BINDING | 2.35 | 8.28 × 10–4 |
| CLASS_A_1_RHODOPSIN_LIKE_RECEPTORS | 2.48 | 2.72 × 10–4 | |
| PEPTIDE_LIGAND_BINDING_RECEPTORS | 2.28 | 1.80 × 10–3 | |
| MET_PROMOTES_CELL_MOTILITY | 2.26 | 2.11 × 10–3 | |
| INTEGRIN_SIGNALING | 2.25 | 2.18 × 10–3 | |
| DEGRADATION_OF_AXIN | –2.40 | 0 | |
| BETA_CATENIN_INDEPENDENT_WNT_SIGNALING | –2.03 | 5.47 × 10–4 | |
| DEGRADATION_OF_BETA_CATENIN_BY_THE_DESTRUCTION_COMPLEX | –2.16 | 1.08 × 10–4 | |
| DEGRADATION_OF_DVL | –2.39 | 0 | |
| SIGNALING_BY_HEDGEHOG | –1.91 | 2.82 × 10–3 | |
| HEDGEHOG_LIGAND_BIOGENESIS | –2.39 | 0 | |
| SIGNALING_BY_NOTCH4 | –2.07 | 3.58 × 10–4 | |
| MAPK6_MAPK4_SIGNALING | –2.14 | 1.29 × 10–4 | |
| REGULATION_OF_RAS_BY_GAPS | –2.27 | 3.79 × 10–5 | |
| REGULATION_OF_PTEN_STABILITY_AND_ACTIVITY | –2.22 | 3.22 × 10–5 |
| GEO/TCGA Acession | Human Intestine Dataset | Enriched Genes in xmrk Intestine | |
|---|---|---|---|
| NES | FDR p-Value | ||
| GSE165512 | Ulcerative colitis, colon | 2.2856 | <0.001 |
| GSE165512 | Crohn’s disease, colon | 2.2315 | <0.001 |
| GSE165512 | Crohn’s disease, ileum | 0.6574 | 0.644 |
| TCGA-COAD | Colon adenocarcinoma | –1.4089 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Lee, A.Q.; Lu, Z.; Sun, Y.; Lu, J.-W.; Ren, Z.; Zhang, N.; Liu, D.; Gong, Z. Systematic Characterization of the Disruption of Intestine during Liver Tumor Progression in the xmrk Oncogene Transgenic Zebrafish Model. Cells 2022, 11, 1810. https://doi.org/10.3390/cells11111810
Li Y, Lee AQ, Lu Z, Sun Y, Lu J-W, Ren Z, Zhang N, Liu D, Gong Z. Systematic Characterization of the Disruption of Intestine during Liver Tumor Progression in the xmrk Oncogene Transgenic Zebrafish Model. Cells. 2022; 11(11):1810. https://doi.org/10.3390/cells11111810
Chicago/Turabian StyleLi, Yan, Ai Qi Lee, Zhiyuan Lu, Yuxi Sun, Jeng-Wei Lu, Ziheng Ren, Na Zhang, Dong Liu, and Zhiyuan Gong. 2022. "Systematic Characterization of the Disruption of Intestine during Liver Tumor Progression in the xmrk Oncogene Transgenic Zebrafish Model" Cells 11, no. 11: 1810. https://doi.org/10.3390/cells11111810






