Making Sense of Quorum Sensing at the Intestinal Mucosal Interface
Abstract
1. Introduction
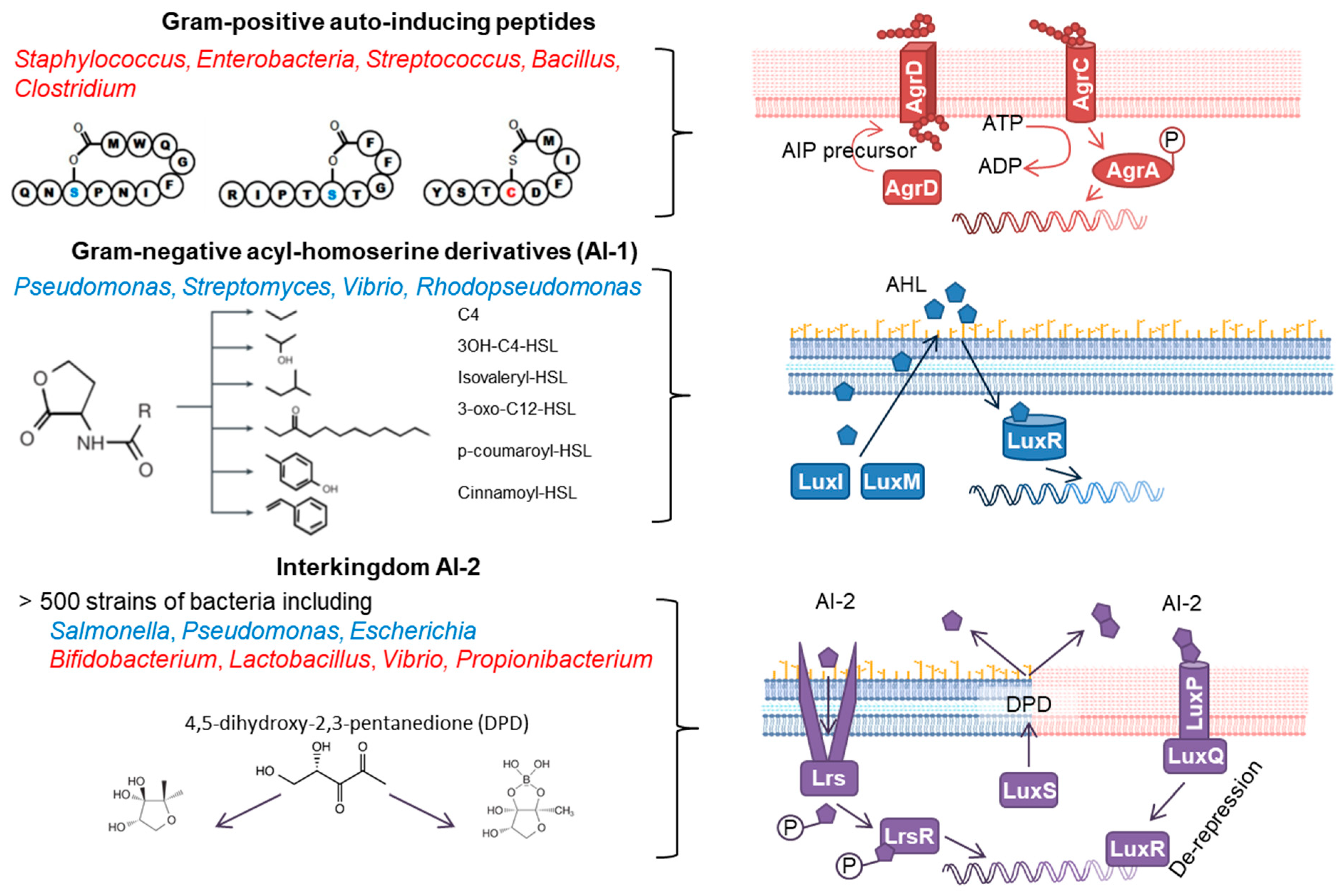
1.1. Bacterial QS Signals
| Bacteria | Positive for Host | Negative for Host | |
|---|---|---|---|
| Toxins, antibiotics | Kill superfluous bacteria with limited viability. | Antimicrobials can inhibit the growth of pathogenic bacteria (novel antibiotics) [20,21]. | Toxins cause severe disease through damaging the intestinal epithelium, activating immune cells and neurons (pain) [22]. |
| Proteases | Increase nutrient availability. | Modulate nutrient pool in the gut through protein degradation for metabolisation/fermentation by bacteria and absorption by the host. | Degrade host mucins and immunoglobulins decreasing host defences [23]. |
| Biofilm formation | Allows motility of otherwise immotile bacteria, provides protection, allows GI colonisation. | Enables the growth and presence of beneficial bacteria (‘niche’) [24,25,26]. | Protects from elimination/targeting by immune cells [27]. |
| Metabolic adaptation | Switch to metabolic pathways using ready-to-use substrates, metabolic slowing [28,29]. | Depletion of nutrients for the growth of pathogenic bacteria, production of inhibitory metabolites [30]. | Depletion of nutrients for the host and adaptation to mucus degradation [31]. |
1.2. Host Molecular and Cellular Targets of Bacterial QS Signals
2. Factors Influencing QS in the Gastrointestinal Tract
2.1. Environmental Conditions (pH)
2.2. Short Chain Fatty Acids
2.3. Dietary Compounds (Secondary Plant Products)
2.4. Host–Gut Microbial Co-Metabolism (Bile Acids)
2.5. Interkingdom Signalling Molecules
2.6. Immunoglobulins
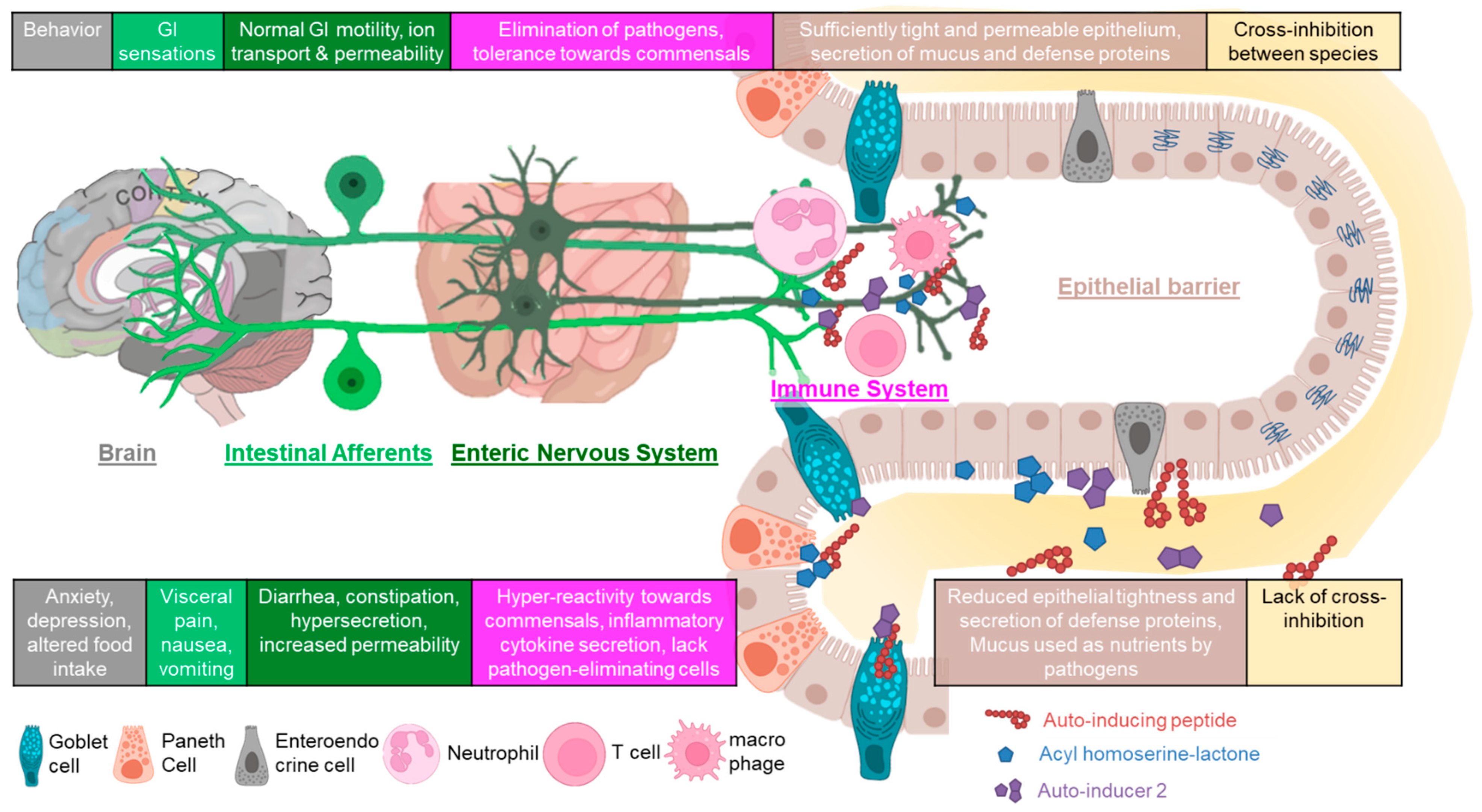
3. QS at the Mucosal Neuroimmune Interface
3.1. Epithelial Barrier
3.2. Immune Modulation
3.3. Gut Intrinsic and Extrinsic Neural Function
4. QS and Association with Gastrointestinal Disease and Dysfunction
5. Perspective
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Chen, C.C.; Wang, L.; Plikus, M.V.; Jiang, T.X.; Murray, P.J.; Ramos, R.; Guerrero-Juarez, C.F.; Hughes, M.W.; Lee, O.K.; Shi, S.; et al. Organ-level quorum sensing directs regeneration in hair stem cell populations. Cell 2015, 161, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Muldoon, J.J.; Chuang, Y.; Bagheri, N.; Leonard, J.N. Macrophages employ quorum licensing to regulate collective activation. Nat. Commun. 2020, 11, 878. [Google Scholar] [CrossRef] [PubMed]
- Brucher, B.L.; Jamall, I.S. Cell-cell communication in the tumor microenvironment, carcinogenesis, and anticancer treatment. Cell Physiol. Biochem. 2014, 34, 213–243. [Google Scholar] [CrossRef] [PubMed]
- Combarnous, Y.; Nguyen, T.M.D. Cell Communications among Microorganisms, Plants, and Animals: Origin, Evolution, and Interplays. Int. J. Mol. Sci. 2020, 21, 8052. [Google Scholar] [CrossRef]
- Qazi, S.; Middleton, B.; Muharram, S.H.; Cockayne, A.; Hill, P.; O’Shea, P.; Chhabra, S.R.; Camara, M.; Williams, P. N-acylhomoserine lactones antagonize virulence gene expression and quorum sensing in Staphylococcus aureus. Infect. Immun. 2006, 74, 910–919. [Google Scholar] [CrossRef]
- Canovas, J.; Baldry, M.; Bojer, M.S.; Andersen, P.S.; Grzeskowiak, P.K.; Stegger, M.; Damborg, P.; Olsen, C.A.; Ingmer, H. Cross-Talk between Staphylococcus aureus and Other Staphylococcal Species via the agr Quorum Sensing System. Front. Microbiol. 2016, 7, 1733. [Google Scholar] [CrossRef]
- Spangler, J.R.; Dean, S.N.; Leary, D.H.; Walper, S.A. Response of Lactobacillus plantarum WCFS1 to the Gram-Negative Pathogen-Associated Quorum Sensing Molecule N-3-Oxododecanoyl Homoserine Lactone. Front. Microbiol. 2019, 10, 715. [Google Scholar] [CrossRef]
- Cook, L.C.; Federle, M.J. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol. Rev. 2014, 38, 473–492. [Google Scholar] [CrossRef]
- Feng, J.; Zong, W.; Wang, P.; Zhang, Z.T.; Gu, Y.; Dougherty, M.; Borovok, I.; Wang, Y. RRNPP-type quorum-sensing systems regulate solvent formation, sporulation and cell motility in Clostridium saccharoperbutylacetonicum. Biotechnol. Biofuels 2020, 13, 84. [Google Scholar] [CrossRef]
- Lingeswaran, A.; Metton, C.; Henry, C.; Monnet, V.; Juillard, V.; Gardan, R. Export of Rgg Quorum Sensing Peptides is Mediated by the PptAB ABC Transporter in Streptococcus Thermophilus Strain LMD-9. Genes 2020, 11, 1096. [Google Scholar] [CrossRef]
- Yi, L.; Dong, X.; Grenier, D.; Wang, K.; Wang, Y. Research progress of bacterial quorum sensing receptors: Classification, structure, function and characteristics. Sci. Total Environ. 2021, 763, 143031. [Google Scholar] [CrossRef] [PubMed]
- Gray, B.; Hall, P.; Gresham, H. Targeting agr- and agr-like quorum sensing systems for development of common therapeutics to treat multiple gram-positive bacterial infections. Sensors 2013, 13, 5130–5166. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Tobias, N.J.; Brehm, J.; Kresovic, D.; Brameyer, S.; Bode, H.B.; Heermann, R. New Vocabulary for Bacterial Communication. ChemBioChem 2020, 21, 759–768. [Google Scholar] [CrossRef]
- Polkade, A.V.; Mantri, S.S.; Patwekar, U.J.; Jangid, K. Quorum Sensing: An Under-Explored Phenomenon in the Phylum Actinobacteria. Front. Microbiol. 2016, 7, 131. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef]
- Mukherjee, S.; Bassler, B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 2019, 17, 371–382. [Google Scholar] [CrossRef]
- Turkina, M.V.; Vikstrom, E. Bacteria-Host Crosstalk: Sensing of the Quorum in the Context of Pseudomonas aeruginosa Infections. J. Innate Immun. 2019, 11, 263–279. [Google Scholar] [CrossRef]
- Fujii, T.; Ingham, C.; Nakayama, J.; Beerthuyzen, M.; Kunuki, R.; Molenaar, D.; Sturme, M.; Vaughan, E.; Kleerebezem, M.; de Vos, W. Two homologous Agr-like quorum-sensing systems cooperatively control adherence, cell morphology, and cell viability properties in Lactobacillus plantarum WCFS1. J. Bacteriol. 2008, 190, 7655–7665. [Google Scholar] [CrossRef]
- Paharik, A.E.; Schreiber, H.L.t.; Spaulding, C.N.; Dodson, K.W.; Hultgren, S.J. Narrowing the spectrum: The new frontier of precision antimicrobials. Genome Med. 2017, 9, 110. [Google Scholar] [CrossRef]
- Collins, F.W.J.; O’Connor, P.M.; O’Sullivan, O.; Gomez-Sala, B.; Rea, M.C.; Hill, C.; Ross, R.P. Bacteriocin Gene-Trait matching across the complete Lactobacillus Pan-genome. Sci. Rep. 2017, 7, 3481. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, S.; Nagao-Kitamoto, H.; Kuffa, P.; Kamada, N. Regulation of virulence: The rise and fall of gastrointestinal pathogens. J. Gastroenterol. 2016, 51, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Amat, C.B.; Motta, J.P.; Fekete, E.; Moreau, F.; Chadee, K.; Buret, A.G. Cysteine Protease-Dependent Mucous Disruptions and Differential Mucin Gene Expression in Giardia duodenalis Infection. Am. J. Pathol. 2017, 187, 2486–2498. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Luo, Y. Bacterial Quorum-Sensing Systems and Their Role in Intestinal Bacteria-Host Crosstalk. Front. Microbiol. 2021, 12, 611413. [Google Scholar] [CrossRef] [PubMed]
- Aoudia, N.; Rieu, A.; Briandet, R.; Deschamps, J.; Chluba, J.; Jego, G.; Garrido, C.; Guzzo, J. Biofilms of Lactobacillus plantarum and Lactobacillus fermentum: Effect on stress responses, antagonistic effects on pathogen growth and immunomodulatory properties. Food Microbiol. 2016, 53, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Luo, X.M.; Liu, J.; Wang, H. Quorum Sensing, Biofilm, and Intestinal Mucosal Barrier: Involvement the Role of Probiotic. Front. Cell Infect. Microbiol. 2020, 10, 538077. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Berends, E.T.M.; Chan, R.; Schwab, E.; Roy, S.; Sen, C.K.; Torres, V.J.; Wozniak, D.J. Staphylococcus aureus biofilms release leukocidins to elicit extracellular trap formation and evade neutrophil-mediated killing. Proc. Natl. Acad. Sci. USA 2018, 115, 7416–7421. [Google Scholar] [CrossRef]
- An, J.H.; Goo, E.; Kim, H.; Seo, Y.S.; Hwang, I. Bacterial.l.l quorum sensing and metabolic slowing in a cooperative population. Proc. Natl. Acad. Sci. USA 2014, 111, 14912–14917. [Google Scholar] [CrossRef]
- Yong, Y.C.; Zhong, J.J. Impacts of quorum sensing on microbial metabolism and human health. Adv. Biochem. Eng. Biotechnol. 2013, 131, 25–61. [Google Scholar] [CrossRef]
- Chowdhury, R.; Pavinski Bitar, P.D.; Keresztes, I.; Condo, A.M., Jr.; Altier, C. A diffusible signal factor of the intestine dictates Salmonella invasion through its direct control of the virulence activator HilD. PLoS Pathog. 2021, 17, e1009357. [Google Scholar] [CrossRef]
- Kalburge, S.S.; Carpenter, M.R.; Rozovsky, S.; Boyd, E.F. Quorum Sensing Regulators Are Required for Metabolic Fitness in Vibrio parahaemolyticus. Infect. Immun. 2017, 85, e00930-16. [Google Scholar] [CrossRef] [PubMed]
- Aymeric, L.; Donnadieu, F.; Mulet, C.; du Merle, L.; Nigro, G.; Saffarian, A.; Berard, M.; Poyart, C.; Robine, S.; Regnault, B.; et al. Colorectal cancer specific conditions promote Streptococcus gallolyticus gut colonization. Proc. Natl. Acad. Sci. USA 2018, 115, E283–E291. [Google Scholar] [CrossRef] [PubMed]
- Christiaen, S.E.; O’Connell Motherway, M.; Bottacini, F.; Lanigan, N.; Casey, P.G.; Huys, G.; Nelis, H.J.; van Sinderen, D.; Coenye, T. Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. PLoS ONE 2014, 9, e98111. [Google Scholar] [CrossRef] [PubMed]
- Lebeer, S.; Claes, I.J.; Verhoeven, T.L.; Shen, C.; Lambrichts, I.; Ceuppens, J.L.; Vanderleyden, J.; De Keersmaecker, S.C. Impact of luxS and suppressor mutations on the gastrointestinal transit of Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 2008, 74, 4711–4718. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Luo, Y.; Cao, X.; Liu, W.; Song, G.; Zhang, Z. LuxS quorum sensing system mediating Lactobacillus plantarum probiotic characteristics. Arch. Microbiol. 2021, 203, 4141–4148. [Google Scholar] [CrossRef]
- Thompson, J.A.; Oliveira, R.A.; Djukovic, A.; Ubeda, C.; Xavier, K.B. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015, 10, 1861–1871. [Google Scholar] [CrossRef]
- Xu, L.; Li, H.; Vuong, C.; Vadyvaloo, V.; Wang, J.; Yao, Y.; Otto, M.; Gao, Q. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect. Immun. 2006, 74, 488–496. [Google Scholar] [CrossRef]
- Husted, A.S.; Trauelsen, M.; Rudenko, O.; Hjorth, S.A.; Schwartz, T.W. GPCR-Mediated Signaling of Metabolites. Cell Metab. 2017, 25, 777–796. [Google Scholar] [CrossRef]
- Cohen, L.J.; Esterhazy, D.; Kim, S.H.; Lemetre, C.; Aguilar, R.R.; Gordon, E.A.; Pickard, A.J.; Cross, J.R.; Emiliano, A.B.; Han, S.M.; et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 2017, 549, 48–53. [Google Scholar] [CrossRef]
- Chen, H.; Nwe, P.-K.; Yang, Y.; Rosen, C.E.; Bielecka, A.A.; Kuchroo, M.; Cline, G.W.; Kruse, A.C.; Ring, A.M.; Crawford, J.M.; et al. A Forward Chemical Genetic Screen Reveals Gut Microbiota Metabolites That Modulate Host Physiology. Cell 2019, 177, 1217–1231.e8. [Google Scholar] [CrossRef]
- Krasulova, K.; Illes, P. Intestinal interplay of quorum sensing molecules and human receptors. Biochimie 2021, 189, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Tizzano, M.; Gulbransen, B.D.; Vandenbeuch, A.; Clapp, T.R.; Herman, J.P.; Sibhatu, H.M.; Churchill, M.E.A.; Silver, W.L.; Kinnamon, S.C.; Finger, T.E. Nasal chemosensory cells use bitter taste signaling to detect irritants and bacterial signals. Proc. Natl. Acad. Sci. USA 2010, 107, 3210–3215. [Google Scholar] [CrossRef] [PubMed]
- Pundir, P.; Liu, R.; Vasavda, C.; Serhan, N.; Limjunyawong, N.; Yee, R.; Zhan, Y.; Dong, X.; Wu, X.; Zhang, Y.; et al. A Connective Tissue Mast-Cell-Specific Receptor Detects Bacterial Quorum-Sensing Molecules and Mediates Antibacterial Immunity. Cell Host Microbe 2019, 26, 114–122.e8. [Google Scholar] [CrossRef] [PubMed]
- Moura-Alves, P.; Puyskens, A.; Stinn, A.; Klemm, M.; Guhlich-Bornhof, U.; Dorhoi, A.; Furkert, J.; Kreuchwig, A.; Protze, J.; Lozza, L.; et al. Host monitoring of quorum sensing du.uring Pseudomonas aeruginosa infection. Science 2019, 366, eaaw1629. [Google Scholar] [CrossRef]
- Marinelli, L.; Martin-Gallausiaux, C.; Bourhis, J.M.; Beguet-Crespel, F.; Blottiere, H.M.; Lapaque, N. Identification of the novel role of butyrate as AhR ligand in human intestinal epithelial cells. Sci. Rep. 2019, 9, 643. [Google Scholar] [CrossRef]
- Sun, M.; Ma, N.; He, T.; Johnston, L.J.; Ma, X. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR). Crit. Rev. Food Sci. Nutr. 2019, 8398, 1760–1768. [Google Scholar] [CrossRef]
- Korecka, A.; Dona, A.; Lahiri, S.; Tett, A.J.; Al-Asmakh, M.; Braniste, V.; D’Arienzo, R.; Abbaspour, A.; Reichardt, N.; Fujii-Kuriyama, Y.; et al. Bidirectional communication between the Aryl hydrocarbon Receptor (AhR) and the microbiome tunes host metabolism. NPJ Biofilms Microbiomes 2016, 2, 16014. [Google Scholar] [CrossRef]
- Natividad, J.M.; Agus, A.; Planchais, J.; Lamas, B.; Jarry, A.C.; Martin, R.; Michel, M.L.; Chong-Nguyen, C.; Roussel, R.; Straube, M.; et al. Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metab. 2018, 28, 737–749.e4. [Google Scholar] [CrossRef]
- Karlsson, T.; Turkina, M.V.; Yakymenko, O.; Magnusson, K.E.; Vikstrom, E. The Pseudomonas aeruginosa N-acylhomoserine lactone quorum sensing molecules target IQGAP1 and modulate epithelial cell migration. PLoS Pathog. 2012, 8, e1002953. [Google Scholar] [CrossRef]
- Kim, H.; White, C.D.; Sacks, D.B. IQGAP1 in microbial pathogenesis: Targeting the actin cytoskeleton. FEBS Lett. 2011, 585, 723–729. [Google Scholar] [CrossRef]
- Meseguer, V.; Alpizar, Y.A.; Luis, E.; Tajada, S.; Denlinger, B.; Fajardo, O.; Manenschijn, J.-A.; Fernández-Peña, C.; Talavera, A.; Kichko, T.; et al. TRPA1 channels mediate acute neurogenic inflammation and pain produced by bacterial endotoxins. Nat. Commun. 2014, 5, 3125. [Google Scholar] [CrossRef] [PubMed]
- Blake, K.J.; Baral, P.; Voisin, T.; Lubkin, A.; Pinho-Ribeiro, F.A.; Adams, K.L.; Roberson, D.P.; Ma, Y.C.; Otto, M.; Woolf, C.J.; et al. Staphylococcus aureus produces pain through pore-forming toxins and neuronal TRPV1 that is silenced by QX-314. Nat. Commun. 2018, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Laabei, M.; Jamieson, W.D.; Yang, Y.; Van Den Elsen, J.; Jenkins, A.T.A. Investigating the lytic activity and structural properties of Staphylococcus aureus phenol soluble modulin (PSM) peptide toxins. Biochim. Biophys. Acta Biomembr. 2014, 1838, 3153–3161. [Google Scholar] [CrossRef] [PubMed]
- Duncan, K.; Carey-Ewend, K.; Vaishnava, S. Spatial analysis of gut microbiome reveals a distinct ecological niche associated with the mucus layer. Gut Microbes 2021, 13, 1874815. [Google Scholar] [CrossRef] [PubMed]
- Ezekwe, E.; Weng, C.; Duncan, J. ADAM10 Cell Surface Expression but Not Activity Is Critical for Staphylococcus aureus α-Hemolysin-Mediated Activation of the NLRP3 Inflammasome in Human Monocytes. Toxins 2016, 8, 95. [Google Scholar] [CrossRef] [PubMed]
- Inoshima, I.; Inoshima, N.; Wilke, G.A.; Powers, M.E.; Frank, K.M.; Wang, Y.; Wardenburg, J.B. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat. Med. 2011, 17, 1310–1314. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Kumar, R.; Singh, B.R. Clostridial Neurotoxins: Structure, Function and Implications to Other Bacterial Toxins. Microorganisms 2021, 9, 2206. [Google Scholar] [CrossRef]
- Singh, A.; Harrison, S.; Schoeniger, J. Gangliosides as Receptors for Biological Toxins: Development of Sensitive Fluoroimmunoassays Using Ganglioside-Bearing Liposomes. Anal. Chem. 2000, 72, 6019–6024. [Google Scholar] [CrossRef]
- Acedo, J.Z.; Chiorean, S.; Vederas, J.C.; van Belkum, M.J. The expanding structural variety among bacteriocins from Gram-positive bacteria. FEMS Microbiol. Rev. 2018, 42, 805–828. [Google Scholar] [CrossRef]
- Dreyer, L.; Smith, C.; Deane, S.M.; Dicks, L.M.T.; van Staden, A.D. Migration of Bacteriocins Across Gastrointestinal Epithelial and Vascular Endothelial Cells, as Determined Using In Vitro Simulations. Sci. Rep. 2019, 9, 11481. [Google Scholar] [CrossRef]
- Soltani, S.; Hammami, R.; Cotter, P.D.; Rebuffat, S.; Said, L.B.; Gaudreau, H.; Bedard, F.; Biron, E.; Drider, D.; Fliss, I. Bacteriocins as a new generation of antimicrobials: Toxicity aspects and regulations. FEMS Microbiol. Rev. 2021, 45, fuaa039. [Google Scholar] [CrossRef] [PubMed]
- Dicks, L.M.T.; Dreyer, L.; Smith, C.; van Staden, A.D. A Review: The Fate of Bacteriocins in the Human Gastro-Intestinal Tract: Do They Cross the Gut-Blood Barrier? Front. Microbiol 2018, 9, 2297. [Google Scholar] [CrossRef] [PubMed]
- Jubrail, J.; Morris, P.; Bewley, M.A.; Stoneham, S.; Johnston, S.A.; Foster, S.J.; Peden, A.A.; Read, R.C.; Marriott, H.M.; Dockrell, D.H. Inability to sustain intraphagolysosomal killing of Staphylococcus aureus predisposes to bacterial persistence in macrophages. Cell. Microbiol. 2016, 18, 80–96. [Google Scholar] [CrossRef]
- Strobel, M.; Pförtner, H.; Tuchscherr, L.; Völker, U.; Schmidt, F.; Kramko, N.; Schnittler, H.J.; Fraunholz, M.J.; Löffler, B.; Peters, G.; et al. Post-invasion events after infection with Staphylococcus aureus are strongly dependent on both the host cell type and the infecting S. aureus strain. Clin. Microbiol. Infect. 2016, 22, 799–809. [Google Scholar] [CrossRef]
- Blattner, S.; Das, S.; Paprotka, K.; Eilers, U.; Krischke, M.; Kretschmer, D.; Remmele, C.W.; Dittrich, M.; Muller, T.; Schuelein-Voelk, C.; et al. Staphylococcus aureus Exploits a Non-ribosomal Cyclic Dipeptide to Modulate Survival within Epithelial Cells and Phagocytes. PLoS Pathog. 2016, 12, e1005857. [Google Scholar] [CrossRef] [PubMed]
- Udayan, S.; Stamou, P.; Crispie, F.; Hickey, A.; Floyd, A.N.; Hsieh, C.S.; Cotter, P.D.; O’Sullivan, O.; Melgar, S.; O’Toole, P.W.; et al. Identification of Gut Bacteria such as Lactobacillus johnsonii that Disseminate to Systemic Tissues of Wild Type and MyD88-/- Mice. Gut Microbes 2022, 14, 2007743. [Google Scholar] [CrossRef]
- Horiuchi, H.; Kamikado, K.; Aoki, R.; Suganuma, N.; Nishijima, T.; Nakatani, A.; Kimura, I. Bifidobacterium animalis subsp. lactis GCL2505 modulates host energy metabolism via the short-chain fatty acid receptor GPR43. Sci. Rep. 2020, 10, 4158. [Google Scholar] [CrossRef]
- Naghmouchi, K.; Belguesmia, Y.; Bendali, F.; Spano, G.; Seal, B.S.; Drider, D. Lactobacillus fermentum: A bacterial species with potential for food preservation and biomedical applications. Crit. Rev. Food Sci. Nutr. 2019, 60, 3387–3399. [Google Scholar] [CrossRef]
- Yang, M.; Meng, F.; Gu, W.; Fu, L.; Zhang, F.; Li, F.; Tao, Y.; Zhang, Z.; Wang, X.; Yang, X.; et al. Influence of Polysaccharides From Polygonatum kingianum on Short-Chain Fatty Acid Production and Quorum Sensing in Lactobacillus faecis. Front. Microbiol. 2021, 12, 758870. [Google Scholar] [CrossRef]
- Abbasiliasi, S.; Ramanan, R.N.; Ibrahim, T.A.T.; Mustafa, S.; Mohamad, R.; Daud, H.H.M.; Ariff, A.B. Effect of Medium Composition and Culture Condition on the Production of Bacteriocin-Like Inhibitory Substances (BLIS) byLactobacillus ParacaseiLA07, a Strain Isolated from Budu. Biotechnol. Biotechnol. Equip. 2014, 25, 2652–2657. [Google Scholar] [CrossRef]
- Ge, J.; Kang, J.; Ping, W. Effect of Acetic Acid on Bacteriocin Production by Gram-Positive Bacteria. J. Microbiol. Biotechnol. 2019, 29, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Lu, F.; Du, H.; Nie, T.; Zhu, X.; Connerton, I.F.; Zhao, H.; Bie, X.; Zhang, C.; Lu, Z.; et al. Acetate and auto-inducing peptide are independent triggers of quorum sensing in Lactobacillus plantarum. Mol. Microbiol. 2021, 116, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Zhao, H.; Nie, T.; Lu, F.; Zhang, C.; Lu, Y.; Lu, Z. Acetate Activates Lactobacillus Bacteriocin Synthesis by Controlling Quorum Sensing. Appl. Env. Microbiol. 2021, 87, e0072021. [Google Scholar] [CrossRef]
- Yeo, S.; Park, H.; Ji, Y.; Park, S.; Yang, J.; Lee, J.; Mathara, J.M.; Shin, H.; Holzapfel, W. Influence of gastrointestinal stress on autoinducer-2 activity of two Lactobacillus species. FEMS Microbiol. Ecol. 2015, 91, fiv065. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Bhawal, S.; Kumari, A.; Kapila, S.; Kapila, R. pH-dependent inhibition of AHL-mediated quorum sensing by cell-free supernatant of lactic acid bacteria in Pseudomonas aeruginosa PAO1. Microb. Pathog. 2020, 142, 104105. [Google Scholar] [CrossRef] [PubMed]
- Mashruwala, A.A.; Bassler, B.L. The Vibrio cholerae Quorum-Sensing Protein VqmA Integrates Cell Density, Environmental, and Host-Derived Cues into the Control of Virulence. mBio 2020, 11, e01572-20. [Google Scholar] [CrossRef]
- Adachi, K.; Ohtani, K.; Kawano, M.; Singh, R.P.; Yousuf, B.; Sonomoto, K.; Shimizu, T.; Nakayama, J. Metabolic dependent and independent pH-drop shuts down VirSR quorum sensing in Clostridium perfringens. J. Biosci. Bioeng. 2018, 125, 525–531. [Google Scholar] [CrossRef]
- Regassa, L.; Novick, R.; Betley, M. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect. Immun. 1992, 60, 3381–3388. [Google Scholar] [CrossRef]
- Scott, D.R.; Marcus, E.A.; Wen, Y.; Singh, S.; Feng, J.; Sachs, G. Cytoplasmic histidine kinase (HP0244)-regulated assembly of urease with UreI, a channel for urea and its metabolites, CO2, NH3, and NH4(+), is necessary for acid survival of Helicobacter pylori. J. Bacteriol. 2010, 192, 94–103. [Google Scholar] [CrossRef]
- Do, H.; Makthal, N.; VanderWal, A.R.; Saavedra, M.O.; Olsen, R.J.; Musser, J.M.; Kumaraswami, M. Environmental pH and peptide signaling control virulence of Streptococcus pyogenes via a quorum-sensing pathway. Nat. Commun. 2019, 10, 2586. [Google Scholar] [CrossRef]
- Nogal, A.; Valdes, A.M.; Menni, C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 2021, 13, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Dass, N.B.; John, A.K.; Bassil, A.K.; Crumbley, C.W.; Shehee, W.R.; Maurio, F.P.; Moore, G.B.; Taylor, C.M.; Sanger, G.J. The relationship between the effects of short-chain fatty acids on intestinal motility in vitro and GPR43 receptor activation. Neurogastroenterol. Motil. 2007, 19, 66–74. [Google Scholar] [CrossRef] [PubMed]
- O’Riordan, K.J.; Collins, M.K.; Moloney, G.M.; Knox, E.G.; Aburto, M.R.; Fulling, C.; Morley, S.J.; Clarke, G.; Schellekens, H.; Cryan, J.F. Short chain fatty acids: Microbial metabolites for gut-brain axis signalling. Mol. Cell Endocrinol. 2022, 546, 111572. [Google Scholar] [CrossRef] [PubMed]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Mirzaei, R.; Dehkhodaie, E.; Bouzari, B.; Rahimi, M.; Gholestani, A.; Hosseini-Fard, S.R.; Keyvani, H.; Teimoori, A.; Karampoor, S. Dual role of microbiota-derived short-chain fatty acids on host and pathogen. Biomed. Pharm. 2022, 145, 112352. [Google Scholar] [CrossRef]
- Nakamura, K.; O’Neill, A.M.; Williams, M.R.; Cau, L.; Nakatsuji, T.; Horswill, A.R.; Gallo, R.L. Short chain fatty acids produced by Cutibacterium acnes inhibit biofilm formation by Staphylococcus epidermidis. Sci. Rep. 2020, 10, 21237. [Google Scholar] [CrossRef]
- Park, T.; Im, J.; Kim, A.R.; Lee, D.; Jeong, S.; Yun, C.H.; Han, S.H. Short-chain fatty acids inhibit the biofilm formation of Streptococcus gordonii through negative regulation of competence-stimulating peptide signaling pathway. J. Microbiol. 2021, 59, 1142–1149. [Google Scholar] [CrossRef]
- Zhu, W.; Gao, J.; Liu, H.; Liu, J.; Jin, T.; Qin, N.; Ren, X.; Xia, X. Antibiofilm effect of sodium butyrate against Vibrio parahaemolyticus. Food Control 2022, 131, 108422. [Google Scholar] [CrossRef]
- Lamas, A.; Regal, P.; Vazquez, B.; Cepeda, A.; Franco, C.M. Short Chain Fatty Acids Commonly Produced by Gut Microbiota Influence Salmonella enterica Motility, Biofilm Formation, and Gene Expression. Antibiotics 2019, 8, 265. [Google Scholar] [CrossRef]
- Chen, Y.; Gozzi, K.; Yan, F.; Chai, Y. Acetic Acid Acts as a Volatile Signal To Stimulate Bacterial Biofilm Formation. mBio 2015, 6, e00392. [Google Scholar] [CrossRef]
- Kalia, V. (Ed.) Biotechnological Applications of Quorum Sensing Inhibitors; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Dingeo, G.; Brito, A.; Samouda, H.; Iddir, M.; La Frano, M.R.; Bohn, T. Phytochemicals as modifiers of gut microbial communities. Food Funct. 2020, 11, 8444–8471. [Google Scholar] [CrossRef] [PubMed]
- Nazzaro, F.; Fratianni, F.; Coppola, R. Quorum sensing and phytochemicals. Int. J. Mol. Sci. 2013, 14, 12607–12619. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.N.; Zimmer, K.R.; Macedo, A.J.; Trentin, D.S. Plant Natural Products Targeting Bacterial Virulence Factors. Chem. Rev. 2016, 116, 9162–9236. [Google Scholar] [CrossRef] [PubMed]
- Ni, N.; Choudhary, G.; Li, M.; Wang, B. Pyrogallol and its analogs can antagonize bacterial quorum sensing in Vibrio harveyi. Bioorg Med. Chem. Lett. 2008, 18, 1567–1572. [Google Scholar] [CrossRef]
- Anju, V.T.; Busi, S.; Ranganathan, S.; Ampasala, D.R.; Kumar, S.; Suchiang, K.; Kumavath, R.; Dyavaiah, M. Sesamin and sesamolin rescues Caenorhabditis elegans from Pseudomonas aeruginosa infection through the attenuation of quorum sensing regulated virulence factors. Microb. Pathog. 2021, 155, 104912. [Google Scholar] [CrossRef]
- Fratianni, F.; De Giulio, A.; Sada, A.; Nazzaro, F. Biochemical characteristics and biological properties of Annurca apple cider. J. Med. Food 2012, 15, 18–23. [Google Scholar] [CrossRef]
- Truchado, P.; Gimenez-Bastida, J.A.; Larrosa, M.; Castro-Ibanez, I.; Espin, J.C.; Tomas-Barberan, F.A.; Garcia-Conesa, M.T.; Allende, A. Inhibition of quorum sensing (QS) in Yersinia enterocolitica by an orange extract rich in glycosylated flavanones. J. Agric. Food Chem. 2012, 60, 8885–8894. [Google Scholar] [CrossRef]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rasamiravaka, T.; Stevigny, C.; Duez, P.; Rajaonson, S.; Diallo, B.; Mol, A.; Baucher, M.; El Jaziri, M. The flavanone naringenin reduces the production of quorum sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Microbiology 2011, 157, 2120–2132. [Google Scholar] [CrossRef]
- Manefield, M.; Rasmussen, T.; Henzter, M.; Andersen, J.; Steinberg, P.; Kjelleberg, S.; Givskov, M. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 2002, 148, 1119–1127. [Google Scholar] [CrossRef]
- Brackman, G.; Defoirdt, T.; Miyamoto, C.; Bossier, P.; Van Calenbergh, S.; Nelis, H.; Coenye, T. Cinnamaldehyde and cinnamaldehyde derivatives reduce virulence in Vibrio spp. by decreasing the DNA-binding activity of the quorum sensing response regulator LuxR. BMC Microbiol. 2008, 8, 149. [Google Scholar] [CrossRef]
- Rivera, M.L.C.; Hassimotto, N.M.A.; Bueris, V.; Sircili, M.P.; de Almeida, F.A.; Pinto, U.M. Effect of Capsicum Frutescens Extract, Capsaicin, and Luteolin on Quorum Sensing Regulated Phenotypes. J. Food Sci. 2019, 84, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.S.; Lee, J.H.; Ryu, S.Y.; Joo, S.W.; Cho, M.H.; Lee, J. Inhibition of Pseudomonas aeruginosa and Escherichia coli O157:H7 biofilm formation by plant metabolite epsilon-viniferin. J. Agric. Food Chem. 2013, 61, 7120–7126. [Google Scholar] [CrossRef] [PubMed]
- Buffie, C.G.; Bucci, V.; Stein, R.R.; McKenney, P.T.; Ling, L.; Gobourne, A.; No, D.; Liu, H.; Kinnebrew, M.; Viale, A.; et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Sorg, J.A.; Sonenshein, A.L. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 2008, 190, 2505–2512. [Google Scholar] [CrossRef]
- Reen, F.J.; Flynn, S.; Woods, D.F.; Dunphy, N.; Chroinin, M.N.; Mullane, D.; Stick, S.; Adams, C.; O’Gara, F. Bile signalling promotes chronic respiratory infections and antibiotic tolerance. Sci. Rep. 2016, 6, 29768. [Google Scholar] [CrossRef]
- Kelly, S.M.; Lanigan, N.; O’Neill, I.J.; Bottacini, F.; Lugli, G.A.; Viappiani, A.; Turroni, F.; Ventura, M.; van Sinderen, D. Bifidobacterial biofilm formation is a multifactorial adaptive phenomenon in response to bile exposure. Sci. Rep. 2020, 10, 11598. [Google Scholar] [CrossRef]
- Foley, M.H.; O’Flaherty, S.; Allen, G.; Rivera, A.J.; Stewart, A.K.; Barrangou, R.; Theriot, C.M. Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization. Proc. Natl. Acad. Sci. USA 2021, 118, e2017709118. [Google Scholar] [CrossRef]
- Karavolos, M.H.; Bulmer, D.M.; Spencer, H.; Rampioni, G.; Schmalen, I.; Baker, S.; Pickard, D.; Gray, J.; Fookes, M.; Winzer, K.; et al. Salmonella Typhi sense host neuroendocrine stress hormones and release the toxin haemolysin E. EMBO Rep. 2011, 12, 252–258. [Google Scholar] [CrossRef]
- Clarke, M.; Hughes, D.; Zhu, C.; Boedeker, E.; Sperandio, V. The QseC sensor kinase: A bacterial adrenergic receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 10420–10425. [Google Scholar] [CrossRef]
- Karavolos, M.H.; Winzer, K.; Williams, P.; Khan, C.M. Pathogen espionage: Multiple bacterial adrenergic sensors eavesdrop on host communication systems. Mol. Microbiol. 2013, 87, 455–465. [Google Scholar] [CrossRef]
- Cambronel, M.; Nilly, F.; Mesguida, O.; Boukerb, A.M.; Racine, P.J.; Baccouri, O.; Borrel, V.; Martel, J.; Fecamp, F.; Knowlton, R.; et al. Influence of Catecholamines (Epinephrine/Norepinephrine) on Biofilm Formation and Adhesion in Pathogenic and Probiotic Strains of Enterococcus faecalis. Front. Microbiol. 2020, 11, 1501. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Russell, R.M.; Pifer, R.; Menezes-Garcia, Z.; Cuesta, S.; Narayanan, S.; MacMillan, J.B.; Sperandio, V. The Serotonin Neurotransmitter Modulates Virulence of Enteric Pathogens. Cell Host Microbe 2020, 28, 41–53.e8. [Google Scholar] [CrossRef] [PubMed]
- Knecht, L.D.; O’Connor, G.; Mittal, R.; Liu, X.Z.; Daftarian, P.; Deo, S.K.; Daunert, S. Serotonin Activates Bacterial Quorum Sensing and Enhances the Virulence of Pseudomonas aeruginosa in the Host. EBioMedicine 2016, 9, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, A.; Gatto, D.; Sainsbury, E.; Harriman, G.; Hengartner, H.; Zinkernagel, R. A Primitive T Cell-Independent Mechanism of Intestinal Mucosal IgA Responses to Commensal Bacteria. Science 2000, 288, 2222–2225. [Google Scholar] [CrossRef]
- Everett, M.L.; Palestrant, D.; Miller, S.E.; Bollinger, R.R.; Parker, W. Immune exclusion and immune inclusion: A new model of host-bacterial interactions in the gut. Clin. Appl. Immunol. Rev. 2004, 4, 321–332. [Google Scholar] [CrossRef]
- Brandtzaeg, P. Secretory IgA: Designed for Anti-Microbial Defense. Front. Immunol. 2013, 4, 222. [Google Scholar] [CrossRef]
- Peterson, D.A.; Planer, J.D.; Guruge, J.L.; Xue, L.; Downey-Virgin, W.; Goodman, A.L.; Seedorf, H.; Gordon, J.I. Characterizing the interactions between a naturally primed immuno.oglo.obulin A and its conserved Bacteroides thetaiotaomicron species-specific epitope in gnotobiotic mice. J. Biol. Chem. 2015, 290, 12630–12649. [Google Scholar] [CrossRef]
- Park, J.; Jagasia, R.; Kaufmann, G.F.; Mathison, J.C.; Ruiz, D.I.; Moss, J.A.; Meijler, M.M.; Ulevitch, R.J.; Janda, K.D. Infection control by antibody disruption of bacterial quorum sensing signaling. Chem. Biol. 2007, 14, 1119–1127. [Google Scholar] [CrossRef]
- Kaufmann, G.F.; Park, J.; Mayorov, A.V.; Kubitz, D.M.; Janda, K.D. Generation of quorum quenching antibodies. Methods Mol. Biol. 2011, 692, 299–311. [Google Scholar] [CrossRef]
- Kaufmann, G.F.; Park, J.; Janda, K.D. Bacterial quorum sensing: A new target for anti-infective immunotherapy. Expert Opin. Biol. 2008, 8, 719–724. [Google Scholar] [CrossRef]
- Hayes, C.L.; Dong, J.; Galipeau, H.J.; Jury, J.; McCarville, J.; Huang, X.; Wang, X.Y.; Naidoo, A.; Anbazhagan, A.N.; Libertucci, J.; et al. Commensal microbiota induces colonic barrier structure and functions that contribute to homeostasis. Sci. Rep. 2018, 8, 14184. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.; Jakobsson, H.E.; Holmen-Larsson, J.; Schutte, A.; Ermund, A.; Rodriguez-Pineiro, A.M.; Arike, L.; Wising, C.; Svensson, F.; Backhed, F.; et al. Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell Host Microbe 2015, 18, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Ohland, C.L.; Macnaughton, W.K. Probiotic bacteria and intestinal epithelial barrier function. Am. J. Physiol. Gastrointest Liver Physiol. 2010, 298, G807–G819. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Cao, H.; Cover, T.L.; Whitehead, R.; Washington, M.K.; Polk, D.B. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 2007, 132, 562–575. [Google Scholar] [CrossRef] [PubMed]
- Gerhard, R.; Schmidt, G.; Hofmann, F.; Aktories, K. Activation of Rho GTPases by Escherichia coli Cytotoxic Necrotizing Factor 1 Increases Intestinal Permeability in Caco-2 Cells. Infect. Immun. 1998, 66, 5125–5131. [Google Scholar] [CrossRef] [PubMed]
- Gerhard, R.; Nottrott, S.; Schoentaube, J.; Tatge, H.; Olling, A.; Just, I. Glucosylation of Rho GTPases by Clostridium difficile toxin A triggers apoptosis in intestinal epithelial cells. J. Med. Microbiol. 2008, 57, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Adiliaghdam, F.; Almpani, M.; Gharedaghi, M.H.; Najibi, M.; Hodin, R.A.; Rahme, L.G. Targeting bacterial quorum sensing shows promise in improving intestinal barrier function following burnsite infection. Mol. Med. Rep. 2019, 19, 4057–4066. [Google Scholar] [CrossRef]
- Cheng, W.; Wang, Z.; Xiong, Y.; Wu, Z.; Tan, X.; Yang, Y.; Zhang, H.; Zhu, X.; Wei, H.; Tao, S. N-(3-oxododecanoyl)-homoserine lactone disrupts intestinal barrier and induces systemic inflammation through perturbing gut microbiome in mice. Sci. Total Environ. 2021, 778, 146347. [Google Scholar] [CrossRef]
- Tao, S.; Xiong, Y.; Wang, Z.; Wu, Y.; Li, N.; Pi, Y.; Han, D.; Zhao, J.; Wang, J. N-Acyl-Homoserine Lactones May Affect the Gut Health of Low-Birth-Weight Piglets by Altering Intestinal Epithelial Cell Barrier Function and Amino Acid Metabolism. J. Nutr. 2021, 151, 1736–1746. [Google Scholar] [CrossRef]
- Paone, P.; Cani, P.D. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut 2020, 69, 2232–2243. [Google Scholar] [CrossRef]
- Alemka, A.; Corcionivoschi, N.; Bourke, B. Defense and adaptation: The complex inter-relationship between Campylobacter jejuni and mucus. Front. Cell Infect. Microbiol. 2012, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Moal, V.L.L.; Sarrazin-Davila, L.E.; Servin, A.L. An experimental study and a randomized, double-blind, placebo-controlled clinical trial to evaluate the antisecretory activity of Lactobacillus acidophilus strain LB against nonrotavirus diarrhea. Pediatrics 2007, 120, e795–e803. [Google Scholar] [CrossRef] [PubMed]
- Caballero-Franco, C.; Keller, K.; De Simone, C.; Chadee, K. The VSL#3 probiotic formula induces mucin gene expression and secretion in colonic epithelial cells. Am. J. Physiol Gastrointest Liver Physiol. 2007, 292, G315–G322. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.O.; Birchenough, G.M.H.; Stahlman, M.; Arike, L.; Johansson, M.E.V.; Hansson, G.C.; Backhed, F. Bifidobacteria or Fiber Protects against Diet-Induced Microbiota-Mediated Colonic Mucus Deterioration. Cell Host Microbe 2018, 23, 27–40.e7. [Google Scholar] [CrossRef]
- Sicard, J.F.; Le Bihan, G.; Vogeleer, P.; Jacques, M.; Harel, J. Interactions of Intestinal Bacteria with Components of the Intestinal Mucus. Front. Cell Infect. Microbiol. 2017, 7, 387. [Google Scholar] [CrossRef]
- Hoffman, C.L.; Lalsiamthara, J.; Aballay, A. Host Mucin Is Exploited by Pseudomonas aeruginosa To Provide Monosaccharides Required for a Successful Infection. mBio 2020, 11, e00060-20. [Google Scholar] [CrossRef]
- Ayabe, T.; Satchell, D.P.; Pesendorfer, P.; Tanabe, H.; Wilson, C.L.; Hagen, S.J.; Ouellette, A.J. Activation of Paneth cell alpha-defensins in mouse small intestine. J. Biol. Chem. 2002, 277, 5219–5228. [Google Scholar] [CrossRef]
- Ayabe, T.; Satchell, D.P.; Wilson, C.L.; Parks, W.C.; Selsted, M.E.; Ouellette, A.J. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 2000, 1, 113–118. [Google Scholar] [CrossRef]
- Cash, H.L.; Whitham, C.V.; Behrendt, C.L.; Hooper, L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 2006, 313, 1126–1130. [Google Scholar] [CrossRef]
- Hapfelmeier, S.; Lawson, M.A.; Slack, E.; Kirundi, J.K.; Stoel, M.; Heikenwalder, M.; Cahenzli, J.; Velykoredko, Y.; Balmer, M.L.; Endt, K.; et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 2010, 328, 1705–1709. [Google Scholar] [CrossRef]
- Putsep, K.; Axelsson, L.G.; Boman, A.; Midtvedt, T.; Normark, S.; Boman, H.G.; Andersson, M. Germ-free and colonized mice generate the same products from enteric prodefensins. J. Biol. Chem. 2000, 275, 40478–40482. [Google Scholar] [CrossRef] [PubMed]
- Martinez Rodriguez, N.R.; Eloi, M.D.; Huynh, A.; Dominguez, T.; Lam, A.H.; Carcamo-Molina, D.; Naser, Z.; Desharnais, R.; Salzman, N.H.; Porter, E. Expansion of Paneth cell population in response to enteric Salmonella enterica serovar Typhimurium infection. Infect. Immun. 2012, 80, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Salzman, N.H.; Chou, M.M.; de Jong, H.; Liu, L.; Porter, E.M.; Paterson, Y. Enteric salmonella infection inhibits Paneth cell antimicrobial peptide expression. Infect. Immun. 2003, 71, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Salzman, N.H.; Underwood, M.A.; Bevins, C.L. Paneth cells, defensins, and the commensal microbiota: A hypothesis on intimate interplay at the intestinal mucosa. Semin. Immunol. 2007, 19, 70–83. [Google Scholar] [CrossRef]
- Holm, A.; Vikstrom, E. Quorum sensing communication between bacteria and human cells: Signals, targets, and functions. Front. Plant. Sci. 2014, 5, 309. [Google Scholar] [CrossRef]
- Aguanno, D.; Coquant, G.; Postal, B.G.; Osinski, C.; Wieckowski, M.; Stockholm, D.; Grill, J.P.; Carriere, V.; Seksik, P.; Thenet, S. The intestinal quorum sensing 3-oxo-C12:2 Acyl homoserine lactone limits cytokine-induced tight junction disruption. Tissue Barriers 2020, 8, 1832877. [Google Scholar] [CrossRef]
- Coquant, G.; Grill, J.-P.; Seksik, P. Impact of N-Acyl-Homoserine Lactones, Quorum Sensing Molecules, on Gut Immunity. Front. Immunol. 2020, 11, 1827. [Google Scholar] [CrossRef]
- Ahrends, T.; Aydin, B.; Matheis, F.; Classon, C.H.; Marchildon, F.; Furtado, G.C.; Lira, S.A.; Mucida, D. Enteric pathogens induce tissue tolerance and prevent neuronal loss from subsequent infections. Cell 2021, 184, 5715–5727.e2. [Google Scholar] [CrossRef]
- Ivanov, II; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef]
- Farkas, A.M.; Panea, C.; Goto, Y.; Nakato, G.; Galan-Diez, M.; Narushima, S.; Honda, K.; Ivanov, II. Induction of Th17 cells by segmented filamentous bacteria in the murine intestine. J. Immunol. Methods 2015, 421, 104–111. [Google Scholar] [CrossRef]
- Medina-Rodriguez, E.M.; Madorma, D.; O’Connor, G.; Mason, B.L.; Han, D.; Deo, S.K.; Oppenheimer, M.; Nemeroff, C.B.; Trivedi, M.H.; Daunert, S.; et al. Identification of a Signaling Mechanism by Which the Microbiome Regulates Th17 Cell-Mediated Depressive-Like Behaviors in Mice. Am. J. Psychiatry 2020, 177, 974–990. [Google Scholar] [CrossRef] [PubMed]
- Ohnmacht, C. Tolerance to the Intestinal Microbiota Mediated by ROR(gammat)(+) Cells. Trends Immunol. 2016, 37, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Ai, T.; Solomon, B.; Hsieh, C. T-cell selection and intestinal homeostasis. Immunol. Rev. 2014, 259, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef]
- Sefik, E.; Geva-Zatorsky, N.; Oh, S.; Konnikova, L.; Zemmour, D.; McGuire, A.M.; Burzyn, D.; Ortiz-Lopez, A.; Lobera, M.; Yang, J.; et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 2015, 349, 993–997. [Google Scholar] [CrossRef]
- Jeon, S.G.; Kayama, H.; Ueda, Y.; Takahashi, T.; Asahara, T.; Tsuji, H.; Tsuji, N.M.; Kiyono, H.; Ma, J.S.; Kusu, T.; et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012, 8, e1002714. [Google Scholar] [CrossRef]
- Sanos, S.L.; Bui, V.L.; Mortha, A.; Oberle, K.; Heners, C.; Johner, C.; Diefenbach, A. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 2009, 10, 83–91. [Google Scholar] [CrossRef]
- Santori, F.R.; Huang, P.; van de Pavert, S.A.; Douglass, E.F., Jr.; Leaver, D.J.; Haubrich, B.A.; Keber, R.; Lorbek, G.; Konijn, T.; Rosales, B.N.; et al. Identification of natural RORgamma ligands that regulate the development of lymphoid cells. Cell Metab. 2015, 21, 286–298. [Google Scholar] [CrossRef]
- Amaudrut, J.; Argiriadi, M.A.; Barth, M.; Breinlinger, E.C.; Bressac, D.; Broqua, P.; Calderwood, D.J.; Chatar, M.; Cusack, K.P.; Gauld, S.B.; et al. Discovery of novel quinoline sulphonamide derivatives as potent, selective and orally active RORgamma inverse agonists. Bioorg Med. Chem. Lett. 2019, 29, 1799–1806. [Google Scholar] [CrossRef]
- Powell, N.; MacDonald, T.T. Recent advances in gut immunology. Parasite Immunol. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.B.; Mazmanian, S.K. The Enteric Network: Interactions between the Immune and Nervous Systems of the Gut. In Immunity; Cell Press: Cambridge, MA, USA, 2017; Volume 46, pp. 910–926. [Google Scholar] [CrossRef]
- Furness, J.B. The enteric nervous system and neurogastroenterology. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Furness, J.B.; Callaghan, B.P.; Rivera, L.R.; Cho, H.J. The enteric nervous system and gastrointestinal innervation: Integrated local and central control. Adv. Exp. Med. Biol. 2014, 817, 39–71. [Google Scholar] [CrossRef] [PubMed]
- Furness, J.B.; Rivera, L.R.; Cho, H.J.; Bravo, D.M.; Callaghan, B. The gut as a sensory organ. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 729–740. [Google Scholar] [CrossRef] [PubMed]
- McVey Neufeld, K.A.; Mao, Y.K.; Bienenstock, J.; Foster, J.A.; Kunze, W.A. The microbiome is essential for normal gut intrinsic primary afferent neuron excitability in the mouse. Neurogastroenterol. Motil. 2013, 25, 183-e88. [Google Scholar] [CrossRef] [PubMed]
- McVey Neufeld, K.A.; Perez-Burgos, A.; Mao, Y.K.; Bienenstock, J.; Kunze, W.A. The gut microbiome restores intrinsic and extrinsic nerve function in germ-free mice accompanied by changes in calbindin. Neurogastroenterol. Motil. 2015, 27, 627–636. [Google Scholar] [CrossRef]
- Aktar, R.; Parkar, N.; Stentz, R.; Baumard, L.; Parker, A.; Goldson, A.; Brion, A.; Carding, S.; Blackshaw, A.; Peiris, M. Human resident gut microbe Bacteroides thetaiotaomicron regulates colonic neuronal innervation and neurogenic function. Gut Microbes 2020, 11, 1745–1757. [Google Scholar] [CrossRef]
- Chandrasekharan, B.; Saeedi, B.J.; Alam, A.; Houser, M.; Srinivasan, S.; Tansey, M.; Jones, R.; Nusrat, A.; Neish, A.S. Interactions Between Commensal Bacteria and Enteric Neurons, via FPR1 Induction of ROS, Increase Gastrointestinal Motility in Mice. Gastroenterology 2019, 157, 179–192.e2. [Google Scholar] [CrossRef]
- Hyland, N.P.; Cryan, J.F. Microbe-host interactions: Influence of the gut microbiota on the enteric nervous system. Dev. Biol. 2016, 417, 182–187. [Google Scholar] [CrossRef]
- Clarke, G.; Grenham, S.; Scully, P.; Fitzgerald, P.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 2013, 18, 666–673. [Google Scholar] [CrossRef]
- Desbonnet, L.; Clarke, G.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. Microbiota is essential for social development in the mouse. Mol. Psychiatry 2014, 19, 146–148. [Google Scholar] [CrossRef] [PubMed]
- Diaz Heijtz, R.; Wang, S.; Anuar, F.; Qian, Y.; Bjorkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, K.M.; Kang, N.; Bienenstock, J.; Foster, J.A. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol. Motil. 2011, 23, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Sudo, N.; Chida, Y.; Aiba, Y.; Sonoda, J.; Oyama, N.; Yu, X.N.; Kubo, C.; Koga, Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J. Physiol. 2004, 558, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Mao, Y.K.; Diorio, C.; Wang, L.; Huizinga, J.D.; Bienenstock, J.; Kunze, W. Lactobacillus reuteri ingestion and IK(Ca) channel blockade have similar effects on rat colon motility and myenteric neurones. Neurogastroenterol. Motil. 2009, 22, 98–107.e3. [Google Scholar] [CrossRef]
- Wu, R.Y.; Pasyk, M.; Wang, B.; Forsythe, P.; Bienenstock, J.; Mao, Y.K.; Sharma, P.; Stanisz, A.M.; Kunze, W.A. Spatiotemporal maps reveal regional differences in the effects on gut motility for Lactobacillus reuteri and rhamnosus strains. Neurogastroenterol. Motil. 2013, 25, e205–e214. [Google Scholar] [CrossRef]
- Liu, C.Y.; Mueller, M.H.; Rogler, G.; Grundy, D.; Kreis, M.E. Differential afferent sensitivity to mucosal lipopolysaccharide from Salmonella typhimurium and Escherichia coli in the rat jejunum. Neurogastroenterol. Motil. 2009, 21, 1335–1342. [Google Scholar] [CrossRef]
- Ochoa-Cortes, F.; Ramos-Lomas, T.; Miranda-Morales, M.; Spreadbury, I.; Ibeakanma, C.; Barajas-Lopez, C.; Vanner, S. Bacterial cell products signal to mouse colonic nociceptive dorsal root ganglia neurons. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G723–G732. [Google Scholar] [CrossRef]
- Mao, Y.K.; Kasper, D.L.; Wang, B.; Forsythe, P.; Bienenstock, J.; Kunze, W.A. Bacteroides fragilis polysaccharide A is necessary and sufficient for acute activation of intestinal sensory neurons. Nat. Commun. 2013, 4, 1465. [Google Scholar] [CrossRef]
- Chiu, I.M.; Heesters, B.A.; Ghasemlou, N.; Von Hehn, C.A.; Zhao, F.; Tran, J.; Wainger, B.; Strominger, A.; Muralidharan, S.; Horswill, A.R.; et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature 2013, 501, 52–57. [Google Scholar] [CrossRef]
- Uhlig, F.; Grundy, L.; Garcia-Caraballo, S.; Brierley, S.M.; Foster, S.J.; Grundy, D. Identification of a Quorum Sensing-Dependent Communication Pathway Mediating Bacteria-Gut-Brain Cross Talk. iScience 2020, 23, 101695. [Google Scholar] [CrossRef] [PubMed]
- Song, O.R.; Kim, H.B.; Jouny, S.; Ricard, I.; Vandeputte, A.; Deboosere, N.; Marion, E.; Queval, C.J.; Lesport, P.; Bourinet, E.; et al. A bacterial toxin with analgesic properties: Hyperpolarization of DRG neurons by mycolactone. Toxins 2017, 9, 227. [Google Scholar] [CrossRef] [PubMed]
- Sessenwein, J.L.; Baker, C.C.; Pradhananga, S.; Maitland, M.E.; Petrof, E.O.; Allen-Vercoe, E.; Noordhof, C.; Reed, D.E.; Vanner, S.J.; Lomax, A.E. Protease-Mediated Suppression of DRG Neuron Excitability by Commensal Bacteria. J. Neurosci. 2017, 37, 11758–11768. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Berezo, T.; Pujo, J.; Martin, P.; Le Faouder, P.; Galano, J.M.; Guy, A.; Knauf, C.; Tabet, J.C.; Tronnet, S.; Barreau, F.; et al. Identification of an analgesic lipopeptide produced by the probiotic Escherichia coli strain Nissle. Nat. Commun. 2017, 8, 1314. [Google Scholar] [CrossRef] [PubMed]
- Fulling, C.; Dinan, T.G.; Cryan, J.F. Gut Microbe to Brain Signaling: What Happens in Vagus. Neuron 2019, 101, 998–1002. [Google Scholar] [CrossRef] [PubMed]
- LaSarre, B.; Federle, M.J. Exploiting Quorum Sensing To Confuse Bacterial Pathogens. Microbiol. Mol. Biol. Rev. 2013, 77, 73–111. [Google Scholar] [CrossRef] [PubMed]
- Wagle, B.R.; Donoghue, A.M.; Shrestha, S.; Upadhyaya, I.; Arsi, K.; Gupta, A.; Liyanage, R.; Rath, N.C.; Donoghue, D.J.; Upadhyay, A. Carvacrol attenuates Campylobacter jejuni colonization factors and proteome critical for persistence in the chicken gut. Poult. Sci. 2020, 99, 4566–4577. [Google Scholar] [CrossRef] [PubMed]
- Joshi, J.R.; Khazanov, N.; Senderowitz, H.; Burdman, S.; Lipsky, A.; Yedidia, I. Plant phenolic volatiles inhibit quorum sensing in pectobacteria and reduce their virulence by potential binding to ExpI and ExpR proteins. Sci. Rep. 2016, 6, 38126. [Google Scholar] [CrossRef]
- Landman, C.; Grill, J.P.; Mallet, J.M.; Marteau, P.; Humbert, L.; Le Balc’h, E.; Maubert, M.A.; Perez, K.; Chaara, W.; Brot, L.; et al. Inter-kingdom effect on epithelial cells of the N-Acyl homoserine lactone 3-oxo-C12:2, a major quorum-sensing molecule from gut microbiota. PLoS ONE 2018, 13, e0202587. [Google Scholar] [CrossRef]
- Raut, N.; Pasini, P.; Daunert, S. Deciphering bacterial universal language by detecting the quorum sensing signal, autoinducer-2, with a whole-cell sensing system. Anal. Chem. 2013, 85, 9604–9609. [Google Scholar] [CrossRef]
- Golinska, E.; Tomusiak, A.; Gosiewski, T.; Wiecek, G.; Machul, A.; Mikolajczyk, D.; Bulanda, M.; Heczko, P.B.; Strus, M. Virulence factors of Enterococcus strains isolated from patients with inflammatory bowel disease. World J. Gastroenterol. 2013, 19, 3562–3572. [Google Scholar] [CrossRef] [PubMed]
- Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Joo, H.S.; Villaruz, A.E.; Glose, K.A.; Fisher, E.L.; Hunt, R.L.; Li, B.; et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 2018, 562, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, L.R.; Jenabzadeh, P. IBD and Bile Acid Absorption: Focus on Pre-clinical and Clinical Observations. Front. Physiol. 2020, 11, 564. [Google Scholar] [CrossRef] [PubMed]
- Press, A.; Hauptmann, I.; Hauptmann, L.; Fuchs, B.; Ewe, K.; Ramadori, G. Gastrointestinal pH profiles in patients with inflammatory bowel disease. Aliment. Pharm. 1998, 12, 673–678. [Google Scholar] [CrossRef]
- Verbeke, F.; De Craemer, S.; Debunne, N.; Janssens, Y.; Wynendaele, E.; Van de Wiele, C.; De Spiegeleer, B. Peptides as Quorum Sensing Molecules: Measurement Techniques and Obtained Levels In vitro and In vivo. Front. Neurosci. 2017, 11, 183. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uhlig, F.; Hyland, N.P. Making Sense of Quorum Sensing at the Intestinal Mucosal Interface. Cells 2022, 11, 1734. https://doi.org/10.3390/cells11111734
Uhlig F, Hyland NP. Making Sense of Quorum Sensing at the Intestinal Mucosal Interface. Cells. 2022; 11(11):1734. https://doi.org/10.3390/cells11111734
Chicago/Turabian StyleUhlig, Friederike, and Niall P. Hyland. 2022. "Making Sense of Quorum Sensing at the Intestinal Mucosal Interface" Cells 11, no. 11: 1734. https://doi.org/10.3390/cells11111734
APA StyleUhlig, F., & Hyland, N. P. (2022). Making Sense of Quorum Sensing at the Intestinal Mucosal Interface. Cells, 11(11), 1734. https://doi.org/10.3390/cells11111734






