Triticale Green Plant Regeneration Is Due to DNA Methylation and Sequence Changes Affecting Distinct Sequence Contexts in the Presence of Copper Ions in Induction Medium
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Cultures
2.2. DNA Extraction and metAFLP Assay
2.3. Quantifying Variation
2.4. Statistics
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 2,4-D | 2,4-Dichlorophenoxyacetic Acid |
| AOX | Alternative Oxidase |
| COX | Cytochrome c Oxidase |
| DMV | Demethylation |
| DNMT | DNA Methyltransferase |
| DNMV | De Novo Methylation |
| ETC | Electron Transport Chain |
| GP | Green Plants |
| GSH | Glutathione |
| IAA | Indole-3-Acetic Acid |
| IM | Induction Medium |
| metAFLP | Methylatio-Sensitive Amplified Fragment Length Polymorphism |
| ROS | Reactive Oxygen Species |
| SAM | S-Adenosyl-L-Methionine |
| SV | Sequence Variation |
| TCIV | Tissue Culture-Induced Variation |
References
- Orłowska, R.; Pachota, K.A.; Machczyńska, J.; Niedziela, A.; Makowska, K.; Zimny, J.; Bednarek, P.T. Improvement of anther cultures conditions using the Taguchi method in three cereal crops. Electron. J. Biotechnol. 2020, 43, 8–15. [Google Scholar] [CrossRef]
- Warchoł, M.; Juzoń, K.; Dziurka, K.; Czyczyło-Mysza, I.; Kapłoniak, K.; Marcińska, I.; Skrzypek, E. The effect of zinc, copper, and silver ions on oat (Avena sativa L.) androgenesis. Plants 2021, 10, 248. [Google Scholar] [CrossRef]
- Sharma, S.S.; Dietz, K.-J. The relationship between metal toxicity and cellular redox imbalance. Trends Plant Sci. 2009, 14, 43–50. [Google Scholar] [CrossRef]
- Pilon, M.; Cohu, C.M.; Ravet, K.; Abdel-Ghany, S.E.; Gaymard, F. Essential transition metal homeostasis in plants. Curr. Opin. Plant Biol. 2009, 12, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-Y.; Chang, C.-L.; Wang, Y.-N.; Fu, L.-M. Microfluidic Mixing: A Review. Int. J. Mol. Sci. 2011, 12, 3263–3287. [Google Scholar] [CrossRef] [PubMed]
- Vanlerberghe, G.C. Alternative oxidase: A mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 2013, 14, 6805–6847. [Google Scholar] [CrossRef] [PubMed]
- Moller, I.M. Plant mitochondria and oxidative stress: Electron transport, nadph turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 561–591. [Google Scholar] [CrossRef] [PubMed]
- Garcia, L.; Welchen, E.; Gonzalez, D.H. Mitochondria and copper homeostasis in plants. Mitochondrion 2014, 19, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Manceau, A.; Simionovici, A.; Lanson, M.; Perrin, J.; Tucoulou, R.; Bohic, S.; Fakra, S.C.; Marcus, M.A.; Bedell, J.P.; Nagy, K.L. Thlaspi arvense binds Cu(II) as a bis-(l-histidinato) complex on root cell walls in an urban ecosystem. Metallomics 2013, 5, 1674–1684. [Google Scholar] [CrossRef]
- Gizatullin, A.; Becker, J.; Islamov, D.; Serov, N.; Schindler, S.; Klimovitskii, A.; Shtyrlin, V. Synthesis and structure of a complex of copper(I) with l-cysteine and chloride ions containing Cu12S6 nanoclusters. Acta Crystallographica Section E 2021, 77, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Burkhead, J.L.; Gogolin Reynolds, K.A.; Abdel-Ghany, S.E.; Cohu, C.M.; Pilon, M. Copper homeostasis. New Phytol. 2009, 182, 799–816. [Google Scholar] [CrossRef]
- Mäkinen, K.; De, S. The significance of methionine cycle enzymes in plant virus infections. Curr. Opin. Plant Biol. 2019, 50, 67–75. [Google Scholar] [CrossRef]
- Roje, S. S-Adenosyl-L-methionine: Beyond the universal methyl group donor. Phytochemistry 2006, 67, 1686–1698. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zou, T.; McCormick, S. S-Adenosylmethionine synthetase 3 is important for pollen tube growth. Plant Physiol. 2016, 172, 244–253. [Google Scholar] [CrossRef]
- Heidari, P.; Mazloomi, F.; Nussbaumer, T.; Barcaccia, G. Insights into the SAM synthetase gene family and its roles in tomato seedlings under abiotic stresses and hormone treatments. Plants 2020, 9, 586. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Glutathione in plants: Biosynthesis and physiological role in environmental stress tolerance. Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2017, 23, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Giovanelli, J.; Mudd, S.H.; Datko, A.H. Homocysteine biosynthesis in green plants. Physiological importance of the transsulfuration pathway in Chlorella sorokiniana growing under steady state conditions with limiting sulfate. J. Biol. Chem. 1978, 253, 5665–5677. [Google Scholar] [CrossRef]
- Meng, J.; Wang, L.; Wang, J.; Zhao, X.; Cheng, J.; Yu, W.; Jin, D.; Li, Q.; Gong, Z. Methionine adenosyltransferase4 mediates DNA and histone methylation. Plant Physiol. 2018, 177, 652–670. [Google Scholar] [CrossRef]
- Solís, M.-T.; Rodríguez-Serrano, M.; Meijón, M.; Cañal, M.-J.; Cifuentes, A.; Risueño, M.C.; Testillano, P.S. DNA methylation dynamics and MET1a-like gene expression changes during stress-induced pollen reprogramming to embryogenesis. J. Exp. Bot. 2012, 63, 6431–6444. [Google Scholar] [CrossRef]
- He, X.-J.; Chen, T.; Zhu, J.-K. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011, 21, 442–465. [Google Scholar] [CrossRef]
- Kumar, S.; Chinnusamy, V.; Mohapatra, T. Epigenetics of modified DNA bases: 5-methylcytosine and beyond. Front. Genet. 2018, 9, 640. [Google Scholar] [CrossRef]
- Lee, D.H.; O’Connor, T.R.; Pfeifer, G.P. Oxidative DNA damage induced by copper and hydrogen peroxide promotes CG→TT tandem mutations at methylated CpG dinucleotides in nucleotide excision repair-deficient cells. Nucleic Acids Res. 2002, 30, 3566–3573. [Google Scholar] [CrossRef]
- Kusmartsev, V.; Drożdż, M.; Schuster-Böckler, B.; Warnecke, T. Cytosine methylation affects the mutability of neighboring nucleotides in germline and soma. Genetics 2020, 214, 809–823. [Google Scholar] [CrossRef]
- Orłowska, R.; Zimny, J.; Bednarek, P.T. Copper ions induce DNA sequence variation in zygotic embryo culture-derived barley regenerants. Front. Plant Sci. 2021, 11, 614837. [Google Scholar] [CrossRef]
- Bednarek, P.T.; Zebrowski, J.; Orłowska, R. Exploring the biochemical origin of DNA sequence variation in barley plants regenerated via in vitro anther culture. Int. J. Mol. Sci. 2020, 21, 5770. [Google Scholar] [CrossRef]
- Bednarek, P.T.; Orłowska, R. Time of in vitro anther culture may moderate action of copper and silver ions that affect the relationship between DNA methylation change and the yield of barley green regenerants. Plants 2020, 9, 1064. [Google Scholar] [CrossRef]
- Ciriolo, M.R.; Civitareale, P.; Carrì, M.T.; De Martino, A.; Galiazzo, F.; Rotilio, G. Purification and characterization of Ag,Zn-superoxide dismutase from Saccharomyces cerevisiae exposed to silver. J. Biol. Chem. 1994, 269, 25783–25787. [Google Scholar] [CrossRef]
- Grozeva, S.; Nankar, A.N. Effect of incubation period and culture medium on pepper anther culture. Indian J. Biotechnol. 2020, 19, 53–59. [Google Scholar]
- Sato, S.; Katoh, N.; Iwai, S.; Hagimori, M. Effect of low temperature pretreatment of buds or inflorescence on isolated microspore culture in Brassica rapa (syn. B. campestris). Breed. Sci. 2002, 52, 23–26. [Google Scholar] [CrossRef]
- Mikuła, A.; Tomiczak, K.; Rybczynski, J.J. Cryopreservation enhances embryogenic capacity of Gentiana cruciata L. suspension culture and maintains (epi)genetic uniformity of regenerants. Plant Cell Rep. 2011, 30, 565–574. [Google Scholar] [CrossRef]
- Machczyńska, J.; Orłowska, R.; Zimny, J.; Bednarek, P.T. Extended metAFLP approach in studies of the tissue culture induced variation (TCIV) in case of triticale. Mol. Breed. 2014, 34, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Orłowska, R.; Bednarek, P.T. Precise evaluation of tissue culture-induced variation during optimisation of in vitro regeneration regime in barley. Plant Mol. Biol. 2020, 103, 33–50. [Google Scholar] [CrossRef]
- Fiuk, A.; Bednarek, P.T.; Rybczyński, J.J. Flow cytometry, HPLC-RP, and metAFLP analyses to assess genetic variability in somatic embryo-derived plantlets of Gentiana pannonica Scop. Plant Mol. Biol. Report. 2010, 28, 413–420. [Google Scholar] [CrossRef]
- Śliwińska, A.A.; Białek, A.; Orłowska, R.; Mańkowski, D.; Sykłowska-Baranek, K.; Pietrosiuk, A. Comparative study of the genetic and biochemical variability of Polyscias filicifolia (Araliaceae) regenerants obtained by indirect and direct somatic embryogenesis as a source of triterpenes. Int. J. Mol. Sci. 2021, 22, 5752. [Google Scholar] [CrossRef]
- Hayes, A.F. Introduction to Mediation, Moderation, and Conditional Process Analysis. A Regression Bases Approach; A Division of Guilford Publications, Inc.: New York, NY, USA, 2018; p. 507. [Google Scholar]
- Bednarek, P.T.; Orłowska, R.; Mańkowski, D.R.; Oleszczuk, S.; Zebrowski, J. Structural Equation Modeling (SEM) analysis of sequence variation and green plant regeneration via anther culture in barley. Cells 2021, 10, 2774. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.J.; Xu, J. Increasing differentiation frequencies in wheat pollen callus. In Cell and Tissue Culture Techniques for Cereal Crop Improvement; Hu, H., Vega, M.R., Eds.; Science Press: Beijing, China, 1983; p. 431. [Google Scholar]
- Chu, C.C. The N6 medium and its applications to anther culture of cereal crops. In Proc. Symp. Plant Tissue Culture; Hu, H., Ed.; Science Press: Peking, China, 1978; pp. 45–50. [Google Scholar]
- Bednarek, P.T.; Orłowska, R.; Koebner, R.M.D.; Zimny, J. Quantification of the tissue-culture induced variation in barley (Hordeum vulgare L.). BMC Plant Biol. 2007, 7, 10. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Faul, F.; Erdfelder, E.; Buchner, A.; Lang, A.-G. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav. Res. Methods 2009, 41, 1149–1160. [Google Scholar] [CrossRef] [PubMed]
- Addinsoft. XLSTAT Statistical and Data Analysis Solution. New York, USA. 2020. Available online: https://www.xlstat.com (accessed on 20 December 2021).
- Yang, S.F.; Hoffman, N.E. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 1984, 35, 155–189. [Google Scholar] [CrossRef]
- Bednarek, P.T.; Orłowska, R. CG demethylation leads to sequence mutations in an anther culture of barley due to the presence of Cu, Ag ions in the medium and culture time. Int. J. Mol. Sci. 2020, 21, 4401. [Google Scholar] [CrossRef]
- Festa, R.A.; Thiele, D.J. Copper: An essential metal in biology. Curr. Biol. 2011, 8, 877–883. [Google Scholar] [CrossRef]
- Cong, W.; Miao, Y.; Xu, L.; Zhang, Y.; Yuan, C.; Wang, J.; Zhuang, T.; Lin, X.; Jiang, L.; Wang, N.; et al. Transgenerational memory of gene expression changes induced by heavy metal stress in rice (Oryza sativa L.). BMC Plant Biol. 2019, 19, 282. [Google Scholar] [CrossRef]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204. [Google Scholar] [CrossRef] [PubMed]
- Kankel, M.W.; Ramsey, D.E.; Stokes, T.L.; Flowers, S.K.; Haag, J.R.; Jeddeloh, J.A.; Riddle, N.C.; Verbsky, M.L.; Richards, E.J. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 2003, 163, 1109–1122. [Google Scholar] [CrossRef]
- Bray, C.M.; West, C.E. DNA repair mechanisms in plants: Crucial sensors and effectors for the maintenance of genome integrity. New Phytol. 2005, 168, 511–528. [Google Scholar] [CrossRef] [PubMed]
- Machczyńska, J.; Orłowska, R.; Mańkowski, D.R.; Zimny, J.; Bednarek, P.T. DNA methylation changes in triticale due to in vitro culture plant regeneration and consecutive reproduction. Plant Cell Tissue Organ Cult. 2014, 119, 289–299. [Google Scholar] [CrossRef]
- Orłowska, R.; Machczyńska, J.; Oleszczuk, S.; Zimny, J.; Bednarek, P.T. DNA methylation changes and TE activity induced in tissue cultures of barley (Hordeum vulgare L.). J. Biol. Res. 2016, 23, 19. [Google Scholar] [CrossRef]
- Valentine, J.S.; de Freitas, D.M. Copper-zinc superoxide dismutase: A unique biological “ligand” for bioinorganic studies. J. Chem. Educ. 1985, 62, 990. [Google Scholar] [CrossRef]
- Roe, J.A.; Peoples, R.; Scholler, D.M.; Valentine, J.S. Silver-binding properties of bovine cuprozinc superoxide dismutase and the overall stability of selected metalion derivatives. J. Am. Chem. Soc. 1990, 112, 1538–1545. [Google Scholar] [CrossRef]
- Çekiç, F.O.; Ekinci, S.; Inal, M.; Ozakca, D. Silver nanoparticles induced genotoxicity and oxidative stress in tomato plants. Turk. J. Biol. 2017, 41, 700–707. [Google Scholar] [CrossRef]
- Bairu, M.W.; Fennell, C.W.; van Staden, J. The effect of plant growth regulators on somaclonal variation in Cavendish banana (Musa AAA cv. ‘Zelig’). Sci. Hortic. 2006, 108, 347–351. [Google Scholar] [CrossRef]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef]
- Schmitz, R.J.; Schultz, M.D.; Lewsey, M.G.; O’Malley, R.C.; Urich, M.A.; Libiger, O.; Schork, N.J.; Ecker, J.R. Transgenerational epigenetic instability is a source of novel methylation variants. Science 2011, 334, 369–373. [Google Scholar] [CrossRef]
- Becker, C.; Hagmann, J.; Müller, J.; Koenig, D.; Stegle, O.; Borgwardt, K.; Weigel, D. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 2011, 480, 245–249. [Google Scholar] [CrossRef]
- Grin, I.; Ishchenko, A.A. An interplay of the base excision repair and mismatch repair pathways in active DNA demethylation. Nucleic Acids Res. 2016, 44, 3713–3727. [Google Scholar] [CrossRef]
- Drohat, A.C.; Coey, C.T. Role of base excision “repair” enzymes in erasing epigenetic marks from DNA. Chem. Rev. 2016, 116, 12711–12729. [Google Scholar] [CrossRef] [PubMed]
- Klungland, A.; Robertson, A.B. Oxidized C5-methyl cytosine bases in DNA: 5-Hydroxymethylcytosine; 5-formylcytosine; and 5-carboxycytosine. Free Radic. Biol. Med. 2017, 107, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Xiong, J.; Jiang, H.-P.; Zheng, S.-J.; Feng, Y.-Q.; Yuan, B.-F. Determination of Oxidation Products of 5-Methylcytosine in Plants by Chemical Derivatization Coupled with Liquid Chromatography/Tandem Mass Spectrometry Analysis. Anal. Chem. 2014, 86, 7764–7772. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Jordan, W.T.; Shi, X.; Hu, L.; He, C.; Schmitz, R.J. TET-mediated epimutagenesis of the Arabidopsis thaliana methylome. Nat. Commun. 2018, 9, 895. [Google Scholar] [CrossRef]
- Shi, D.Q.; Ali, I.; Tang, J.; Yang, W.C. New Insights into 5hmC DNA Modification: Generation, Distribution and Function. Front. Genet. 2017, 8, 100. [Google Scholar] [CrossRef]
- Domb, K.; Katz, A.; Yaari, R.; Kaisler, E.; Nguyen, V.H.; Hong, U.V.T.; Griess, O.; Heskiau, K.G.; Ohad, N.; Zemach, A. Non-CG methylation is superior to CG methylation in genome regulation. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zemach, A.; Kim, M.Y.; Silva, P.; Rodrigues, J.A.; Dotson, B.; Brooks, M.D.; Zilberman, D. Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. USA 2010, 107, 18729–18734. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; McDaniel, I.E.; Silva, P.; Zilberman, D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 2010, 328, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Kim, M.Y.; Vickers, M.; Park, J.-S.; Hyun, Y.; Okamoto, T.; Zilberman, D.; Fischer, R.L.; Feng, X.; Choi, Y. DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc. Natl. Acad. Sci. USA 2016, 113, 15138–15143. [Google Scholar] [CrossRef]
- Nebert, D.W.; Vasiliou, V. Analysis of the glutathione S-transferase (GST) gene family. Hum. Genom. 2004, 1, 460. [Google Scholar] [CrossRef] [PubMed]
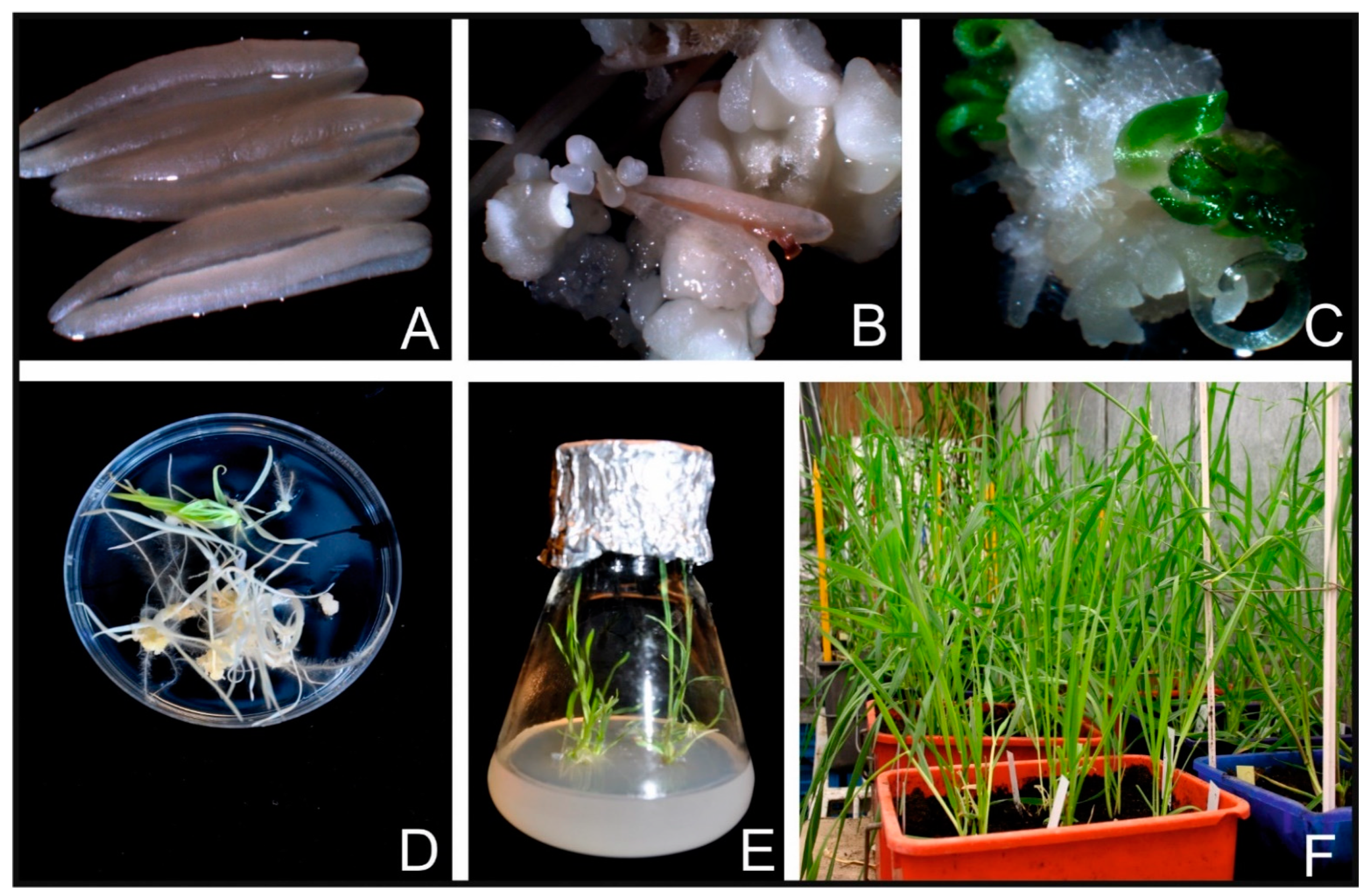
| No. | Trial | metAFLP Characteristics 1 (%) | In Vitro Conditions | GP | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SV | DNMV | DMV | Sequence CHH | Sequence CHG | Sequence CG | ||||||||||||
| SV | DNMV | DMV | SV | DNMV | DMV | SV | DNMV | DMV | Cu(II) (μM) | Ag(I) (μM) | Time (Days) | ||||||
| 1 | M1 | 43.30 | 3.34 | 5.02 | 8.66 | 0.37 | 0.74 | 23.34 | 1.62 | 2.92 | 11.35 | 1.33 | 1.33 | 0.1 | 0 | 35 | 0.91 |
| 2 | M1 | 43.62 | 3.18 | 5.01 | 8.66 | 0.37 | 0.74 | 23.49 | 1.62 | 2.92 | 11.51 | 1.17 | 1.33 | 0.1 | 0 | 35 | 0.91 |
| 3 | M1 | 43.61 | 3.18 | 5.18 | 8.66 | 0.37 | 0.74 | 23.49 | 1.62 | 2.92 | 11.51 | 1.17 | 1.50 | 0.1 | 0 | 35 | 0.91 |
| 4 | M1 | 43.61 | 3.18 | 5.18 | 8.66 | 0.37 | 0.74 | 23.49 | 1.62 | 2.92 | 11.51 | 1.17 | 1.50 | 0.1 | 0 | 35 | 0.91 |
| 5 | M1 | 43.62 | 3.18 | 5.01 | 8.66 | 0.37 | 0.74 | 23.49 | 1.62 | 2.92 | 11.51 | 1.17 | 1.33 | 0.1 | 0 | 35 | 0.91 |
| 6 | M2 | 43.61 | 3.18 | 5.18 | 8.66 | 0.37 | 0.74 | 23.49 | 1.62 | 2.92 | 11.51 | 1.17 | 1.50 | 0.1 | 10 | 42 | 0.87 |
| 7 | M2 | 43.45 | 3.18 | 5.18 | 8.66 | 0.37 | 0.74 | 23.34 | 1.62 | 2.92 | 11.51 | 1.17 | 1.50 | 0.1 | 10 | 42 | 0.87 |
| 8 | M2 | 43.33 | 3.17 | 5.00 | 8.52 | 0.36 | 0.73 | 23.34 | 1.62 | 2.92 | 11.51 | 1.17 | 1.33 | 0.1 | 10 | 42 | 0.87 |
| 9 | M2 | 43.46 | 3.01 | 5.18 | 8.50 | 0.37 | 0.74 | 23.49 | 1.62 | 2.92 | 11.51 | 1.00 | 1.50 | 0.1 | 10 | 42 | 0.87 |
| 10 | M3 | 43.61 | 3.18 | 5.18 | 8.64 | 0.37 | 0.92 | 23.49 | 1.62 | 2.92 | 11.51 | 1.17 | 1.33 | 0.1 | 60 | 49 | 1.52 |
| 11 | M3 | 43.61 | 3.17 | 5.35 | 8.64 | 0.37 | 0.92 | 23.49 | 1.62 | 2.92 | 11.51 | 1.17 | 1.50 | 0.1 | 60 | 49 | 1.52 |
| 12 | M3 | 43.97 | 3.19 | 5.37 | 8.79 | 0.37 | 0.93 | 23.69 | 1.62 | 2.92 | 11.52 | 1.17 | 1.50 | 0.1 | 60 | 49 | 1.52 |
| 13 | M3 | 43.81 | 3.19 | 5.37 | 8.79 | 0.37 | 0.93 | 23.53 | 1.62 | 2.92 | 11.52 | 1.17 | 1.50 | 0.1 | 60 | 49 | 1.52 |
| 14 | M3 | 43.81 | 3.36 | 5.04 | 8.79 | 0.56 | 0.75 | 23.53 | 1.62 | 2.92 | 11.53 | 1.17 | 1.34 | 0.1 | 60 | 49 | 1.52 |
| 15 | M4 | 43.49 | 3.53 | 5.04 | 8.76 | 0.75 | 0.75 | 23.37 | 1.62 | 2.92 | 11.37 | 1.17 | 1.34 | 5 | 60 | 42 | 0.71 |
| 16 | M4 | 43.66 | 3.36 | 5.04 | 8.79 | 0.56 | 0.75 | 23.53 | 1.62 | 2.92 | 11.37 | 1.17 | 1.34 | 5 | 60 | 42 | 0.71 |
| 17 | M4 | 43.29 | 3.34 | 5.18 | 8.64 | 0.55 | 0.73 | 23.34 | 1.62 | 2.92 | 11.35 | 1.17 | 1.50 | 5 | 60 | 42 | 0.71 |
| 18 | M4 | 43.13 | 3.34 | 5.18 | 8.64 | 0.55 | 0.73 | 23.18 | 1.62 | 2.92 | 11.35 | 1.17 | 1.50 | 5 | 60 | 42 | 0.71 |
| 19 | M5 | 43.30 | 3.18 | 5.18 | 8.64 | 0.55 | 0.73 | 23.18 | 1.62 | 2.92 | 11.51 | 1.00 | 1.50 | 5 | 0 | 49 | 2.38 |
| 20 | M5 | 43.13 | 3.34 | 5.18 | 8.64 | 0.55 | 0.73 | 23.02 | 1.62 | 2.92 | 11.51 | 1.17 | 1.50 | 5 | 0 | 49 | 2.38 |
| 21 | M5 | 43.49 | 3.53 | 5.04 | 8.76 | 0.75 | 0.75 | 23.21 | 1.62 | 2.92 | 11.53 | 1.17 | 1.34 | 5 | 0 | 49 | 2.38 |
| 22 | M5 | 43.49 | 3.53 | 5.04 | 8.76 | 0.75 | 0.75 | 23.21 | 1.62 | 2.92 | 11.53 | 1.17 | 1.34 | 5 | 0 | 49 | 2.38 |
| 23 | M5 | 43.49 | 3.53 | 5.04 | 8.76 | 0.75 | 0.75 | 23.21 | 1.62 | 2.92 | 11.53 | 1.17 | 1.34 | 5 | 0 | 49 | 2.38 |
| 24 | M5 | 43.32 | 3.53 | 5.21 | 8.76 | 0.75 | 0.75 | 23.06 | 1.62 | 2.92 | 11.52 | 1.17 | 1.50 | 5 | 0 | 49 | 2.38 |
| 25 | M5 | 43.33 | 3.53 | 5.04 | 8.76 | 0.75 | 0.75 | 23.06 | 1.62 | 2.92 | 11.53 | 1.17 | 1.34 | 5 | 0 | 49 | 2.38 |
| 26 | M5 | 43.32 | 3.53 | 5.21 | 8.76 | 0.75 | 0.75 | 23.06 | 1.62 | 2.92 | 11.52 | 1.17 | 1.50 | 5 | 0 | 49 | 2.38 |
| 27 | M5 | 43.48 | 3.53 | 5.20 | 8.76 | 0.75 | 0.75 | 23.21 | 1.62 | 2.92 | 11.52 | 1.17 | 1.50 | 5 | 0 | 49 | 2.38 |
| 28 | M5 | 44.89 | 3.64 | 5.03 | 8.91 | 0.76 | 0.76 | 24.04 | 2.02 | 2.86 | 11.96 | 0.87 | 1.39 | 5 | 0 | 49 | 2.38 |
| 29 | M6 | 43.19 | 3.52 | 5.19 | 8.63 | 0.73 | 0.73 | 23.06 | 1.62 | 2.92 | 11.52 | 1.17 | 1.50 | 5 | 10 | 35 | 1.17 |
| 30 | M6 | 43.19 | 3.52 | 5.19 | 8.63 | 0.73 | 0.73 | 23.06 | 1.62 | 2.92 | 11.52 | 1.17 | 1.50 | 5 | 10 | 35 | 1.17 |
| 31 | M6 | 43.31 | 3.50 | 5.16 | 8.48 | 0.72 | 0.72 | 23.34 | 1.62 | 2.92 | 11.51 | 1.17 | 1.50 | 5 | 10 | 35 | 1.17 |
| 32 | M6 | 43.16 | 3.33 | 5.17 | 8.48 | 0.72 | 0.72 | 23.18 | 1.62 | 2.92 | 11.51 | 1.00 | 1.50 | 5 | 10 | 35 | 1.17 |
| 33 | M6 | 43.16 | 3.33 | 5.00 | 8.50 | 0.54 | 0.72 | 23.18 | 1.62 | 2.92 | 11.51 | 1.17 | 1.33 | 5 | 10 | 35 | 1.17 |
| 34 | M7 | 43.15 | 3.33 | 5.33 | 8.48 | 0.54 | 0.90 | 23.18 | 1.62 | 2.92 | 11.51 | 1.17 | 1.50 | 10 | 10 | 49 | 3.79 |
| 35 | M7 | 43.36 | 3.35 | 5.19 | 8.65 | 0.55 | 0.74 | 23.21 | 1.62 | 2.92 | 11.52 | 1.17 | 1.50 | 10 | 10 | 49 | 3.79 |
| 36 | M7 | 43.20 | 3.35 | 5.19 | 8.65 | 0.55 | 0.74 | 23.06 | 1.62 | 2.92 | 11.52 | 1.17 | 1.50 | 10 | 10 | 49 | 3.79 |
| 37 | M8 | 43.17 | 3.36 | 5.21 | 8.62 | 0.56 | 0.75 | 23.06 | 1.62 | 2.92 | 11.52 | 1.17 | 1.50 | 10 | 60 | 35 | 4.24 |
| 38 | M8 | 42.88 | 3.35 | 5.19 | 8.49 | 0.55 | 0.74 | 22.90 | 1.62 | 2.92 | 11.52 | 1.17 | 1.50 | 10 | 60 | 35 | 4.24 |
| 39 | M8 | 43.04 | 3.35 | 5.19 | 8.49 | 0.55 | 0.74 | 23.06 | 1.62 | 2.92 | 11.52 | 1.17 | 1.50 | 10 | 60 | 35 | 4.24 |
| 40 | M8 | 43.36 | 3.35 | 5.19 | 8.65 | 0.55 | 0.74 | 23.21 | 1.62 | 2.92 | 11.52 | 1.17 | 1.50 | 10 | 60 | 35 | 4.24 |
| 41 | M9 | 42.88 | 3.35 | 5.19 | 8.49 | 0.55 | 0.74 | 22.90 | 1.62 | 2.92 | 11.52 | 1.17 | 1.50 | 10 | 0 | 42 | 6.06 |
| 42 | M9 | 43.01 | 3.36 | 5.21 | 8.49 | 0.55 | 0.74 | 23.03 | 1.63 | 2.94 | 11.52 | 1.17 | 1.50 | 10 | 0 | 42 | 6.06 |
| 43 | M9 | 43.35 | 3.19 | 5.04 | 8.65 | 0.55 | 0.74 | 23.06 | 1.62 | 2.92 | 11.67 | 1.01 | 1.35 | 10 | 0 | 42 | 6.06 |
| 44 | M9 | 42.88 | 3.52 | 5.03 | 8.65 | 0.55 | 0.74 | 22.90 | 1.62 | 2.92 | 11.36 | 1.34 | 1.34 | 10 | 0 | 42 | 6.06 |
| Mean | 43.40 | 3.34 | 5.15 | 8.65 | 0.55 | 0.76 | 23.27 | 1.63 | 2.92 | 11.51 | 1.15 | 14.11 | 2.29 | ||||
| SD | 0.34 | 0.15 | 0.10 | 0.11 | 0.14 | 0.06 | 0.24 | 0.06 | 0.01 | 0.09 | 0.08 | 0.10 | 1.64 | ||||
| Simple Mediation (IV → M → DV) 1 | Statistics | |||
|---|---|---|---|---|
| Path a | Path b and c’ | Path c | Indirect Effect | |
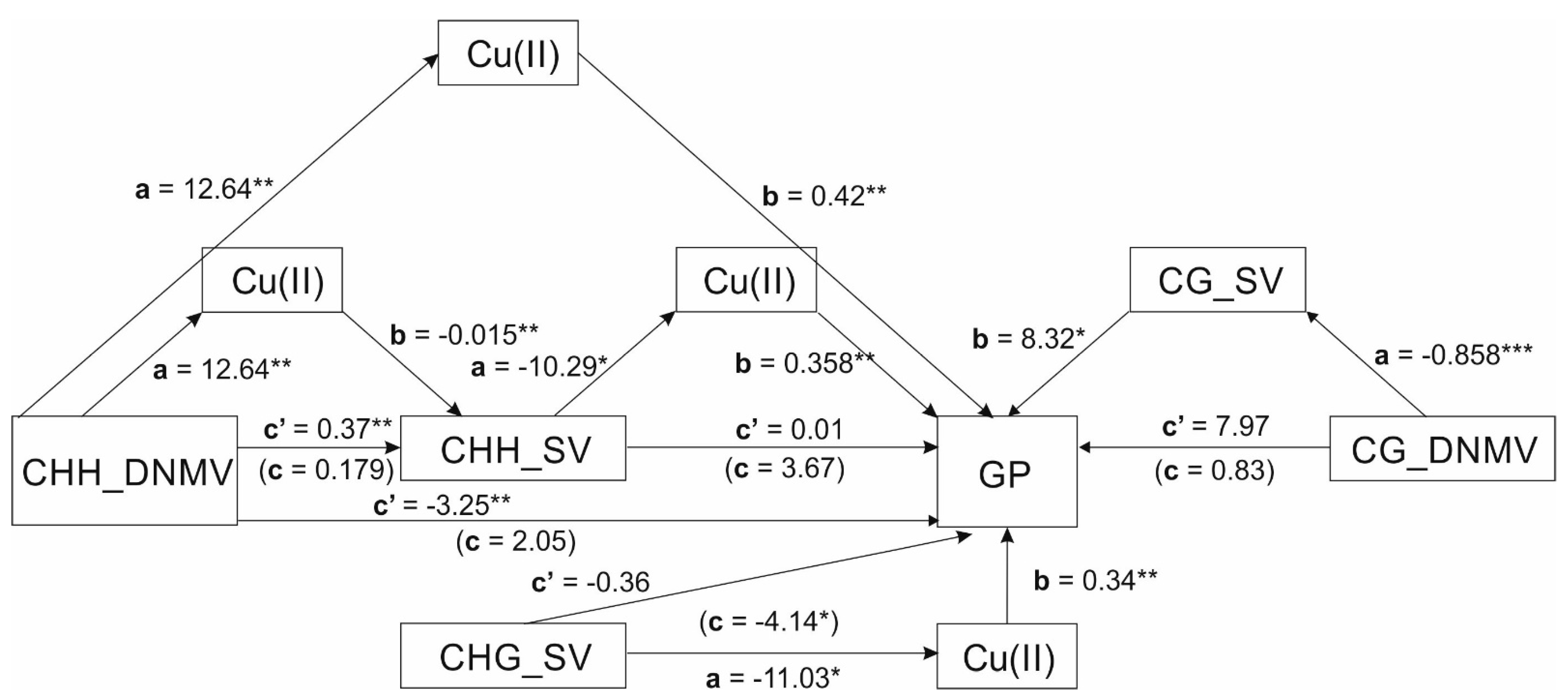
| DNMV → Cu → SV | F(1,42) = 17.4334, MSE = 11.4947, p = 0.0001, R2 = 0.2046: | F(2.41) = 15.8139, MSE = 0.0803, p < 0.0001, R2 = 0.3389 | F(1.42) = 0.0246, MSE = 0.1183, p = 0.8762, R2 = 0.0015 | B = −0.6771, se = 0.2239, CI[−1.1570;−0.2897] |
| (a) B = 11.4812, se = 2.7498, t(42) = 4.1753, p = 0.0001, 95%CI[5.9319;17.0305], β = 0.4523 | (b) B = −0.0590, se = 0.0144, t(41) = −4.0885, p = 0.0002, 95%CI[−0.0881;−0.0298], β = −0.6512 | (c) B = −0.0899, se = 0.5736, t(42) = −0.1567, p = 0.8762, 95%CI[−1.2474;1.0676], β = −0.0391 | ||
| (c’) B = 0.5872, se = 0.6351, t(41) = 0.9245, p = 0.3606, 95%CI[−0.6955;1.8699], β = 0.2554 | ||||
| CHH_DNMV → Cu → CHH_SV | F(1,42) = 26.874, MSE = 10.985, p < 0.0001, R2 = 0.2398 | F(2.41) = 7.7133, MSE = 0.0087, p = 0.0014, R2 = 0.2837 | F(1.42) = 2.4947, MSE = 0.0111, p = 0.1217, R2 = 0.0590 | B = −0.1966, se = 0.0637, CI[−0.3311;−0.0792] |
| (a) B = 12.6377, se = 2.4378, t(42) = 5.184, p < 0.0001, 95%CI[7 0.7179;17.5575], β = 0.4897 | (b) B = −0.0156, se = 0.0041, t(41) = −3.7754, p = 0.0005, 95%CI[−0.0239;−0.0072], β = −0.5438 | (c) B = 0.1792, se = 0.1135, t(42) = 1.5795, p = 0.1217, 95%CI[−0.0498;0.4083], β = 0.2428 | ||
| (c’) B = 0.3758, se = 0.1251, t(41) = 3.0038, p = 0.0045, 95%CI[0.1231;0.628], β = 0.5091 | ||||
| CHH_DNMV → Cu → GP | F(1,42) = 26.8740, MSE = 10.9850, p < 0.0001, R2 = 0.2398 | F(2.41) = 54.9096, MSE = 0.7443, p < 0.0001, R2 = 0.7363 | F(1.42) = 3.9786, MSE = 2.6641, p = 0.0526, R2 = 0.0332 | B = 5.3075, se = 1.2321, CI[3.0808;8.0131] |
| (a) B = 12.6377, se = 2.4378, t(42) = 5.1840, p < 0.0001, 95%CI[7.7179;17.5575], β = 0.4897 | (b) B = 0.4200, se = 0.0468, t(41) = 8.9768, p < 0.0001, 95%CI[0.3255;0.5145], β = 0.9618 | (c) B = 2.0519, se = 1.0287, t(42) = 1.9946, p = 0.0526, 95%CI[−0.0241;4.127], β = 0.1821 | ||
| (c’) B = −3.2556, se = 0.9286, t(41) = −3.5060, p = 0.0011, 95%CI[−5.1309;−1.3803], β = −0.2889 | ||||
| CHH_SV → Cu → GP | F(1,42) = 4.5538, MSE = 13.1980, p = 0.0387, R2 = 0.0867 | F(2.41) = 42.4471, MSE = 0.9234, p < 0.0001, R2 = 0.6729 | F(1.42) = 2.3942, MSE = 2.5955, p = 0.1293, R2 = 0.0581 | B = −3.6875, se = 1.7244, CI[−7.0939;−0.4042] |
| (a) B = −10.2925, se = 4.8232, t(42) = −2.1340, p = 0.0387, 95%CI[−20.0261;−0.5588], β = −0.2944 | (b) B = 0.3583, se = 0.0389, t(41) = 9.2089, p < 0.0001, 95%CI[0.2797;0.4368], β = 0.8205 | (c) B = −3.6780, se = 2.3770, t(42) = −1.5473, p = 0.1293, 95%CI[−8.4752;1.1191], β = −0.2410 | ||
| (c’) B = 0.0094, se = 1.4308, t(41) = 0.0066, p = 0.9948, 95%CI[−2.8802;2.8990], β = 0.0006 | ||||
| CHG_SV → Cu → GP | F(1,42) = 7.1830, MSE = 7.5428, p = 0.0105, R2 = 0.6914 | F(2.41) = 43.5602, MSE = 0.9196, p < 0.0001, R2 = 0.6742 | F(1.42) = 6.1337, MSE = 1.7838, p = 0.0174, R2 = 0.3526 | B = −3.7818, se = 1.1871, CI[−6.5858;−2.0545] |
| (a) B = −11.0336, se = 4.1169, t(42) = −2.6801, p = 0.0105, 95%CI[−19.3419;−2.7254], β = −0.6914 | (b) B = 0.3428, se = 0.0432, t(41) = 7.9321, p < 0.0001, 95%CI[0.2555;0.4300], β = 0.7849 | (c) B = −4.1381, se = 1.67090, t(42) = −2.4766, p = 0.0174, 95%CI[−7.5100;−0.7661], β = 0.5938 | ||
| (c’) B = −0.3563, se = 0.7581, t(41) = −0.4700, p = 0.6409, 95%CI[−1.8874;1.1748], β = −0.0511 | ||||
| CG_DNMV → CG_SV → GP | F(1,42) = 4.5366, MSE = 0.0045, p = 0.0391, R2 = 0.4927 | F(2.41) = 3.3356, MSE = 2.5015, p = 0.0455, R2 = 0.1138 | F(1.42) = 0.0336, MSE = 2.7514, p = 0.8554, R2 = 0.0015 | B = −7.1420, se = 3.3000, CI[−13.8242;−1.1299] |
| (a) B = −0.8583, se = 0.4030, t(42) = −2.1299, p = 0.0391, 95%CI[−1.6716;−0.0451 ], β = −0.7019 | (b) B = 8.3208, se = 4.0779, t(41) = 2.0405, p = 0.0478, 95%CI[0.0853;16.5564], β = 0.4705 | (c) B = 0.8307, se = 4.5290, t(42) = 0.1834, p = 0.8554, 95%CI[−8.3093;9.9707], β = 0.0384 | ||
| (c’) B = 7.9727, se = 4.7204, t(41) = 1.6890, p = 0.0988, 95%CI[−1.5605;17.5058], β = 0.3687 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orłowska, R.; Pachota, K.A.; Androsiuk, P.; Bednarek, P.T. Triticale Green Plant Regeneration Is Due to DNA Methylation and Sequence Changes Affecting Distinct Sequence Contexts in the Presence of Copper Ions in Induction Medium. Cells 2022, 11, 84. https://doi.org/10.3390/cells11010084
Orłowska R, Pachota KA, Androsiuk P, Bednarek PT. Triticale Green Plant Regeneration Is Due to DNA Methylation and Sequence Changes Affecting Distinct Sequence Contexts in the Presence of Copper Ions in Induction Medium. Cells. 2022; 11(1):84. https://doi.org/10.3390/cells11010084
Chicago/Turabian StyleOrłowska, Renata, Katarzyna Anna Pachota, Piotr Androsiuk, and Piotr Tomasz Bednarek. 2022. "Triticale Green Plant Regeneration Is Due to DNA Methylation and Sequence Changes Affecting Distinct Sequence Contexts in the Presence of Copper Ions in Induction Medium" Cells 11, no. 1: 84. https://doi.org/10.3390/cells11010084
APA StyleOrłowska, R., Pachota, K. A., Androsiuk, P., & Bednarek, P. T. (2022). Triticale Green Plant Regeneration Is Due to DNA Methylation and Sequence Changes Affecting Distinct Sequence Contexts in the Presence of Copper Ions in Induction Medium. Cells, 11(1), 84. https://doi.org/10.3390/cells11010084






