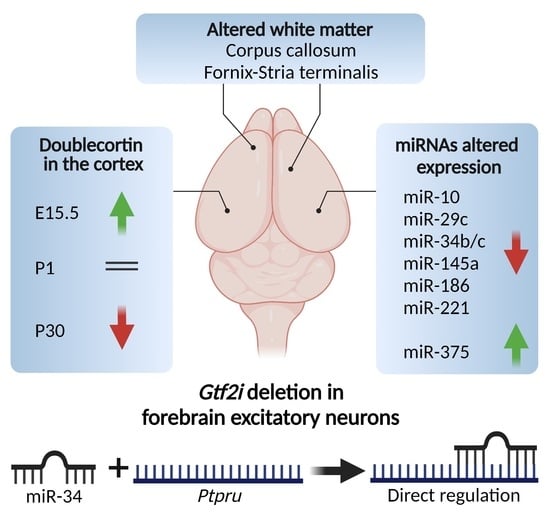
Altered White Matter and microRNA Expression in a Murine Model Related to Williams Syndrome Suggests That miR-34b/c Affects Brain Development via Ptpru and Dcx Modulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Genotyping
2.3. Cortex Extraction
2.4. RNA Extraction
2.5. Small RNA Sequencing
2.6. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.6.1. mRNA Expression
2.6.2. miRNA Expression
2.7. Western Blotting
2.8. Cell Culture
2.9. Cloning and Site-Directed Mutagenesis
2.10. Cloning and Mutagenesis of Human PTPRU 3’UTR
2.11. Cloning and Mutagenesis of Human RHEBL1 3’UTR
2.12. Cloning of Human pre-miR-34c
2.13. Plasmid Transfections
2.14. Dual Luciferase Assay
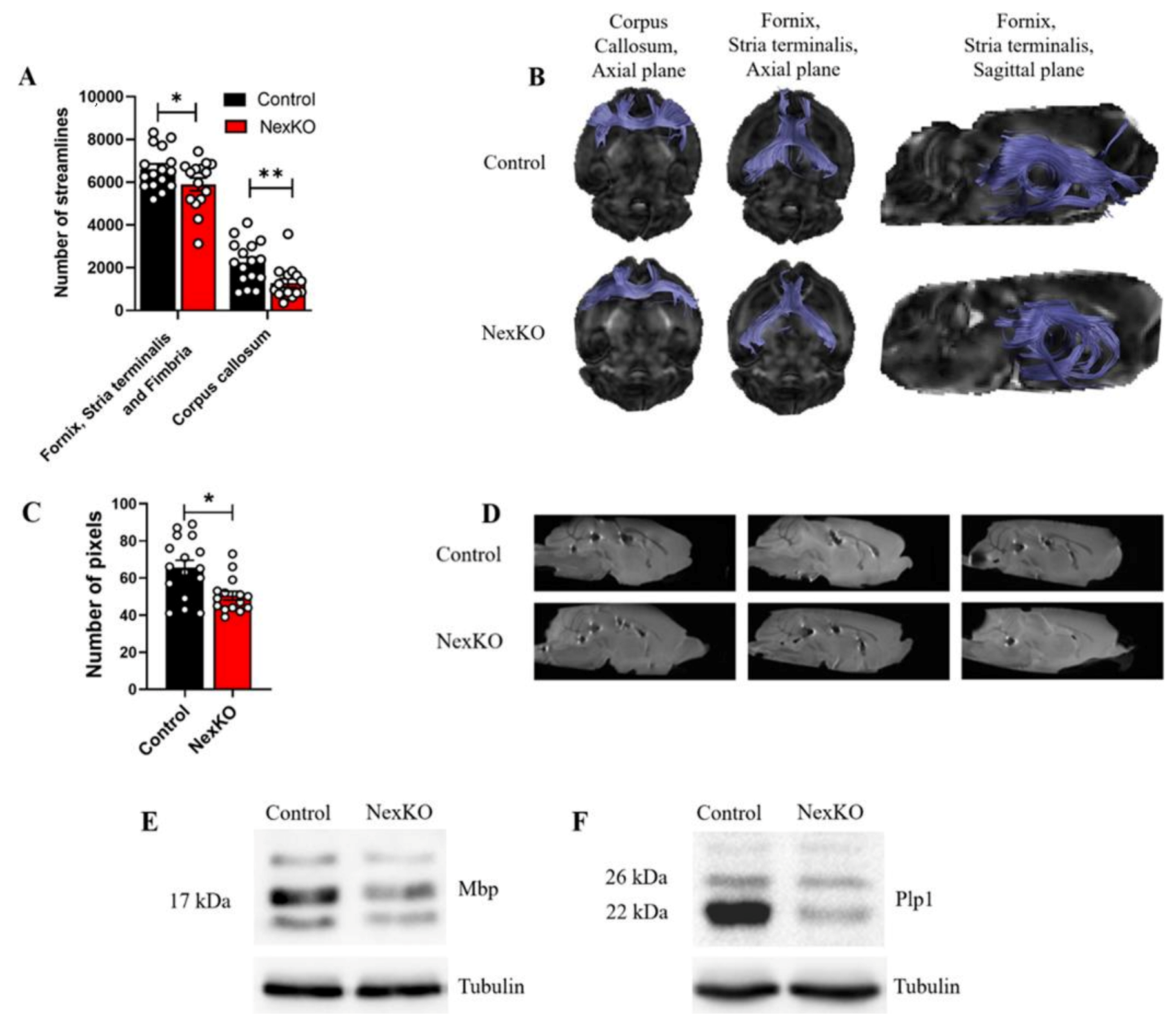
2.15. Diffusion MRI, Fiber Tracking, and Analysis
- Anisotropic smoothing with a 0.48 mm Gaussian kernel. This procedure de-noises the data and benefits the fiber tracking procedure;
- Motion and distortion correction to correct for possible motion- and susceptibility-induced artifacts;
- Transformation into atlas space via nonlinear registration and extraction of atlas space FA and MD per mouse brain;
- Whole-brain fiber tracking with 0.16 mm × 0.16 mm × 0.16 mm seed voxel resolution; minimal FA and stropping criteria for tracking: FA > 0.05; maximal 30° tracking angle was allowed. Tracking step size: 0.16 mm;
- Tracking was conducted from two seed regions of interest (ROIs): genu of the corpus callosum (CC) identified on a mid-sagittal plane; and capturing limbic outputs such as the fimbria/fornix fibers and the stria terminalis in the coronal plane, 0.5 mm before the level of the anterior commissure;
- The reconstructed number of fibers was taken for statistical analysis between groups.
2.16. Statistics
3. Results
3.1. White Matter Microstructure, Tract Connectivity, and Myelin Deficits in the Brains of 1-Month-Old NexKO Mice
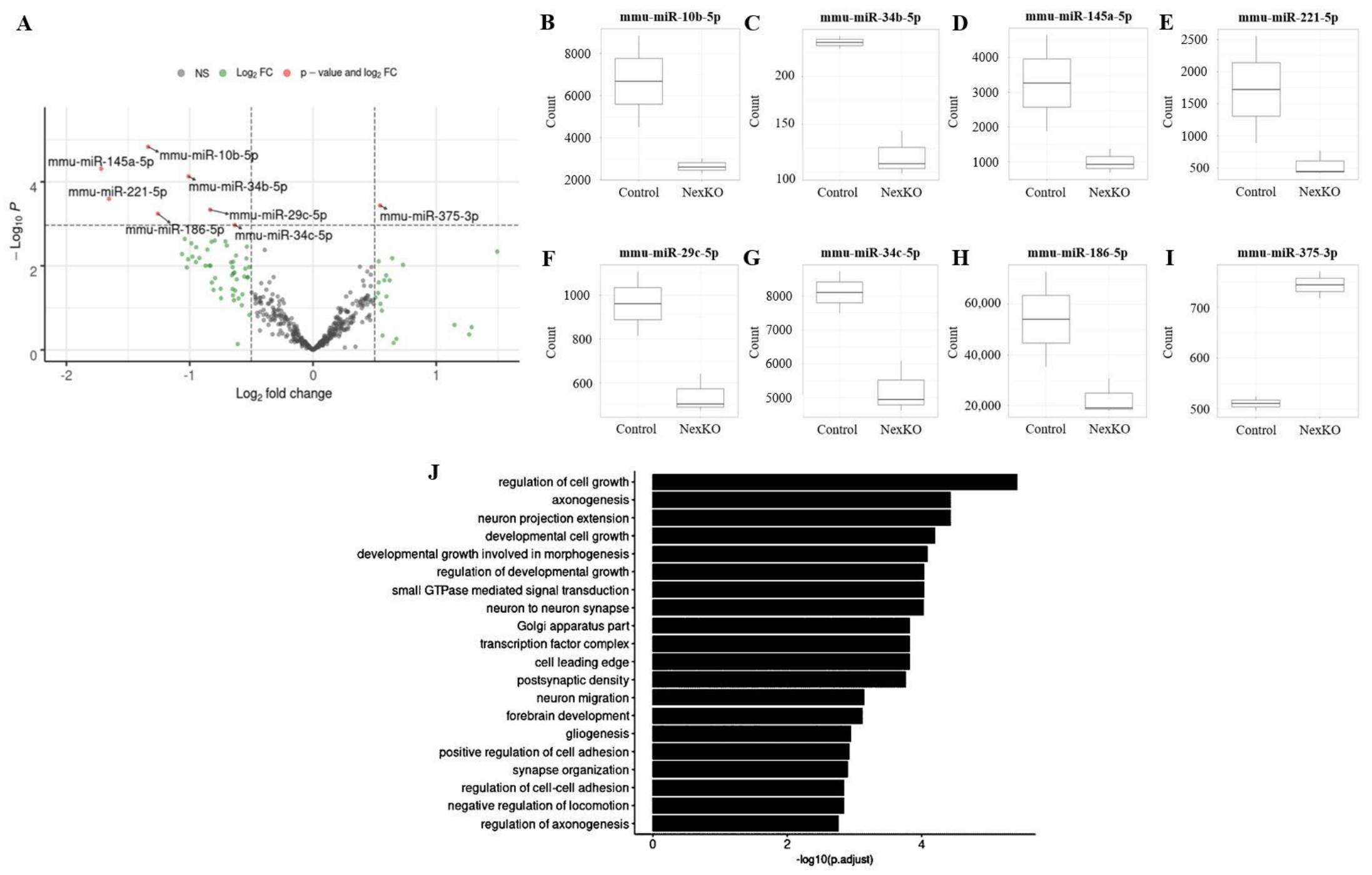
3.2. miRNA and mRNA Expression Is Altered in the Cortices of 1-Month-Old NexKO Mice
3.3. miR-34b/c-5p Expression Levels Are Downregulated in the Cortices of 1-Month-Old NexKO Mice
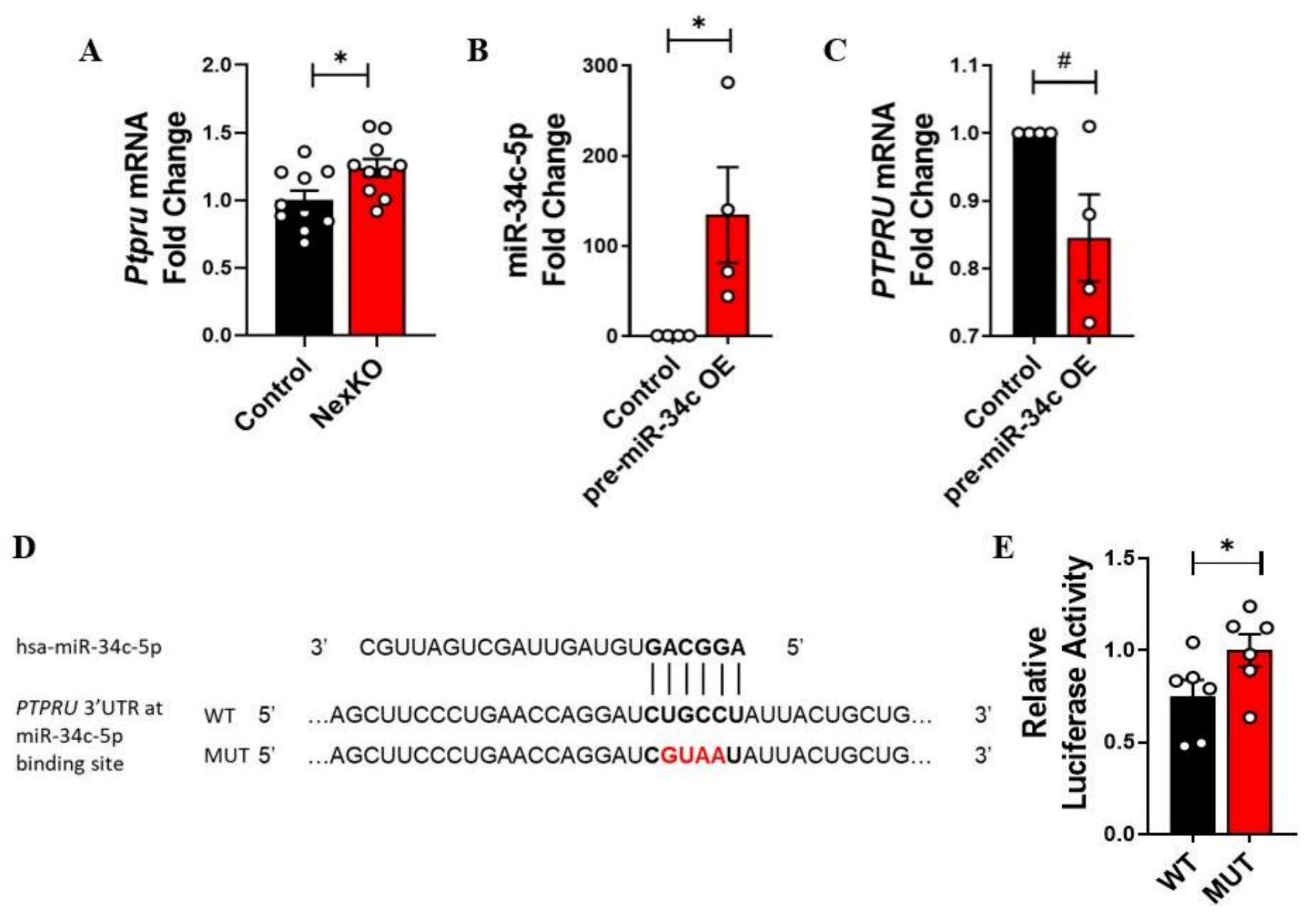
3.4. PTPRU Is Directly Regulated by hsa-miR-34c-5p
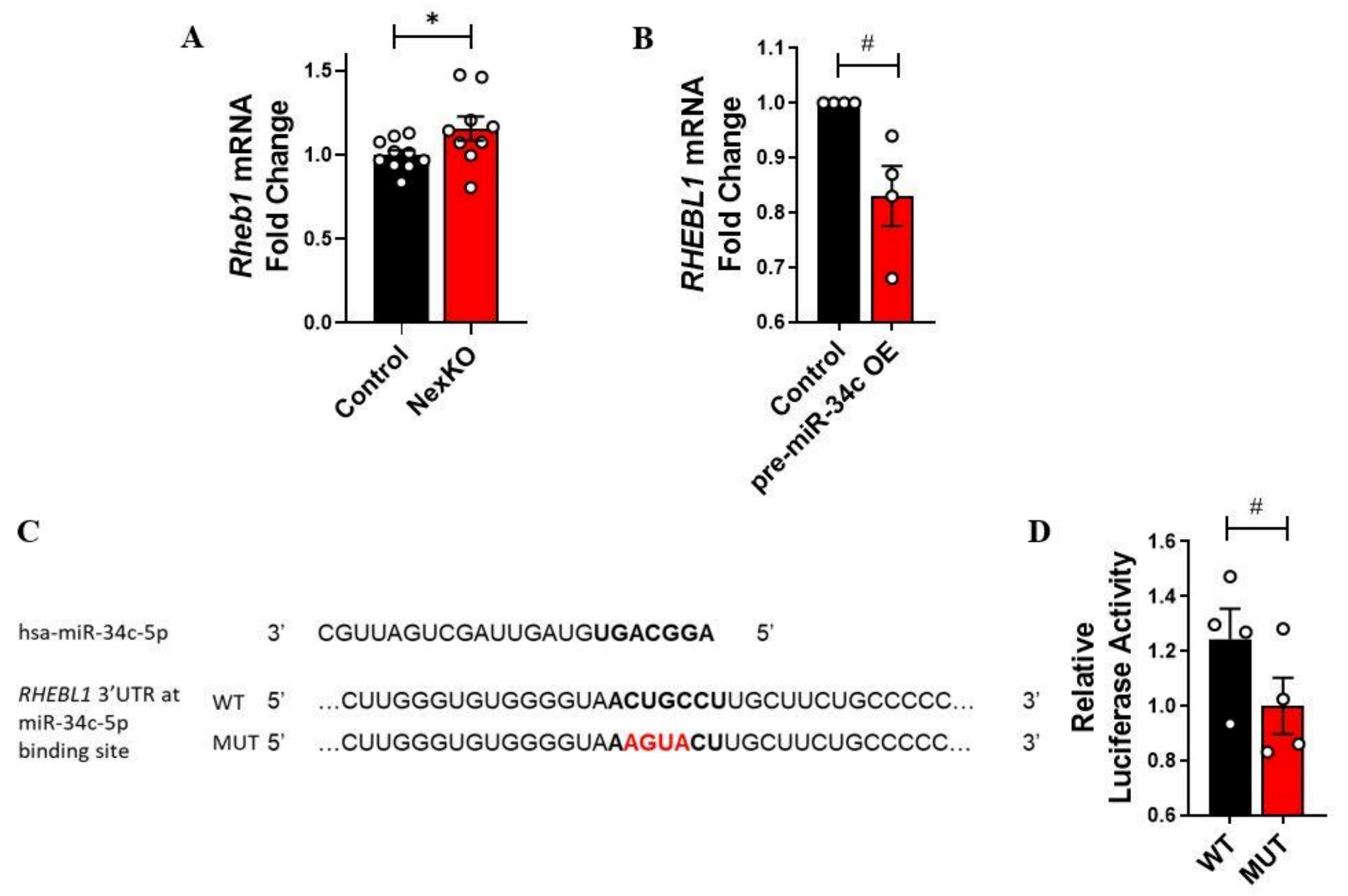
3.5. RAS Homolog Enriched in Brain-Like Protein 1 (RHEBL1) Is Not Directly Regulated by hsa-mir-34c-5p
3.6. Doublecortin (Dcx)—A Target of miR-34—Was Differentially Expressed in the Cortices of NexKO Mice Compared to Controls and Across Development
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Osborne, L.R.; Mervis, C.B. 7q11.23 Deletion and Duplication. Curr. Opin. Genet. Dev. 2021, 68, 41–48. [Google Scholar] [CrossRef]
- Pober, B.R. Williams–Beuren Syndrome. N. Engl. J. Med. 2010, 362, 239–252. [Google Scholar] [CrossRef]
- Barak, B.; Feng, G. Neurobiology of Social Behavior Abnormalities in Autism and Williams Syndrome. Nat. Neurosci. 2016, 19, 647–655. [Google Scholar] [CrossRef]
- Mervis, C.B.; Robinson, B.F.; Bertrand, J.; Morris, C.A.; Klein-Tasman, B.P.; Armstrong, S.C. The Williams Syndrome Cognitive Profile. Brain Cogn. 2000, 44, 604–628. [Google Scholar] [CrossRef]
- Mervis, C.B.; John, A.E. Cognitive and Behavioral Characteristics of Children with Williams Syndrome: Implications for Intervention Approaches. Am. J. Med. Genet. Part C Semin. Med. Genet. 2010, 154C, 229–248. [Google Scholar] [CrossRef]
- Mervis, C.B.; Velleman, S.L. Children with Williams Syndrome: Language, Cognitive, and Behavioral Characteristics and Their Implications for Intervention. Perspect Lang Learn. Educ 2011, 18, 98–107. [Google Scholar] [CrossRef]
- Mervis, C.B.; Kistler, D.J.; John, A.E.; Morris, C.A. Longitudinal Assessment of Intellectual Abilities of Children with Williams Syndrome: Multilevel Modeling of Performance on the Kaufman Brief Intelligence Test—Second Edition. Am. J. Intellect. Dev. Disabil. 2012, 117, 134–155. [Google Scholar] [CrossRef]
- Mervis, C.B.; Pitts, C.H. Children with Williams Syndrome: Developmental Trajectories for Intellectual Abilities, Vocabulary Abilities, and Adaptive Behavior. Am. J. Med. Genet. Part C Semin. Med. Genet. 2015, 169, 158–171. [Google Scholar] [CrossRef]
- Shaikh, S.; Waxler, J.L.; Lee, H.; Grinke, K.; Garry, J.; Pober, B.R.; Stanley, T.L. Glucose and Lipid Metabolism, Bone Density, and Body Composition in Individuals with Williams Syndrome. Clin. Endocrinol. 2018, 89, 596–604. [Google Scholar] [CrossRef]
- Stanley, T.L.; Leong, A.; Pober, B.R. Growth, Body Composition, and Endocrine Issues in Williams Syndrome. Curr. Opin. Endocrinol. Diabetes Obes. 2021, 28, 64–74. [Google Scholar] [CrossRef]
- Meyer-Lindenberg, A.; Kohn, P.; Mervis, C.B.; Kippenhan, J.S.; Olsen, R.K.; Morris, C.A.; Berman, K.F. Neural Basis of Genetically Determined Visuospatial Construction Deficit in Williams Syndrome. Neuron 2004, 43, 623–631. [Google Scholar] [CrossRef]
- Mervis, C.B. Language and Literacy Development of Children with Williams Syndrome. Top Lang Disord 2009, 29, 149–169. [Google Scholar] [CrossRef]
- Pober, B.R.; Morris, C.A. Diagnosis and Management of Medical Problems in Adults with Williams–Beuren Syndrome. Am. J. Med. Genet. Part C Semin. Med. Genet. 2007, 145C, 280–290. [Google Scholar] [CrossRef]
- Lugo, M.; Wong, Z.C.; Billington, C.J.; Parrish, P.C.R.; Muldoon, G.; Liu, D.; Pober, B.R.; Kozel, B.A. Social, Neurodevelopmental, Endocrine, and Head Size Differences Associated with Atypical Deletions in Williams–Beuren Syndrome. Am. J. Med. Genet. Part A 2020, 182, 1008–1020. [Google Scholar] [CrossRef]
- John, A.E.; Mervis, C.B. Sensory Modulation Impairments in Children with Williams Syndrome. Am. J. Med. Genet. Part C Semin. Med. Genet. 2010, 154C, 266–276. [Google Scholar] [CrossRef]
- Kozel, B.A.; Barak, B.; Kim, C.A.; Mervis, C.B.; Osborne, L.R.; Porter, M.; Pober, B.R. Williams Syndrome. Nat. Rev. Dis. Primers. 2021, 7, 1–22. [Google Scholar] [CrossRef]
- Leyfer, O.; Woodruff-Borden, J.; Mervis, C.B. Anxiety Disorders in Children with Williams Syndrome, Their Mothers, and Their Siblings: Implications for the Etiology of Anxiety Disorders. J. Neurodev. Disord. 2009, 1, 4–14. [Google Scholar] [CrossRef][Green Version]
- Woodruff-Borden, J.; Kistler, D.J.; Henderson, D.R.; Crawford, N.A.; Mervis, C.B. Longitudinal Course of Anxiety in Children and Adolescents with Williams Syndrome. Am. J. Med. Genet. Part C Semin. Med. Genet. 2010, 154C, 277–290. [Google Scholar] [CrossRef]
- Jabbi, M.; Kippenhan, J.S.; Kohn, P.; Marenco, S.; Mervis, C.B.; Morris, C.A.; Meyer-Lindenberg, A.; Berman, K.F. The Williams Syndrome Chromosome 7q11.23 Hemideletion Confers Hypersocial, Anxious Personality Coupled with Altered Insula Structure and Function. Proc. Natl. Acad. Sci. USA 2012, 109, E860–E866. [Google Scholar] [CrossRef]
- Mervis, C.B.; Becerra, A.M. Language and Communicative Development in Williams Syndrome. Ment. Retard. Dev. Disabil. Res. Rev. 2007, 13, 3–15. [Google Scholar] [CrossRef]
- Fan, C.C.; Brown, T.T.; Bartsch, H.; Kuperman, J.M.; Hagler, D.J.; Schork, A.; Searcy, Y.; Bellugi, U.; Halgren, E.; Dale, A.M. Williams Syndrome-Specific Neuroanatomical Profile and Its Associations with Behavioral Features. NeuroImage Clin. 2017, 15, 343–347. [Google Scholar] [CrossRef]
- Kippenhan, J.S.; Olsen, R.K.; Mervis, C.B.; Morris, C.A.; Kohn, P.; Meyer-Lindenberg, A.; Berman, K.F. Genetic Contributions to Human Gyrification: Sulcal Morphometry in Williams Syndrome. J. Neurosci. 2005, 25, 7840–7846. [Google Scholar] [CrossRef]
- Thompson, P.M.; Lee, A.D.; Dutton, R.A.; Geaga, J.A.; Hayashi, K.M.; Eckert, M.A.; Bellugi, U.; Galaburda, A.M.; Korenberg, J.R.; Mills, D.L.; et al. Abnormal Cortical Complexity and Thickness Profiles Mapped in Williams Syndrome. J. Neurosci. 2005, 25, 4146–4158. [Google Scholar] [CrossRef]
- Reiss, A.L.; Eckert, M.A.; Rose, F.E.; Karchemskiy, A.; Kesler, S.; Chang, M.; Reynolds, M.F.; Kwon, H.; Galaburda, A. An Experiment of Nature: Brain Anatomy Parallels Cognition and Behavior in Williams Syndrome. J. Neurosci. 2004, 24, 5009–5015. [Google Scholar] [CrossRef]
- Meyer-Lindenberg, A.; Hariri, A.R.; Munoz, K.E.; Mervis, C.B.; Mattay, V.S.; Morris, C.A.; Berman, K.F. Neural Correlates of Genetically Abnormal Social Cognition in Williams Syndrome. Nat. Neurosci. 2005, 8, 991–993. [Google Scholar] [CrossRef]
- Meyer-Lindenberg, A.; Mervis, C.B.; Faith Berman, K. Neural Mechanisms in Williams Syndrome: A Unique Window to Genetic Influences on Cognition and Behaviour. Nat. Rev. Neurosci. 2006, 7, 380–393. [Google Scholar] [CrossRef]
- Marenco, S.; Siuta, M.A.; Kippenhan, J.S.; Grodofsky, S.; Chang, W.; Kohn, P.; Mervis, C.B.; Morris, C.A.; Weinberger, D.R.; Meyer-Lindenberg, A.; et al. Genetic Contributions to White Matter Architecture Revealed by Diffusion Tensor Imaging in Williams Syndrome. Proc. Natl. Acad. Sci. USA 2007, 104, 15117–15122. [Google Scholar] [CrossRef]
- Campbell, L.E.; Daly, E.; Toal, F.; Stevens, A.; Azuma, R.; Karmiloff-Smith, A.; Murphy, D.G.M.; Murphy, K.C. Brain Structural Differences Associated with the Behavioural Phenotype in Children with Williams Syndrome. Brain Res. 2009, 1258, 96–107. [Google Scholar] [CrossRef]
- Arlinghaus, L.R.; Thornton-Wells, T.A.; Dykens, E.M.; Anderson, A.W. Alterations in Diffusion Properties of White Matter in Williams Syndrome. Magn. Reson. Imaging 2011, 29, 1165–1174. [Google Scholar] [CrossRef]
- Avery, S.N.; Thornton-Wells, T.A.; Anderson, A.W.; Blackford, J.U. White Matter Integrity Deficits in Prefrontal–Amygdala Pathways in Williams Syndrome. NeuroImage 2012, 59, 887–894. [Google Scholar] [CrossRef][Green Version]
- Haas, B.W.; Barnea-Goraly, N.; Sheau, K.E.; Yamagata, B.; Ullas, S.; Reiss, A.L. Altered Microstructure Within Social-Cognitive Brain Networks During Childhood in Williams Syndrome. Cereb. Cortex 2014, 24, 2796–2806. [Google Scholar] [CrossRef]
- Chailangkarn, T.; Muotri, A.R. Modeling Williams Syndrome with Induced Pluripotent Stem Cells. Neurogenesis 2017, 4, e1283187. [Google Scholar] [CrossRef]
- Gagliardi, C.; Arrigoni, F.; Nordio, A.; De Luca, A.; Peruzzo, D.; Decio, A.; Leemans, A.; Borgatti, R. A Different Brain: Anomalies of Functional and Structural Connections in Williams Syndrome. Front Neurol 2018, 9, 721. [Google Scholar] [CrossRef]
- Nir, A.; Barak, B. White Matter Alterations in Williams Syndrome Related to Behavioral and Motor Impairments. Glia 2021, 69, 5–19. [Google Scholar] [CrossRef]
- Gothelf, D.; Searcy, Y.M.; Reilly, J.; Lai, P.T.; Lanre-Amos, T.; Mills, D.; Korenberg, J.R.; Galaburda, A.; Bellugi, U.; Reiss, A.L. Association between Cerebral Shape and Social Use of Language in Williams Syndrome. Am. J. Med. Genet. Part A 2008, 146A, 2753–2761. [Google Scholar] [CrossRef]
- Tomaiuolo, F.; Di Paola, M.; Caravale, B.; Vicari, S.; Petrides, M.; Caltagirone, C. Morphology and Morphometry of the Corpus Callosum in Williams Syndrome: A T1-Weighted MRI Study. NeuroReport 2002, 13, 2281–2284. [Google Scholar] [CrossRef]
- Porter, M.A.; Coltheart, M.; Langdon, R. The Neuropsychological Basis of Hypersociability in Williams and Down Syndrome. Neuropsychologia 2007, 45, 2839–2849. [Google Scholar] [CrossRef]
- Mobbs, D.; Eckert, M.A.; Mills, D.; Korenberg, J.; Bellugi, U.; Galaburda, A.M.; Reiss, A.L. Frontostriatal Dysfunction During Response Inhibition in Williams Syndrome. Biol. Psychiatry 2007, 62, 256–261. [Google Scholar] [CrossRef]
- Little, K.; Riby, D.M.; Janes, E.; Clark, F.; Fleck, R.; Rodgers, J. Heterogeneity of Social Approach Behaviour in Williams Syndrome: The Role of Response Inhibition. Res. Dev. Disabil. 2013, 34, 959–967. [Google Scholar] [CrossRef]
- Frigerio, E.; Burt, D.M.; Gagliardi, C.; Cioffi, G.; Martelli, S.; Perrett, D.I.; Borgatti, R. Is Everybody Always My Friend? Perception of Approachability in Williams Syndrome. Neuropsychologia 2006, 44, 254–259. [Google Scholar] [CrossRef]
- Berg, J.S.; Brunetti-Pierri, N.; Peters, S.U.; Kang, S.-H.L.; Fong, C.-T.; Salamone, J.; Freedenberg, D.; Hannig, V.L.; Prock, L.A.; Miller, D.T.; et al. Speech Delay and Autism Spectrum Behaviors Are Frequently Associated with Duplication of the 7q11.23 Williams-Beuren Syndrome Region. Genet. Med. 2007, 9, 427–441. [Google Scholar] [CrossRef]
- Merla, G.; Brunetti-Pierri, N.; Micale, L.; Fusco, C. Copy Number Variants at Williams–Beuren Syndrome 7q11.23 Region. Hum. Genet. 2010, 128, 3–26. [Google Scholar] [CrossRef]
- Doyle, T.F.; Bellugi, U.; Korenberg, J.R.; Graham, J. “Everybody in the World Is My Friend” Hypersociability in Young Children with Williams Syndrome. Am. J. Med. Genet. Part A 2004, 124A, 263–273. [Google Scholar] [CrossRef]
- Roy, A.L. Biochemistry and Biology of the Inducible Multifunctional Transcription Factor TFII-I: 10years Later. Gene 2012, 492, 32–41. [Google Scholar] [CrossRef]
- Morris, C.A.; Mervis, C.B.; Hobart, H.H.; Gregg, R.G.; Bertrand, J.; Ensing, G.J.; Sommer, A.; Moore, C.A.; Hopkin, R.J.; Spallone, P.A.; et al. GTF2I Hemizygosity Implicated in Mental Retardation in Williams Syndrome: Genotype–Phenotype Analysis of Five Families with Deletions in the Williams Syndrome Region. Am. J. Med. Genet. Part A 2003, 123A, 45–59. [Google Scholar] [CrossRef]
- van Hagen, J.M.; van der Geest, J.N.; van der Giessen, R.S.; Lagers-van Haselen, G.C.; Eussen, H.J.F.M.M.; Gille, J.J.P.; Govaerts, L.C.P.; Wouters, C.H.; de Coo, I.F.M.; Hoogenraad, C.C.; et al. Contribution of CYLN2 and GTF2IRD1 to Neurological and Cognitive Symptoms in Williams Syndrome. Neurobiol. Dis. 2007, 26, 112–124. [Google Scholar] [CrossRef]
- Osborne, L.R. Animal Models of Williams Syndrome. Am. J. Med. Genet. Part C Semin. Med. Genet. 2010, 154C, 209–219. [Google Scholar] [CrossRef]
- Mervis, C.B.; Dida, J.; Lam, E.; Crawford-Zelli, N.A.; Young, E.J.; Henderson, D.R.; Onay, T.; Morris, C.A.; Woodruff-Borden, J.; Yeomans, J.; et al. Duplication of GTF2I Results in Separation Anxiety in Mice and Humans. Am. J. Hum. Genet. 2012, 90, 1064–1070. [Google Scholar] [CrossRef]
- Orellana, C.; Bernabeu, J.; Monfort, S.; Roselló, M.; Oltra, S.; Ferrer, I.; Quiroga, R.; Martínez-Garay, I.; Martínez, F. Duplication of the Williams–Beuren Critical Region: Case Report and Further Delineation of the Phenotypic Spectrum. J. Med. Genet. 2008, 45, 187–189. [Google Scholar] [CrossRef]
- Depienne, C.; Heron, D.; Betancur, C.; Benyahia, B.; Trouillard, O.; Bouteiller, D.; Verloes, A.; LeGuern, E.; Leboyer, M.; Brice, A. Autism, Language Delay and Mental Retardation in a Patient with 7q11 Duplication. J. Med. Genet. 2007, 44, 452–458. [Google Scholar] [CrossRef]
- Beunders, G.; van de Kamp, J.M.; Veenhoven, R.H.; van Hagen, J.M.; Nieuwint, A.W.M.; Sistermans, E.A. A Triplication of the Williams–Beuren Syndrome Region in a Patient with Mental Retardation, a Severe Expressive Language Delay, Behavioural Problems and Dysmorphisms. J. Med. Genet. 2010, 47, 271–275. [Google Scholar] [CrossRef]
- Enkhmandakh, B.; Makeyev, A.V.; Erdenechimeg, L.; Ruddle, F.H.; Chimge, N.-O.; Tussie-Luna, M.I.; Roy, A.L.; Bayarsaihan, D. Essential Functions of the Williams-Beuren Syndrome-Associated TFII-I Genes in Embryonic Development. Proc. Natl. Acad. Sci. USA 2009, 106, 181–186. [Google Scholar] [CrossRef]
- Sakurai, T.; Dorr, N.P.; Takahashi, N.; McInnes, L.A.; Elder, G.A.; Buxbaum, J.D. Haploinsufficiency of Gtf2i, a Gene Deleted in Williams Syndrome, Leads to Increases in Social Interactions. Autism Res. 2011, 4, 28–39. [Google Scholar] [CrossRef]
- Lucena, J.; Pezzi, S.; Aso, E.; Valero, M.C.; Carreiro, C.; Dubus, P.; Sampaio, A.; Segura, M.; Barthelemy, I.; Zindel, M.Y.; et al. Essential Role of the N-Terminal Region of TFII-I in Viability and Behavior. BMC Med. Genet. 2010, 11, 61. [Google Scholar] [CrossRef]
- Gandal, M.J.; Leppa, V.; Won, H.; Parikshak, N.N.; Geschwind, D.H. The Road to Precision Psychiatry: Translating Genetics into Disease Mechanisms. Nat. Neurosci. 2016, 19, 1397–1407. [Google Scholar] [CrossRef]
- Barak, B.; Zhang, Z.; Liu, Y.; Nir, A.; Trangle, S.S.; Ennis, M.; Levandowski, K.M.; Wang, D.; Quast, K.; Boulting, G.L.; et al. Neuronal Deletion of Gtf2i, Associated with Williams Syndrome, Causes Behavioral and Myelin Alterations Rescuable by a Remyelinating Drug. Nat. Neurosci. 2019, 22, 700–708. [Google Scholar] [CrossRef]
- Hayes, K.C. The Use of 4-Aminopyridine (Fampridine) in Demyelinating Disorders. CNS Drug Rev. 2004, 10, 295–316. [Google Scholar] [CrossRef]
- Liu, J.; Dupree, J.L.; Gacias, M.; Frawley, R.; Sikder, T.; Naik, P.; Casaccia, P. Clemastine Enhances Myelination in the Prefrontal Cortex and Rescues Behavioral Changes in Socially Isolated Mice. J. Neurosci. 2016, 36, 957–962. [Google Scholar] [CrossRef]
- Cree, B.A.C.; Niu, J.; Hoi, K.K.; Zhao, C.; Caganap, S.D.; Henry, R.G.; Dao, D.Q.; Zollinger, D.R.; Mei, F.; Shen, Y.-A.A.; et al. Clemastine Rescues Myelination Defects and Promotes Functional Recovery in Hypoxic Brain Injury. Brain 2018, 141, 85–98. [Google Scholar] [CrossRef]
- Rajman, M.; Schratt, G. MicroRNAs in Neural Development: From Master Regulators to Fine-Tuners. Development 2017, 144, 2310–2322. [Google Scholar] [CrossRef]
- Hsieh, J.; Zhao, X. Genetics and Epigenetics in Adult Neurogenesis. Cold Spring Harb. Perspect. Biol. 2016, 8, a018911. [Google Scholar] [CrossRef]
- Shibata, M.; Nakao, H.; Kiyonari, H.; Abe, T.; Aizawa, S. MicroRNA-9 Regulates Neurogenesis in Mouse Telencephalon by Targeting Multiple Transcription Factors. J. Neurosci. 2011, 31, 3407–3422. [Google Scholar] [CrossRef]
- Paridaen, J.T.; Huttner, W.B. Neurogenesis during Development of the Vertebrate Central Nervous System. EMBO Rep. 2014, 15, 351–364. [Google Scholar] [CrossRef]
- Giorgi Silveira, R.; Perelló Ferrúa, C.; do Amaral, C.C.; Fernandez Garcia, T.; de Souza, K.B.; Nedel, F. MicroRNAs Expressed in Neuronal Differentiation and Their Associated Pathways: Systematic Review and Bioinformatics Analysis. Brain Res. Bull. 2020, 157, 140–148. [Google Scholar] [CrossRef]
- Shvarts-Serebro, I.; Sheinin, A.; Gottfried, I.; Adler, L.; Schottlender, N.; Ashery, U.; Barak, B. MiR-128 as a Regulator of Synaptic Properties in 5xFAD Mice Hippocampal Neurons. J. Mol. Neurosci. 2021. [Google Scholar] [CrossRef]
- Barak, B.; Shvarts-Serebro, I.; Modai, S.; Gilam, A.; Okun, E.; Michaelson, D.M.; Mattson, M.P.; Shomron, N.; Ashery, U. Opposing Actions of Environmental Enrichment and Alzheimer’s Disease on the Expression of Hippocampal MicroRNAs in Mouse Models. Transl. Psychiatry 2013, 3, e304. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.-P.; et al. An Estimate of the Total Number of True Human MiRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The Widespread Regulation of MicroRNA Biogenesis, Function and Decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many Roads to Maturity: MicroRNA Biogenesis Pathways and Their Regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef]
- Hammond, S.M. An Overview of MicroRNAs. Adv. Drug. Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef]
- Xu, X.-L.; Zong, R.; Li, Z.; Biswas, M.H.U.; Fang, Z.; Nelson, D.L.; Gao, F.-B. FXR1P But Not FMRP Regulates the Levels of Mammalian Brain-Specific MicroRNA-9 and MicroRNA-124. J. Neurosci. 2011, 31, 13705–13709. [Google Scholar] [CrossRef]
- Esteller, M. Non-Coding RNAs in Human Disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Kimura, R.; Swarup, V.; Tomiwa, K.; Gandal, M.J.; Parikshak, N.N.; Funabiki, Y.; Nakata, M.; Awaya, T.; Kato, T.; Iida, K.; et al. Integrative Network Analysis Reveals Biological Pathways Associated with Williams Syndrome. J. Child Psychol. Psychiatry 2019, 60, 585–598. [Google Scholar] [CrossRef]
- Stark, K.L.; Xu, B.; Bagchi, A.; Lai, W.-S.; Liu, H.; Hsu, R.; Wan, X.; Pavlidis, P.; Mills, A.A.; Karayiorgou, M.; et al. Altered Brain MicroRNA Biogenesis Contributes to Phenotypic Deficits in a 22q11-Deletion Mouse Model. Nat. Genet. 2008, 40, 751–760. [Google Scholar] [CrossRef]
- Schepici, G.; Cavalli, E.; Bramanti, P.; Mazzon, E. Autism Spectrum Disorder and MiRNA: An Overview of Experimental Models. Brain Sci. 2019, 9, 265. [Google Scholar] [CrossRef]
- Nakata, M.; Kimura, R.; Funabiki, Y.; Awaya, T.; Murai, T.; Hagiwara, M. MicroRNA Profiling in Adults with High-Functioning Autism Spectrum Disorder. Mol. Brain 2019, 12, 82. [Google Scholar] [CrossRef]
- Goebbels, S.; Bormuth, I.; Bode, U.; Hermanson, O.; Schwab, M.H.; Nave, K.-A. Genetic Targeting of Principal Neurons in Neocortex and Hippocampus of NEX-Cre Mice. Genes 2006, 44, 611–621. [Google Scholar] [CrossRef]
- Truett, G.E.; Heeger, P.; Mynatt, R.L.; Truett, A.A.; Walker, J.A.; Warman, M.L. Preparation of PCR-Quality Mouse Genomic DNA with Hot Sodium Hydroxide and Tris (HotSHOT). BioTechniques 2000, 29, 52–54. [Google Scholar] [CrossRef]
- Tunster, S.J. Genetic Sex Determination of Mice by Simplex PCR. Biol. Sex Differ. 2017, 8. [Google Scholar] [CrossRef]
- Russell, P.H.; Vestal, B.; Shi, W.; Rudra, P.D.; Dowell, R.; Radcliffe, R.; Saba, L.; Kechris, K. MiR-MaGiC Improves Quantification Accuracy for Small RNA-Seq. BMC Res. Notes 2018, 11, 296. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ru, Y.; Kechris, K.J.; Tabakoff, B.; Hoffman, P.; Radcliffe, R.A.; Bowler, R.; Mahaffey, S.; Rossi, S.; Calin, G.A.; Bemis, L.; et al. The MultiMiR R Package and Database: Integration of MicroRNA–Target Interactions along with Their Disease and Drug Associations. Nucleic Acids Res. 2014, 42, e133. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative C T Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Sanger, F.; Coulson, A.R. A Rapid Method for Determining Sequences in DNA by Primed Synthesis with DNA Polymerase. J. Mol. Biol. 1975, 94, 441–448. [Google Scholar] [CrossRef]
- Mohammadipoor-Ghasemabad, L.; Sangtarash, M.H.; Sheibani, V.; Sasan, H.A.; Esmaeili-Mahani, S. Hippocampal MicroRNA-191a-5p Regulates BDNF Expression and Shows Correlation with Cognitive Impairment Induced by Paradoxical Sleep Deprivation. Neuroscience 2019, 414, 49–59. [Google Scholar] [CrossRef]
- Leemans, A.; Sijbers, J.; Jeurissen, B.; Jones, D.K. ExploreDTI: A Graphical Toolbox for Processing, Analyzing, and Visualizing Diffusion MR Data. Proc. Intl. Soc. Magn. Reson. Med. 2009, 17. [Google Scholar]
- Wang, L.; Liu, W.; Zhang, Y.; Hu, Z.; Guo, H.; Lv, J.; Du, H. Dexmedetomidine Had Neuroprotective Effects on Hippocampal Neuronal Cells via Targeting LncRNA SHNG16 Mediated MicroRNA-10b-5p/BDNF Axis. Mol. Cell. Biochem. 2020, 469, 41–51. [Google Scholar] [CrossRef]
- Gao, S.; Li, C.; Xu, Y.; Chen, S.; Zhao, Y.; Chen, L.; Jiang, Y.; Liu, Z.; Fan, R.; Sun, L.; et al. Differential Expression of MicroRNAs in TM3 Leydig Cells of Mice Treated with Brain-Derived Neurotrophic Factor. Cell Biochem. Funct. 2017, 35, 364–371. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Wang, T.-Y.; Lu, R.-B.; Wang, L.-J.; Chang, C.-H.; Chiang, Y.-C.; Tsai, K.-W. Peripheral BDNF Correlated with MiRNA in BD-II Patients. J. Psychiatr. Res. 2021, 136, 184–189. [Google Scholar] [CrossRef]
- Renu, C.S.; Li, Y.C.; Hong, W.; Gautam, S.; Feng, R.T. Neuronal Development-Related MiRNAs as Biomarkers for Alzheimer’s Disease, Depression, Schizophrenia and Ionizing Radiation Exposure. Curr. Med. Chem. 2020, 28, 19–52. [Google Scholar]
- Silva, M.M.; Rodrigues, B.; Fernandes, J.; Santos, S.D.; Carreto, L.; Santos, M.A.S.; Pinheiro, P.; Carvalho, A.L. MicroRNA-186-5p Controls GluA2 Surface Expression and Synaptic Scaling in Hippocampal Neurons. Proc. Natl. Acad. Sci. USA 2019, 116, 5727–5736. [Google Scholar] [CrossRef]
- Jauhari, A.; Singh, T.; Singh, P.; Parmar, D.; Yadav, S. Regulation of MiR-34 Family in Neuronal Development. Mol. Neurobiol. 2018, 55, 936–945. [Google Scholar] [CrossRef]
- Mollinari, C.; Racaniello, M.; Berry, A.; Pieri, M.; de Stefano, M.C.; Cardinale, A.; Zona, C.; Cirulli, F.; Garaci, E.; Merlo, D. MiR-34a Regulates Cell Proliferation, Morphology and Function of Newborn Neurons Resulting in Improved Behavioural Outcomes. Cell Death Dis. 2015, 6, e1622. [Google Scholar] [CrossRef]
- Venø, M.T.; Venø, S.T.; Rehberg, K.; van Asperen, J.V.; Clausen, B.H.; Holm, I.E.; Pasterkamp, R.J.; Finsen, B.; Kjems, J. Cortical Morphogenesis during Embryonic Development Is Regulated by MiR-34c and MiR-204. Front. Mol. Neurosci. 2017, 10. [Google Scholar] [CrossRef]
- Jauhari, A.; Yadav, S. MiR-34 and MiR-200: Regulator of Cell Fate Plasticity and Neural Development. Neuromol. Med. 2019, 21, 97–109. [Google Scholar] [CrossRef]
- Wu, J.; Bao, J.; Kim, M.; Yuan, S.; Tang, C.; Zheng, H.; Mastick, G.S.; Xu, C.; Yan, W. Two MiRNA Clusters, MiR-34b/c and MiR-449, Are Essential for Normal Brain Development, Motile Ciliogenesis, and Spermatogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, E2851–E2857. [Google Scholar] [CrossRef]
- De Gregorio, R.; Pulcrano, S.; De Sanctis, C.; Volpicelli, F.; Guatteo, E.; von Oerthel, L.; Latagliata, E.C.; Esposito, R.; Piscitelli, R.M.; Perrone-Capano, C.; et al. MiR-34b/c Regulates Wnt1 and Enhances Mesencephalic Dopaminergic Neuron Differentiation. Stem Cell Rep. 2018, 10, 1237–1250. [Google Scholar] [CrossRef]
- Hsu, S.-D.; Lin, F.-M.; Wu, W.-Y.; Liang, C.; Huang, W.-C.; Chan, W.-L.; Tsai, W.-T.; Chen, G.-Z.; Lee, C.-J.; Chiu, C.-M.; et al. MiRTarBase: A Database Curates Experimentally Validated MicroRNA-Target Interactions. Nucleic Acids Res. 2011, 39, D163–D169. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics Enrichment Tools: Paths toward the Comprehensive Functional Analysis of Large Gene Lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- The Gene Ontology Resource: 20 Years and Still GOing Strong. Nucleic Acids Res 2019, 47, D330–D338. [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER Version 14: More Genomes, a New PANTHER GO-Slim and Improvements in Enrichment Analysis Tools. Nucleic Acids Res 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Carbon, S.; Ireland, A.; Mungall, C.J.; Shu, S.; Marshall, B.; Lewis, S. the AmiGO Hub; the Web Presence Working Group AmiGO: Online Access to Ontology and Annotation Data. Bioinformatics 2009, 25, 288–289. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates That Thousands of Human Genes Are MicroRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Besco, J.; Popesco, M.C.; Davuluri, R.V.; Frostholm, A.; Rotter, A. Genomic Structure and Alternative Splicing of Murine R2B Receptor Protein Tyrosine Phosphatases (PTPκ, μ, ρ and PCP-2). BMC Genomics 2004, 5, 14. [Google Scholar] [CrossRef]
- Zou, J.; Zhou, L.; Du, X.-X.; Ji, Y.; Xu, J.; Tian, J.; Jiang, W.; Zou, Y.; Yu, S.; Gan, L.; et al. Rheb1 Is Required for MTORC1 and Myelination in Postnatal Brain Development. Dev. Cell 2011, 20, 97–108. [Google Scholar] [CrossRef]
- Zou, Y.; Jiang, W.; Wang, J.; Li, Z.; Zhang, J.; Bu, J.; Zou, J.; Zhou, L.; Yu, S.; Cui, Y.; et al. Oligodendrocyte Precursor Cell-Intrinsic Effect of Rheb1 Controls Differentiation and Mediates MTORC1-Dependent Myelination in Brain. J. Neurosci. 2014, 34, 15764–15778. [Google Scholar] [CrossRef]
- Hay, I.M.; Fearnley, G.W.; Rios, P.; Köhn, M.; Sharpe, H.J.; Deane, J.E. The Receptor PTPRU Is a Redox Sensitive Pseudophosphatase. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Craig, S.E.L.; Brady-Kalnay, S.M. Regulation of Development and Cancer by the R2B Subfamily of RPTPs and the Implications of Proteolysis. Semin. Cell Dev. Biol. 2015, 37, 108–118. [Google Scholar] [CrossRef]
- Jacobs, F.M.J.; van der Linden, A.J.A.; Wang, Y.; von Oerthel, L.; Sul, H.S.; Burbach, J.P.H.; Smidt, M.P. Identification of Dlk1, Ptpru and Klhl1 as Novel Nurr1 Target Genes in Meso-Diencephalic Dopamine Neurons. Development 2009, 136, 2363–2373. [Google Scholar] [CrossRef]
- Gu, J.; Zhang, Z.; Lang, T.; Ma, X.; Yang, L.; Xu, J.; Tian, C.; Han, K.; Qiu, J. PTPRU, As A Tumor Suppressor, Inhibits Cancer Stemness By Attenuating Hippo/YAP Signaling Pathway. Onco. Targets Ther. 2019, 12, 8095–8104. [Google Scholar] [CrossRef]
- Chilton, J.K.; Stoker, A.W. Expression of Receptor Protein Tyrosine Phosphatases in Embryonic Chick Spinal Cord. Mol. Cell. Neurosci. 2000, 16, 470–480. [Google Scholar] [CrossRef]
- Sommer, L.; Rao, M.; Anderson, D.J. RPTPδ and the Novel Protein Tyrosine Phosphatase RPTPψ Are Expressed in Restricted Regions of the Developing Central Nervous System. Dev. Dyn. 1997, 208, 48–61. [Google Scholar] [CrossRef]
- Luzzati, F.; Bonfanti, L.; Fasolo, A.; Peretto, P. DCX and PSA-NCAM Expression Identifies a Population of Neurons Preferentially Distributed in Associative Areas of Different Pallial Derivatives and Vertebrate Species. Cereb Cortex 2009, 19, 1028–1041. [Google Scholar] [CrossRef]
- Reiner, O. LIS1 and DCX: Implications for Brain Development and Human Disease in Relation to Microtubules. Scientifica (Cairo) 2013, 2013. [Google Scholar] [CrossRef]
- Hanrahan, J.; Blenis, J. Rheb Activation of MTOR and S6K1 Signaling. In Methods in Enzymology; Regulators and Effectors of Small GTPases: Ras Family; Academic Press: San Diego, CA, USA, 2006; Volume 407, pp. 542–555. [Google Scholar]
- Yuan, J.; Shan, Y.; Chen, X.; Tang, W.; Luo, K.; Ni, J.; Wan, B.; Yu, L. Identification and Characterization of RHEBL1, a Novel Member of Ras Family, Which Activates Transcriptional Activities of NF-Kappa, B. Mol. Biol. Rep. 2005, 32, 205–214. [Google Scholar] [CrossRef]
- Francis, F.; Koulakoff, A.; Boucher, D.; Chafey, P.; Schaar, B.; Vinet, M.-C.; Friocourt, G.; McDonnell, N.; Reiner, O.; Kahn, A.; et al. Doublecortin Is a Developmentally Regulated, Microtubule-Associated Protein Expressed in Migrating and Differentiating Neurons. Neuron 1999, 23, 247–256. [Google Scholar] [CrossRef]
- Shahsavani, M.; Pronk, R.J.; Falk, R.; Lam, M.; Moslem, M.; Linker, S.B.; Salma, J.; Day, K.; Schuster, J.; Anderlid, B.-M.; et al. An in Vitro Model of Lissencephaly: Expanding the Role of DCX during Neurogenesis. Mol. Psychiatry 2018, 23, 1674–1684. [Google Scholar] [CrossRef]
- Deuel, T.A.S.; Liu, J.S.; Corbo, J.C.; Yoo, S.-Y.; Rorke-Adams, L.B.; Walsh, C.A. Genetic Interactions between Doublecortin and Doublecortin-like Kinase in Neuronal Migration and Axon Outgrowth. Neuron 2006, 49, 41–53. [Google Scholar] [CrossRef]
- Bai, J.; Ramos, R.L.; Ackman, J.B.; Thomas, A.M.; Lee, R.V.; LoTurco, J.J. RNAi Reveals Doublecortin Is Required for Radial Migration in Rat Neocortex. Nat. Neurosci. 2003, 6, 1277–1283. [Google Scholar] [CrossRef]
- Assaf, Y.; Pasternak, O. Diffusion Tensor Imaging (DTI)-Based White Matter Mapping in Brain Research: A Review. J. Mol. Neurosci 2008, 34, 51–61. [Google Scholar] [CrossRef]
- Fields, R.D. White Matter in Learning, Cognition and Psychiatric Disorders. Trends Neurosci. 2008, 31, 361–370. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Klempin, F.; Kronenberg, G.; Cheung, G.; Kettenmann, H.; Kempermann, G. Properties of Doublecortin-(DCX)-Expressing Cells in the Piriform Cortex Compared to the Neurogenic Dentate Gyrus of Adult Mice. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Pajevic, S.; Basser, P.J.; Fields, R.D. Role of Myelin Plasticity in Oscillations and Synchrony of Neuronal Activity. Neuroscience 2014, 276, 135–147. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most Mammalian MRNAs Are Conserved Targets of MicroRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. MiRBase: Annotating High Confidence MicroRNAs Using Deep Sequencing Data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. MiRBase: Integrating MicroRNA Annotation and Deep-Sequencing Data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. MiRBase: Tools for MicroRNA Genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. MiRBase: MicroRNA Sequences, Targets and Gene Nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. The MicroRNA Registry. Nucleic Acids Res. 2004, 32, D109–D111. [Google Scholar] [CrossRef]
- Pomper, N.; Liu, Y.; Hoye, M.L.; Dougherty, J.D.; Miller, T.M. CNS MicroRNA Profiles: A Database for Cell Type Enriched MicroRNA Expression across the Mouse Central Nervous System. Sci. Rep. 2020, 10, 4921. [Google Scholar] [CrossRef]
- Hoye, M.L.; Koval, E.D.; Wegener, A.J.; Hyman, T.S.; Yang, C.; O’Brien, D.R.; Miller, R.L.; Cole, T.; Schoch, K.M.; Shen, T.; et al. MicroRNA Profiling Reveals Marker of Motor Neuron Disease in ALS Models. J. Neurosci. 2017, 37, 5574–5586. [Google Scholar] [CrossRef]
- He, M.; Liu, Y.; Wang, X.; Zhang, M.Q.; Hannon, G.J.; Huang, Z.J. Cell-Type-Based Analysis of MicroRNA Profiles in the Mouse Brain. Neuron 2012, 73, 35–48. [Google Scholar] [CrossRef]
- Ortega, F.; Gascón, S.; Masserdotti, G.; Deshpande, A.; Simon, C.; Fischer, J.; Dimou, L.; Chichung Lie, D.; Schroeder, T.; Berninger, B. Oligodendrogliogenic and Neurogenic Adult Subependymal Zone Neural Stem Cells Constitute Distinct Lineages and Exhibit Differential Responsiveness to Wnt Signalling. Nat. Cell Biol. 2013, 15, 602–613. [Google Scholar] [CrossRef]
- Hoseth, E.Z.; Krull, F.; Dieset, I.; Mørch, R.H.; Hope, S.; Gardsjord, E.S.; Steen, N.E.; Melle, I.; Brattbakk, H.-R.; Steen, V.M.; et al. Exploring the Wnt Signaling Pathway in Schizophrenia and Bipolar Disorder. Translational Psychiatry 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Rokavec, M.; Li, H.; Jiang, L.; Hermeking, H. The P53/MiR-34 Axis in Development and Disease. J. Mol. Cell Biol. 2014, 6, 214–230. [Google Scholar] [CrossRef]
- Navarro, F.; Lieberman, J. MiR-34 and P53: New Insights into a Complex Functional Relationship. PLoS ONE 2015, 10, e0132767. [Google Scholar] [CrossRef]
- Hermeking, H. The MiR-34 Family in Cancer and Apoptosis. Cell Death Differ. 2010, 17, 193–199. [Google Scholar] [CrossRef]
- Fededa, J.P.; Esk, C.; Mierzwa, B.; Stanyte, R.; Yuan, S.; Zheng, H.; Ebnet, K.; Yan, W.; Knoblich, J.A. MicroRNA-34/449 Controls Mitotic Spindle Orientation during Mammalian Cortex Development. EMBO J. 2016, 35, 2386–2398. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Klein, B.; Mrowetz, H.; Kreutzer, C.; Rotheneichner, P.; Zaunmair, P.; Lange, S.; Coras, R.; Couillard-Despres, S.; Rivera, F.J.; Aigner, L. DCX+ Neuronal Progenitors Contribute to New Oligodendrocytes during Remyelination in the Hippocampus. Sci. Rep. 2020, 10, 20095. [Google Scholar] [CrossRef]
- Li, C.; Xie, Z.; Xing, Z.; Zhu, H.; Zhou, W.; Xie, S.; Zhang, Z.; Li, M.-H. The Notch Signaling Pathway Regulates Differentiation of NG2 Cells into Oligodendrocytes in Demyelinating Diseases. Cell Mol. Neurobiol. 2021. [Google Scholar] [CrossRef]
- Caracci, M.O.; Avila, M.E.; Espinoza-Cavieres, F.A.; López, H.R.; Ugarte, G.D.; De Ferrari, G.V. Wnt/β-Catenin-Dependent Transcription in Autism Spectrum Disorders. Front. Mol. Neurosci. 2021, 14, 263. [Google Scholar] [CrossRef]
- Yan, H.-X.; Yang, W.; Zhang, R.; Chen, L.; Tang, L.; Zhai, B.; Liu, S.-Q.; Cao, H.-F.; Man, X.-B.; Wu, H.-P.; et al. Protein-Tyrosine Phosphatase PCP-2 Inhibits β-Catenin Signaling and Increases E-Cadherin-Dependent Cell Adhesion. J. Biol. Chem. 2006, 281, 15423–15433. [Google Scholar] [CrossRef]
- Badde, A.; Schulte, D. A Role for Receptor Protein Tyrosine Phosphatase λ in Midbrain Development. J. Neurosci. 2008, 28, 6152–6164. [Google Scholar] [CrossRef]
- Tian, Q.; Smart, J.L.; Clement, J.H.; Wang, Y.; Derkatch, A.; Schubert, H.; Danilchik, M.V.; Marks, D.L.; Fedorov, L.M. RHEB1 Expression in Embryonic and Postnatal Mouse. Histochem. Cell Biol. 2016, 145, 561–572. [Google Scholar] [CrossRef]
- Justice, M.J.; Dhillon, P. Using the Mouse to Model Human Disease: Increasing Validity and Reproducibility. Dis. Model. Mech. 2016, 9, 101–103. [Google Scholar] [CrossRef]
- De Schutter, E. Fallacies of Mice Experiments. Neuroinform 2019, 17, 181–183. [Google Scholar] [CrossRef]
- Boulanger, J.J.; Messier, C. Doublecortin in Oligodendrocyte Precursor Cells in the Adult Mouse Brain. Front. Neurosci. 2017, 11, 143. [Google Scholar] [CrossRef]

| Name of Primer | Sequence |
|---|---|
| Nex-Cre fwd | GAGTCCTGGAATCAGTCTTTTTC |
| Nex-Cre rev | AGAATGTGGAGTAGGGTGAC |
| Nex-Cre KO | CCGCATAACCAGTGAAACAG |
| Name of Primer | Sequence |
|---|---|
| Rmb31x/y fwd | CACCTTAAGAACAAGCCAATACA |
| Rmb31x/y rev | GGCTTGTCCTGAAAACATTTGG |
| Origin | Name of Primers | Sequence |
|---|---|---|
| Gapdh | Gapdh fwd | GCCTTCCGTGTTCCTACC |
| Gapdh rev | CCTCAGTGTAGCCCAAGATG | |
| Ptpru | Ptpru fwd | GTGGACAAGTGGCAGGCAGA |
| Ptpru rev | CAGGCTGTGACAGCGGATCA | |
| Rheb1 | Rheb1 fwd | TTGTTGATTCCTACGATCCAACCA |
| Rheb1 rev | CCGCTGTGTCTACAAGCTGAAGATG | |
| Dcx | Dcx fwd | CATCACAGAAGCGATCAAACTGGA |
| Dcx rev | CAGGACCACAAGCAATGAACACA |
| Origin | Name of Primers | Sequence |
|---|---|---|
| Β-ACTIN | Has-β-actin fwd | CCTGGACTTCGAGCAAGAGATGG |
| Has-β-actin rev | TGGAGTTGAAGGTAGTTTCGTGGATG | |
| PTPRU | Has-ptpru fwd | ACCTGTACCGCTGTGTGTCCCA |
| Has-ptpru rev | GGAGTTGGTGTTGAGCTGGATGA | |
| RHEBL1 | Has-rhebl1 fwd | GATAGTGACTCTTGGCAAAGATGAGTT |
| Has-rhebl1 rev | TGGACCCCAATGATGAATGAA |
| miRNA | Mature miRNA Sequence | Thermo Fisher Scientific Assay ID |
|---|---|---|
| U6 snRNA | GTGCTCGCTTCGGCAGCACATATACTAAAATTGGAACGATACAGAGAAGATTAGCATGGCCCCTGCGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTT | 001973 |
| hsa-miR-34c-5p | AGGCAGUGUAGUUAGCUGAUUGC | 478052_mir |
| Purpose | Name of Primers | Sequence * |
|---|---|---|
| WT | XhoI-PTPRU 3’UTR fwd | TACATCGCTCGAGTTGGCAGGGATGAGTGAGGC |
| NotI-PTPRU 3’UTR rev | TAGCGGCCGCCGAGGTGACTTCATTCTGCAACA | |
| Mutant | mut-PTPRU 3’UTR fwd | CAAAATATCTCAGGGGCTGCAGGGTTACTGTGG GAGGAGGGCGCTGCAGTTCCCC |
| mut-PTPRU 3’UTR rev | GGGGAACTGCAGCGCCCTCCTCCCACAGTAACC CTGCAGCCCCTGAGATATTTTG |
| Purpose | Name of Primers | Sequence * |
|---|---|---|
| WT | XhoI-RHEBL1 3’UTR fwd | TACATCGCTCGAGCCATCTCATGTGAGCCCTTGG |
| NotI-RHEBL1 3’UTR rev | TAGCGGCCGCGCCAGTGTCCATGAGAGGTCCT | |
| Mutant | mut-RHEBL1 3’UTR fwd | CCGGGGGCAGAAGCAAGTACTTTACCCCACACC CAAGGGC |
| mut-RHEBL1 3’UTR rev | GCCCTTGGGTGTGGGGTAAAGTACTTGCTTCTGC CCCCGG |
| Name of Primers | Sequence * |
|---|---|
| BamHI-pre-miR- 34c fwd | TGCGGATCCCTCAACCAATGAATTGCCTGCC |
| pre-miR- 34c rev | CCACGCACATTGATGATGCACA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grad, M.; Nir, A.; Levy, G.; Trangle, S.S.; Shapira, G.; Shomron, N.; Assaf, Y.; Barak, B. Altered White Matter and microRNA Expression in a Murine Model Related to Williams Syndrome Suggests That miR-34b/c Affects Brain Development via Ptpru and Dcx Modulation. Cells 2022, 11, 158. https://doi.org/10.3390/cells11010158
Grad M, Nir A, Levy G, Trangle SS, Shapira G, Shomron N, Assaf Y, Barak B. Altered White Matter and microRNA Expression in a Murine Model Related to Williams Syndrome Suggests That miR-34b/c Affects Brain Development via Ptpru and Dcx Modulation. Cells. 2022; 11(1):158. https://doi.org/10.3390/cells11010158
Chicago/Turabian StyleGrad, Meitar, Ariel Nir, Gilad Levy, Sari Schokoroy Trangle, Guy Shapira, Noam Shomron, Yaniv Assaf, and Boaz Barak. 2022. "Altered White Matter and microRNA Expression in a Murine Model Related to Williams Syndrome Suggests That miR-34b/c Affects Brain Development via Ptpru and Dcx Modulation" Cells 11, no. 1: 158. https://doi.org/10.3390/cells11010158
APA StyleGrad, M., Nir, A., Levy, G., Trangle, S. S., Shapira, G., Shomron, N., Assaf, Y., & Barak, B. (2022). Altered White Matter and microRNA Expression in a Murine Model Related to Williams Syndrome Suggests That miR-34b/c Affects Brain Development via Ptpru and Dcx Modulation. Cells, 11(1), 158. https://doi.org/10.3390/cells11010158