Human and Mouse Eosinophils Differ in Their Ability to Biosynthesize Eicosanoids, Docosanoids, the Endocannabinoid 2-Arachidonoyl-glycerol and Its Congeners
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Ethics
2.3. Isolation of Human Eosinophils
2.4. Isolation/Differentiation of Mouse Eosinophils
2.5. Isolation of Mouse Eosinophils from Bronchoalveolar Lavage (BAL)
2.6. Flow Cytometry
2.7. Cell Stimulations
2.8. Extraction and Quantification of Lipid Mediators by LC-MS/MS
2.9. Statistical Analysis
3. Results
3.1. Characterization of Mouse Bone Marrow-Derived Eosinophils
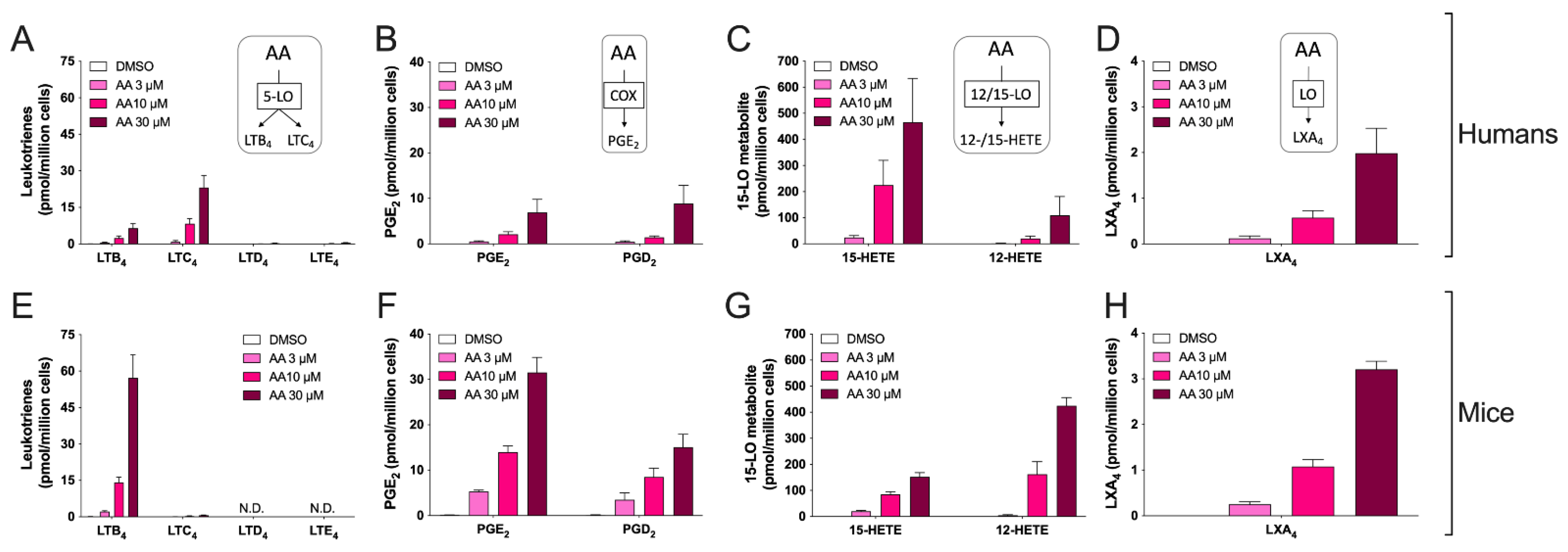
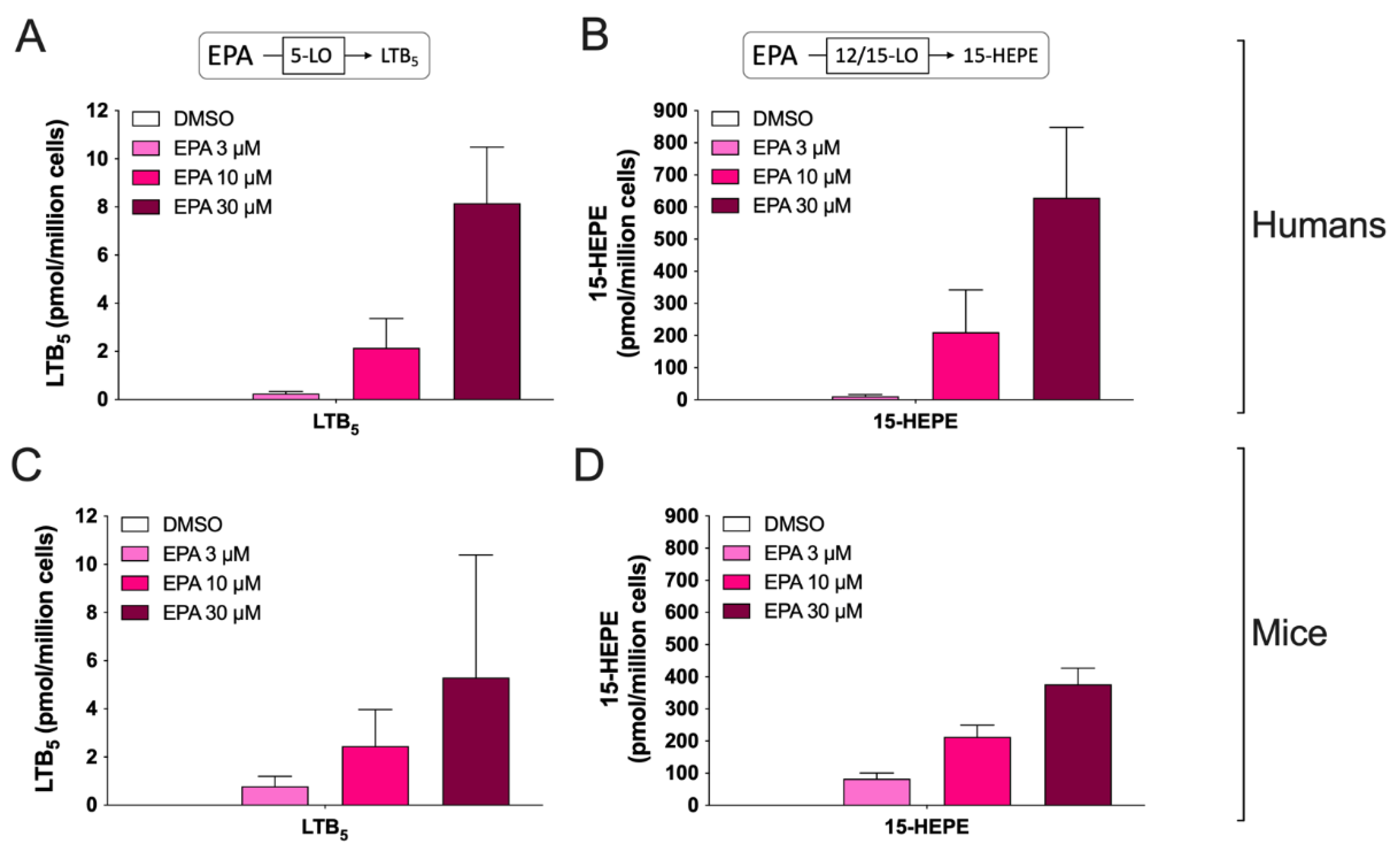
3.2. Biosynthesis of Eicosanoids by hEOS and mEOS
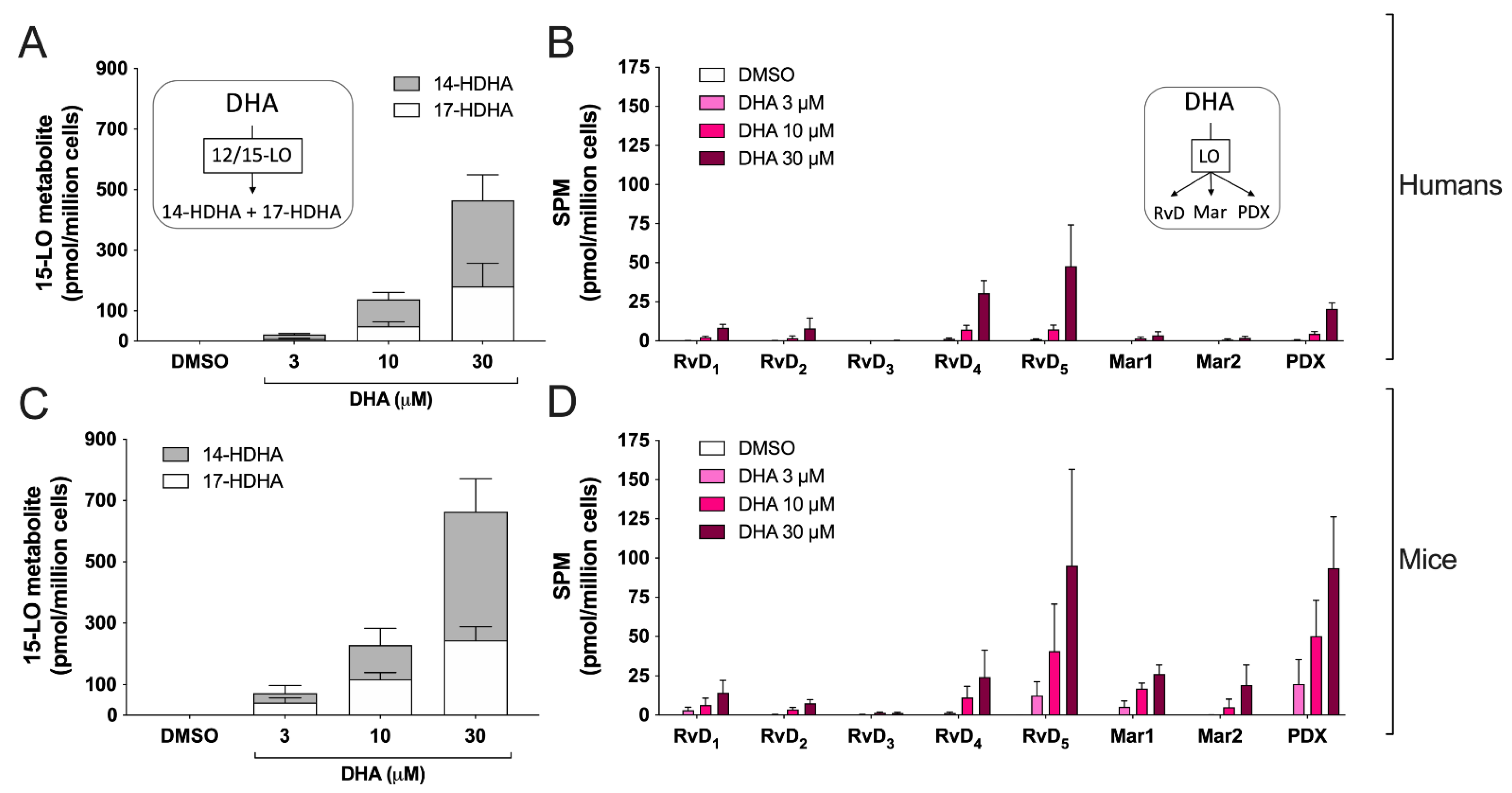
3.3. Biosynthesis of Docosanoids by hEOS and mEOS
3.4. Biosynthesis of the Endocannabinoid 2-AG and Its Congeners by hEOS and mEOS
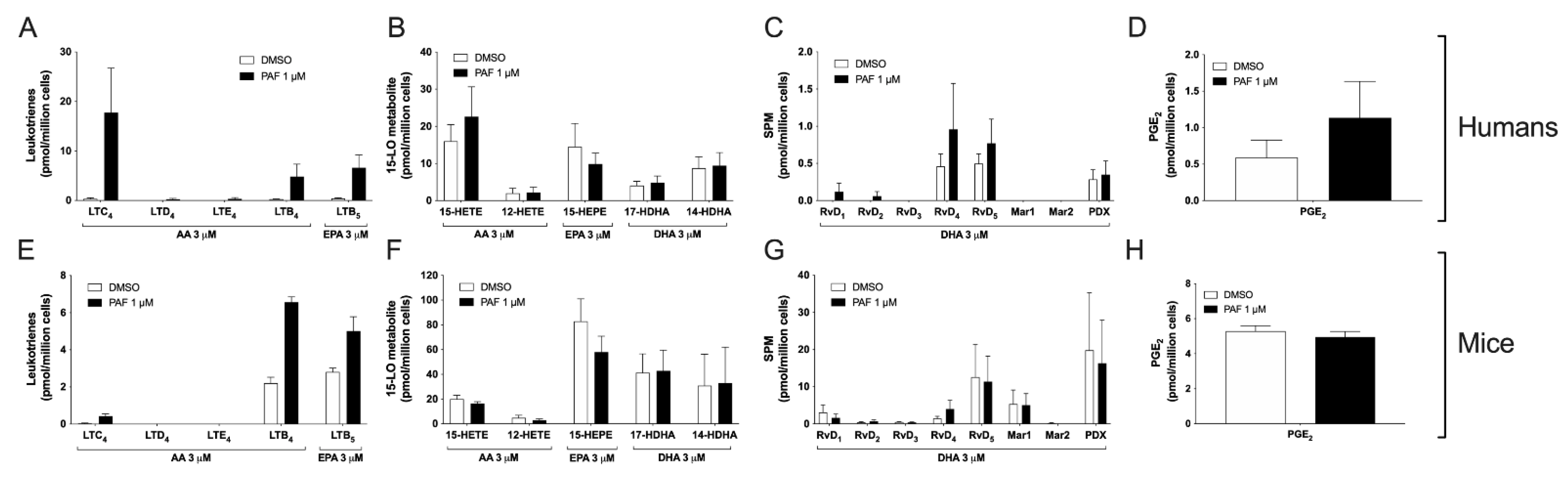
3.5. Impact of the Activation of Cells by PAF in hEOS and mEOS
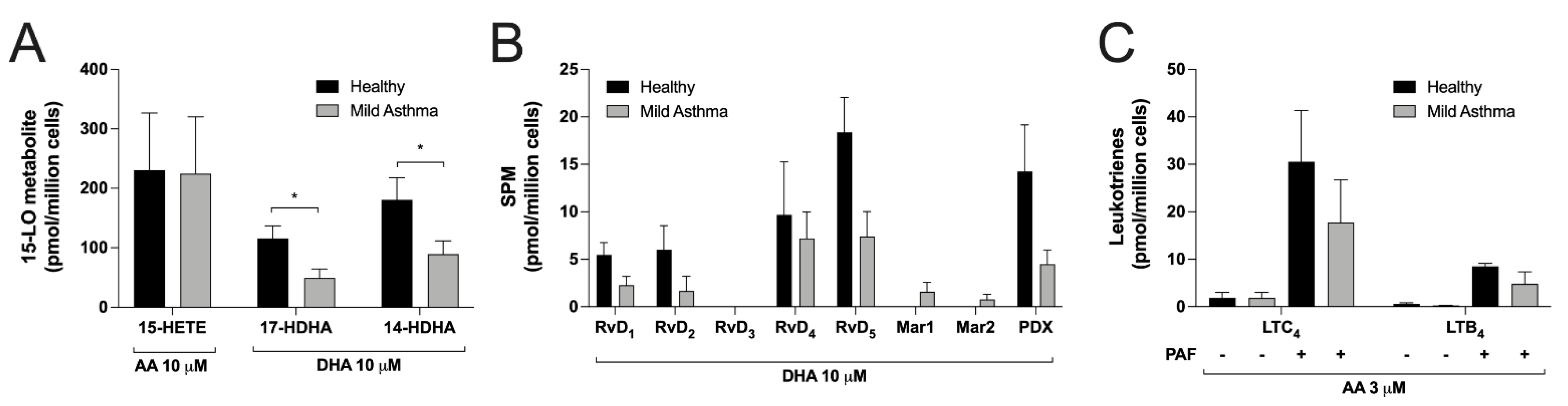
3.6. Differences between Eosinophils Isolated from Mild Asthmatics and Healthy Subjects
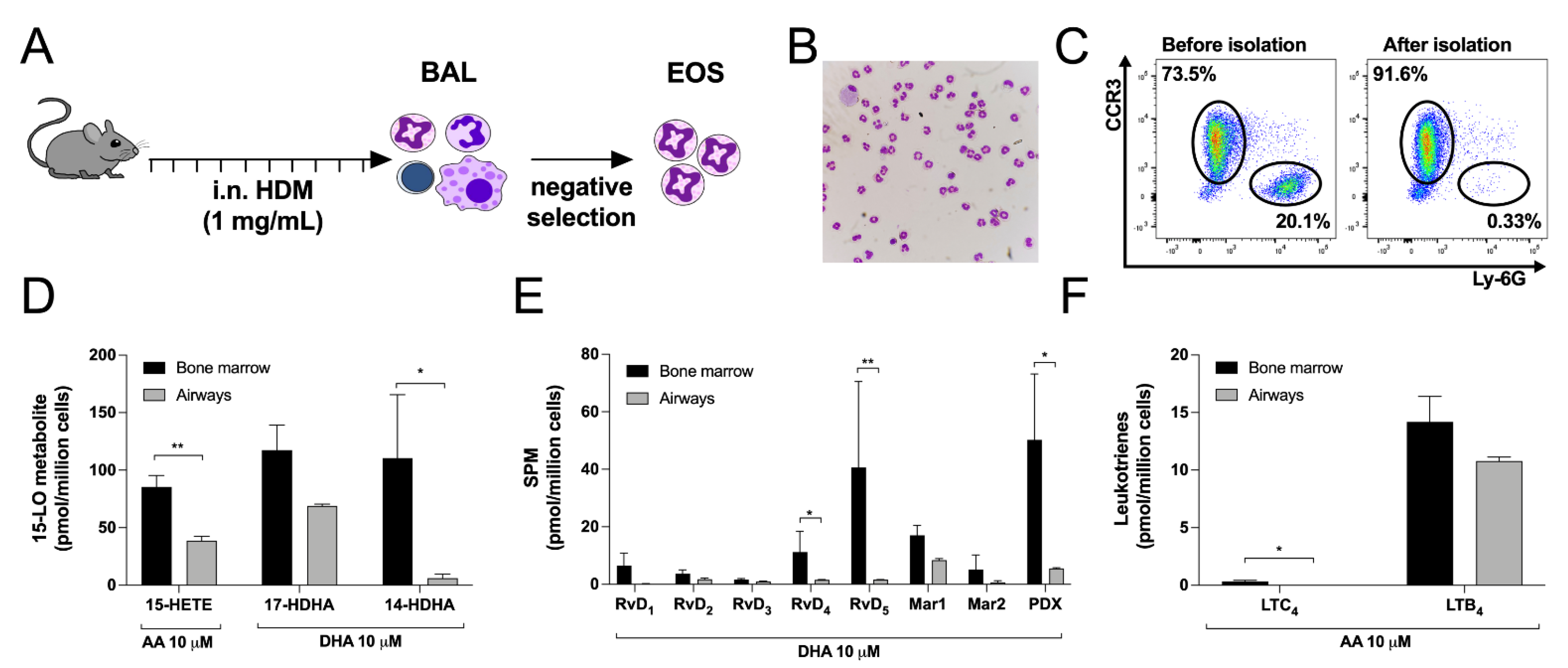
3.7. Differences between Bone Marrow-Derived and Lung Eosinophils from Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bousquet, J.; Jeffery, P.K.; Busse, W.W.; Johnson, M.; Vignola, A.M. Asthma: From bronchoconstriction to airways inflammation and remodeling. Am. J. Respir. Crit. Care Med. 2000, 161, 1720–1745. [Google Scholar] [CrossRef] [Green Version]
- Barnes, P.J. Immunology of asthma and chronic obstructive pulmonary disease. Nat. Rev. Immunol. 2008, 8, 183–192. [Google Scholar] [CrossRef] [PubMed]
- McBrien, C.N.; Menzies-Gow, A. The Biology of Eosinophils and Their Role in Asthma. Front. Med. 2017, 4, 93. [Google Scholar] [CrossRef] [PubMed]
- Choby, G.; Lee, S. Pharmacotherapy for the treatment of asthma: Current treatment options and future directions. Int. Forum Allergy Rhinol. 2015, 5 (Suppl. 1), S35–S40. [Google Scholar] [CrossRef] [PubMed]
- Terawaki, K.; Yokomizo, T.; Nagase, T.; Toda, A.; Taniguchi, M.; Hashizume, K.; Yagi, T.; Shimizu, T. Absence of Leukotriene B4Receptor 1 Confers Resistance to Airway Hyperresponsiveness and Th2-Type Immune Responses. J. Immunol. 2005, 175, 4217–4225. [Google Scholar] [CrossRef] [Green Version]
- Henderson, W.R., Jr.; Lewis, D.B.; Albert, R.K.; Zhang, Y.; Lamm, W.J.; Chiang, G.K.; Jones, F.; Eriksen, P.; Tien, Y.T.; Jonas, M.; et al. The importance of leukotrienes in airway inflammation in a mouse model of asthma. J. Exp. Med. 1996, 184, 1483–1494. [Google Scholar] [CrossRef]
- Singh, R.K.; Tandon, R.; Dastidar, S.G.; Ray, A. A review on leukotrienes and their receptors with reference to asthma. J. Asthma 2013, 50, 922–931. [Google Scholar] [CrossRef]
- Evans, D.J.; Barnes, P.J.; Spaethe, S.M.; Van Alstyne, E.L.; Mitchell, M.I.; O’Connor, B.J. Effect of a leukotriene B4 receptor antagonist, LY293111, on allergen induced responses in asthma. Thorax 1996, 51, 1178–1184. [Google Scholar] [CrossRef] [Green Version]
- LaRose, M.-C.; Archambault, A.-S.; Provost, V.; LaViolette, M.; Flamand, N. Regulation of Eosinophil and Group 2 Innate Lymphoid Cell Trafficking in Asthma. Front. Med. 2017, 4, 136. [Google Scholar] [CrossRef] [Green Version]
- Kytikova, O.; Novgorodtseva, T.; Denisenko, Y.; Antonyuk, M.; Gvozdenko, T. Pro-Resolving Lipid Mediators in the Pathophysiology of Asthma. Medicina 2019, 55, 284. [Google Scholar] [CrossRef] [Green Version]
- Turcotte, C.; Chouinard, F.; Lefebvre, J.S.; Flamand, N. Regulation of inflammation by cannabinoids, the endocannabinoids 2-arachidonoyl-glycerol and arachidonoyl-ethanolamide, and their metabolites. J. Leukoc. Biol. 2015, 97, 1049–1070. [Google Scholar] [CrossRef]
- Turcotte, C.; Blanchet, M.-R.; LaViolette, M.; Flamand, N. The CB2 receptor and its role as a regulator of inflammation. Cell. Mol. Life Sci. 2016, 73, 4449–4470. [Google Scholar] [CrossRef] [Green Version]
- Serhan, C.N.; Levy, B.D. Resolvins in inflammation: Emergence of the pro-resolving superfamily of mediators. J. Clin. Investig. 2018, 128, 2657–2669. [Google Scholar] [CrossRef] [PubMed]
- Larose, M.-C.; Turcotte, C.; Chouinard, F.; Ferland, C.; Martin, C.; Provost, V.; Laviolette, M.; Flamand, N. Mechanisms of human eosinophil migration induced by the combination of IL-5 and the endocannabinoid 2-arachidonoyl-glycerol. J. Allergy Clin. Immunol. 2014, 133, 1480–1482.e1–3. [Google Scholar] [CrossRef] [PubMed]
- Dyer, K.D.; Moser, J.M.; Czapiga, M.; Siegel, S.J.; Percopo, C.M.; Rosenberg, H.F. Functionally Competent Eosinophils Differentiated Ex Vivo in High Purity from Normal Mouse Bone Marrow. J. Immunol. 2008, 181, 4004–4009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Archambault, A.-S.; Poirier, S.; Lefebvre, J.-S.; Robichaud, P.-P.; LaRose, M.-C.; Turcotte, C.; Martin, C.; Provost, V.; Boudreau, L.H.; McDonald, P.P.; et al. 20-Hydroxy- and 20-carboxy-leukotriene (LT) B4 downregulate LTB4 -mediated responses of human neutrophils and eosinophils. J. Leukoc. Biol. 2019, 105, 1131–1142. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Rothenberg, M. Bone Marrow Derived Eosinophil Cultures. Bio-Protocol 2014, 4. [Google Scholar] [CrossRef] [Green Version]
- Xia, L.-X.; Hua, W.; Jin, Y.; Tian, B.-P.; Qiu, Z.-W.; Zhang, C.; Che, L.-Q.; Zhou, H.-B.; Wu, Y.-F.; Huang, H.-Q.; et al. Eosinophil differentiation in the bone marrow is promoted by protein tyrosine phosphatase SHP2. Cell Death Dis. 2016, 7, e2175. [Google Scholar] [CrossRef] [Green Version]
- Lu, T.X.; Lim, E.-J.; Besse, J.A.; Itskovich, S.; Plassard, A.J.; Fulkerson, P.C.; Aronow, B.J.; Rothenberg, M.E. MiR-223 deficiency increases eosinophil progenitor proliferation. J. Immunol. 2013, 190, 1576–1582. [Google Scholar] [CrossRef] [Green Version]
- Willebrand, R.; Dietschmann, A.; Nitschke, L.; Krappmann, S.; Voehringer, D. Murine eosinophil development and allergic lung eosinophilia are largely dependent on the signaling adaptor GRB2. Eur. J. Immunol. 2018, 48, 1786–1795. [Google Scholar] [CrossRef] [Green Version]
- Wen, T.; Besse, J.A.; Mingler, M.K.; Fulkerson, P.C.; Rothenberg, M.E. Eosinophil adoptive transfer system to directly evaluate pulmonary eosinophil trafficking in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 6067–6072. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.J.; Jacobsen, E.A.; Ochkur, S.I.; McGarry, M.P.; Condjella, R.M.; Doyle, A.D.; Luo, H.; Zellner, K.R.; Protheroe, C.A.; Willetts, L.; et al. Human versus mouse eosinophils: “That which we call an eosinophil, by any other name would stain as red”. J. Allergy Clin. Immunol. 2012, 130, 572–584. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Lanckacker, E.; Dentener, M.; Bracke, K.; Provoost, S.; De Grove, K.; Brusselle, G.; Wouters, E.; Maes, T.; Joos, G. Aggravation of Allergic Airway Inflammation by Cigarette Smoke in Mice Is CD44-Dependent. PLoS ONE 2016, 11, e0151113. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.H.; Sun, A.H.; Ojcius, D.M.; Hu, W.L.; Ge, Y.M.; Lin, X.A.; Li, L.J.; Pan, J.P.; Yan, J. Eosinophils from Murine Lamina Propria Induce Differentiation of Naive T Cells into Regulatory T Cells via TGF-beta1 and Retinoic Acid. PLoS ONE 2015, 10, e0142881. [Google Scholar]
- Stevens, W.W.; Kim, T.S.; Pujanauski, L.M.; Hao, X.; Braciale, T.J. Detection and quantitation of eosinophils in the murine respiratory tract by flow cytometry. J. Immunol. Methods 2007, 327, 63–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macri, C.; Pang, E.S.; Patton, T.; O’Keeffe, M. Dendritic cell subsets. Semin. Cell Dev. Biol. 2018, 84, 11–21. [Google Scholar] [CrossRef]
- Turcotte, C.; Zarini, S.; Jean, S.; Martin, C.; Murphy, R.C.; Marsolais, D.; LaViolette, M.; Blanchet, M.-R.; Flamand, N. The Endocannabinoid Metabolite Prostaglandin E2 (PGE2)-Glycerol Inhibits Human Neutrophil Functions: Involvement of Its Hydrolysis into PGE2 and EP Receptors. J. Immunol. 2017, 198, 3255–3263. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.P.; Cuthbert, M.F.; Dunlop, L.S. Effects of Inhaled Prostaglandins E1, E2 and F2α on the Airway Resistance of Healthy and Asthmatic Man. Clin. Sci. Mol. Med. 1975, 48, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Säfholm, J.; Manson, M.; Bood, J.; Delin, I.; Orre, A.-C.; Bergman, P.; Al-Ameri, M.; Dahlén, S.-E.; Adner, M. Prostaglandin E2 inhibits mast cell–dependent bronchoconstriction in human small airways through the E prostanoid subtype 2 receptor. J. Allergy Clin. Immunol. 2015, 136, 1232–1239.e1. [Google Scholar] [CrossRef]
- Rittchen, S.; Heinemann, A. Therapeutic Potential of Hematopoietic Prostaglandin D2 Synthase in Allergic Inflammation. Cells 2019, 8, 619. [Google Scholar] [CrossRef] [Green Version]
- Morley, J.; Bray, M.; Jones, R.; Nugteren, D.; van Dorp, D. Prostaglandin and thromboxane production by human and guinea-pig macrophages and leucocytes. Prostaglandins 1979, 17, 729–736. [Google Scholar] [CrossRef]
- Foegh, M.L.; Maddox, Y.T.; Ramwell, P.W. Human Peritoneal Eosinophils and Formation of Arachidonate Cyclooxygenase Products. Scand. J. Immunol. 1986, 23, 599–603. [Google Scholar] [CrossRef]
- Chu, H.W.; Balzar, S.; Westcott, J.Y.; Trudeau, J.B.; Sun, Y.; Conrad, D.J.; Wenzel, S.E. Expression and activation of 15-lipoxygenase pathway in severe asthma: Relationship to eosinophilic phenotype and collagen deposition. Clin. Exp. Allergy 2002, 32, 1558–1565. [Google Scholar] [CrossRef] [PubMed]
- Profita, M.; Sala, A.; Riccobono, L.; Paternò, A.; Mirabella, A.; Bonanno, A.; Guerrera, D.; Pace, E.; Bonsignore, G.; Bousquet, J.; et al. 15-Lipoxygenase expression and 15(S)-hydroxyeicoisatetraenoic acid release and reincorporation in induced sputum of asthmatic subjects. J. Allergy Clin. Immunol. 2000, 105, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.; Kuhn, H.; Heydeck, D. Structural and functional biology of arachidonic acid 15-lipoxygenase-1 (ALOX15). Gene 2015, 573, 1–32. [Google Scholar] [CrossRef]
- Kutzner, L.; Goloshchapova, K.; Heydeck, D.; Stehling, S.; Kuhn, H.; Schebb, N.H. Mammalian ALOX15 orthologs exhibit pronounced dual positional specificity with docosahexaenoic acid. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2017, 1862, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Hajek, A.R.; Lindley, A.R.; Favoreto, S., Jr.; Carter, R.; Schleimer, R.P.; Kuperman, D.A. 12/15-Lipoxygenase deficiency protects mice from allergic airways inflammation and increases secretory IgA levels. J. Allergy Clin. Immunol. 2008, 122, 633–639.e3. [Google Scholar] [CrossRef] [Green Version]
- Andersson, C.K.; Claesson, H.E.; Rydell-Tormanen, K.; Swedmark, S.; Hallgren, A.; Erjefalt, J.S. Mice lacking 12/15-lipoxygenase have attenuated airway allergic inflammation and remodeling. Am. J. Respir. Cell Mol. Biol. 2008, 39, 648–656. [Google Scholar] [CrossRef]
- Johannesson, M.; Backman, L.; Claesson, H.E.; Forsell, P.K.A. Cloning, purification and characterization of non-human primate 12/15-lipoxygenases. Prostaglandins Leukot. Essent. Fat. Acids 2010, 82, 121–129. [Google Scholar] [CrossRef]
- Archambault, A.-S.; Turcotte, C.; Martin, C.; Provost, V.; Larose, M.-C.; Laprise, C.; Chakir, J.; Bissonnette, É.; Laviolette, M.; Bossé, Y.; et al. Comparison of eight 15-lipoxygenase (LO) inhibitors on the biosynthesis of 15-LO metabolites by human neutrophils and eosinophils. PLoS ONE 2018, 13, e0202424. [Google Scholar] [CrossRef] [Green Version]
- Isobe, Y.; Arita, M.; Matsueda, S.; Iwamoto, R.; Fujihara, T.; Nakanishi, H.; Taguchi, R.; Masuda, K.; Sasaki, K.; Urabe, D.; et al. Identification and Structure Determination of Novel Anti-inflammatory Mediator Resolvin E3, 17,18-Dihydroxyeicosapentaenoic Acid. J. Biol. Chem. 2012, 287, 10525–10534. [Google Scholar] [CrossRef] [Green Version]
- Di Marzo, V. New approaches and challenges to targeting the endocannabinoid system. Nat. Rev. Drug Discov. 2018, 17, 623–639. [Google Scholar] [CrossRef] [PubMed]
- Turcotte, C.; Dumais, É.; Archambault, A.; Martin, C.; Blanchet, M.; Bissonnette, É.; Boulet, L.; Laviolette, M.; Di Marzo, V.; Flamand, N. Human leukocytes differentially express endocannabinoid-glycerol lipases and hydrolyze 2-arachidonoyl-glycerol and its metabolites from the 15-lipoxygenase and cyclooxygenase pathways. J. Leukoc. Biol. 2019, 106, 1337–1347. [Google Scholar] [CrossRef] [PubMed]
- Turcotte, C.; Archambault, A.; Dumais, É.; Martin, C.; Blanchet, M.; Bissonnette, E.; Ohashi, N.; Yamamoto, K.; Itoh, T.; Laviolette, M.; et al. Endocannabinoid hydrolysis inhibition unmasks that unsaturated fatty acids induce a robust biosynthesis of 2-arachidonoyl-glycerol and its congeners in human myeloid leukocytes. FASEB J. 2020, 34, 4253–4265. [Google Scholar] [CrossRef] [Green Version]
- Krump, E.; Pouliot, M.; Naccache, P.H.; Borgeat, P. Leukotriene synthesis in calcium-depleted human neutrophils: Arachidonic acid release correlates with calcium influx. Biochem. J. 1995, 310 Pt 2, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Bernatchez, E.; Gold, M.J.; Langlois, A.; Lemay, A.-M.; Brassard, J.; Flamand, N.; Marsolais, D.; McNagny, K.M.; Blanchet, M.-R. Pulmonary CD103 expression regulates airway inflammation in asthma. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 308, L816–L826. [Google Scholar] [CrossRef] [Green Version]
- Pal, K.; Feng, X.; Steinke, J.W.; Burdick, M.D.; Shim, Y.; Sung, S.-S.; Teague, W.G.; Borish, L. Leukotriene A4 Hydrolase Activation and Leukotriene B4 Production by Eosinophils in Severe Asthma. Am. J. Respir. Cell Mol. Biol. 2019, 60, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Watkins, A.D.; A Hatfield, C.; Fidler, S.F.; E Winterrowd, G.; Brashler, J.R.; Sun, F.F.; Taylor, B.M.; Vonderfecht, S.L.; A Conder, G.; Holgate, S.T.; et al. Phenotypic analysis of airway eosinophils and lymphocytes in a Th-2-driven murine model of pulmonary inflammation. Am. J. Respir. Cell Mol. Biol. 1996, 15, 20–34. [Google Scholar] [CrossRef]
- de Andres, B.; Del Pozo, V.; Martin, E.; Palomino, P.; Lahoz, C. Release of O2- and LTC4 by murine eosinophils: Role of intra- and extracellular calcium. Immunology 1990, 69, 271–276. [Google Scholar] [PubMed]
- Lam, B.K.; Penrose, J.F.; Rokach, J.; Xu, K.; Baldasaro, M.H.; Austen, K.F. Molecular cloning, expression and characterization of mouse leukotriene C4 synthase. Eur. J. Biochem. 1996, 238, 606–612. [Google Scholar] [CrossRef]
- Hirata, H.; Arima, M.; Fukushima, Y.; Honda, K.; Sugiyama, K.; Tokuhisa, T.; Fukuda, T. Over-expression of the LTC4 synthase gene in mice reproduces human aspirin-induced asthma. Clin. Exp. Allergy 2011, 41, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.R.; Gerard, N.P.; Galli, S.J.; Drazen, J.M. Pulmonary responses to bronchoconstrictor agonists in the mouse. J. Appl. Physiol. 1988, 64, 2318–2323. [Google Scholar] [CrossRef] [PubMed]
- Turk, J.; Rand, T.H.; Maas, R.L.; Lawson, J.A.; Brash, A.R.; Roberts, L.; Colley, D.G.; Oates, J.A. Identification of lipoxygenase products from arachidonic acid metabolism in stimulated murine eosinophils. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1983, 750, 78–90. [Google Scholar] [CrossRef]
- Powell, W.S.; Rokach, J. Biosynthesis, biological effects, and receptors of hydroxyeicosatetraenoic acids (HETEs) and oxoeicosatetraenoic acids (oxo-ETEs) derived from arachidonic acid. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2015, 1851, 340–355. [Google Scholar] [CrossRef] [Green Version]
- Kristjansson, R.P.; Benonisdottir, S.; Davidsson, O.B.; Oddsson, A.; Tragante, V.; Sigurdsson, J.K.; Stefansdottir, L.; Jonsson, S.; Jensson, B.O.; Arthur, J.G.; et al. A loss-of-function variant in ALOX15 protects against nasal polyps and chronic rhinosinusitis. Nat. Genet. 2019, 51, 267–276. [Google Scholar] [CrossRef]
- Miyata, J.; Yokokura, Y.; Moro, K.; Arai, H.; Fukunaga, K.; Arita, M. 12/15-Lipoxygenase Regulates IL-33-Induced Eosinophilic Airway Inflammation in Mice. Front. Immunol. 2021, 12, 687192. [Google Scholar] [CrossRef] [PubMed]
- Feltenmark, S.; Gautam, N.; Brunnstrom, A.; Griffiths, W.; Backman, L.; Edenius, C.; Lindbom, L.; Bjorkholm, M.; Claesson, H.-E. Eoxins are proinflammatory arachidonic acid metabolites produced via the 15-lipoxygenase-1 pathway in human eosinophils and mast cells. Proc. Natl. Acad. Sci. USA 2008, 105, 680–685. [Google Scholar] [CrossRef] [Green Version]
- Archambault, A.; Zaid, Y.; Rakotoarivelo, V.; Turcotte, C.; Doré, É.; Dubuc, I.; Martin, C.; Flamand, O.; Amar, Y.; Cheikh, A.; et al. High levels of eicosanoids and docosanoids in the lungs of intubated COVID-19 patients. FASEB J. 2021, 35, e21666. [Google Scholar] [CrossRef] [PubMed]
- Miyata, J.; Fukunaga, K.; Kawashima, Y.; Watanabe, T.; Saitoh, A.; Hirosaki, T.; Araki, Y.; Kikawada, T.; Betsuyaku, T.; Ohara, O.; et al. Dysregulated fatty acid metabolism in nasal polyp-derived eosinophils from patients with chronic rhinosinusitis. Allergy 2019, 74, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Miyata, J.; Fukunaga, K.; Iwamoto, R.; Isobe, Y.; Niimi, K.; Takamiya, R.; Takihara, T.; Tomomatsu, K.; Suzuki, Y.; Oguma, T.; et al. Dysregulated synthesis of protectin D1 in eosinophils from patients with severe asthma. J. Allergy Clin. Immunol. 2013, 131, 353–360.e2. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Weng, Q.-Y.; Zhou, L.-R.; Cao, C.; Li, F.; Wu, Y.-F.; Wu, Y.-P.; Li, M.; Hu, Y.; Shen, J.-X.; et al. Homeostatic and early-recruited CD101− eosinophils suppress endotoxin-induced acute lung injury. Eur. Respir. J. 2020, 56, 1902354. [Google Scholar] [CrossRef] [PubMed]

| Lipid | Internal Standard Used | Q1 → Q3 | Retention Time (min) | Detection Limit (fmol) |
|---|---|---|---|---|
| LTB4-d4 | - | 339.30 → 197.20 | 7.39 | - |
| 15-HETE-d8 | - | 327.20 → 226.20 | 10.20 | - |
| 1-AG-d5 | - | 384.50 → 287.20 | 12.64 | - |
| RvD2-d5 | - | 380.40 → 175.20 | 5.31 | - |
| PGE2-d4 | - | 355.20 → 275.35 | 5.04 | - |
| LTC4 | LTB4-d4 | 624.30 → 272.20 | 5.69 | 50 |
| LTD4 | LTB4-d4 | 495.30 → 176.85 | 5.38 | 50 |
| LTE4 | LTB4-d4 | 438.30 → 333.20 | 6.28 | 50 |
| LTB4 | LTB4-d4 | 335.30 → 195.25 | 7.47 | 50 |
| LTB5 | LTB4-d4 | 333.40 → 195.25 | 6.62 | 50 |
| 15-HETE | 15-HETE-d8 | 319.40 → 219.30 | 9.76 | 50 |
| 12-HETE | 15-HETE-d8 | 319.10 → 179.25 | 10.11 | 50 |
| 15-HEPE | 15-HETE-d8 | 317.40 → 219.25 | 9.05 | 50 |
| 17-HDHA | 15-HETE-d8 | 343.50 → 281.30 | 10.43 | 50 |
| 14-HDHA | 15-HETE-d8 | 343.50 → 281.30 | 10.59 | 50 |
| 2-AG | 1-AG-d5 | 379.30 → 287.25 | 12.54 | 25 |
| 2-DHG | 1-AG-d5 | 403.20 → 311.20 | 12.57 | 50 |
| 2-EPG | 1-AG-d5 | 377.10 → 285.25 | 11.74 | 50 |
| RvD1 | RvD2-d5 | 375.40 → 141.10 | 5.36 | 50 |
| RvD2 | RvD2-d5 | 375.40 → 175.20 | 5.36 | 50 |
| RvD3 | RvD2-d5 | 375.40 → 147.20 | 5.15 | 50 |
| RvD4 | RvD2-d5 | 375.40 → 101.05 | 6.34 | 50 |
| RvD5 | RvD2-d5 | 359.40 → 199.25 | 7.51 | 50 |
| RvE1 | RvD2-d5 | 349.30 → 195.20 | 3.53 | 50 |
| Maresin 1 | RvD2-d5 | 359.40 → 177.25 | 7.42 | 50 |
| Maresin 2 | RvD2-d5 | 359.40 → 221.05 | 8.20 | 50 |
| PDX | RvD2-d5 | 359.30 → 153.15 | 7.48 | 50 |
| 6-keto-PGF1α | PGE2-d4 | 369.30 → 163.10 | 3.42 | 50 |
| PGE2 | PGE2-d4 | 351.20 → 271.15 | 5.08 | 5 |
| PGD2 | PGE2-d4 | 351.30 → 271.20 | 5.31 | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Archambault, A.-S.; Brassard, J.; Bernatchez, É.; Martin, C.; Di Marzo, V.; Laviolette, M.; Boulet, L.-P.; Blanchet, M.-R.; Flamand, N. Human and Mouse Eosinophils Differ in Their Ability to Biosynthesize Eicosanoids, Docosanoids, the Endocannabinoid 2-Arachidonoyl-glycerol and Its Congeners. Cells 2022, 11, 141. https://doi.org/10.3390/cells11010141
Archambault A-S, Brassard J, Bernatchez É, Martin C, Di Marzo V, Laviolette M, Boulet L-P, Blanchet M-R, Flamand N. Human and Mouse Eosinophils Differ in Their Ability to Biosynthesize Eicosanoids, Docosanoids, the Endocannabinoid 2-Arachidonoyl-glycerol and Its Congeners. Cells. 2022; 11(1):141. https://doi.org/10.3390/cells11010141
Chicago/Turabian StyleArchambault, Anne-Sophie, Julyanne Brassard, Émilie Bernatchez, Cyril Martin, Vincenzo Di Marzo, Michel Laviolette, Louis-Philippe Boulet, Marie-Renée Blanchet, and Nicolas Flamand. 2022. "Human and Mouse Eosinophils Differ in Their Ability to Biosynthesize Eicosanoids, Docosanoids, the Endocannabinoid 2-Arachidonoyl-glycerol and Its Congeners" Cells 11, no. 1: 141. https://doi.org/10.3390/cells11010141
APA StyleArchambault, A.-S., Brassard, J., Bernatchez, É., Martin, C., Di Marzo, V., Laviolette, M., Boulet, L.-P., Blanchet, M.-R., & Flamand, N. (2022). Human and Mouse Eosinophils Differ in Their Ability to Biosynthesize Eicosanoids, Docosanoids, the Endocannabinoid 2-Arachidonoyl-glycerol and Its Congeners. Cells, 11(1), 141. https://doi.org/10.3390/cells11010141






