A Novel Type III Endosome Transmembrane Protein, TEMP
Abstract
:1. Introduction
2. Results and Discussion
2.1. Bioinformatic Analysis of TEMP

2.2. TEMP, is a Type III Transmembrane Protein

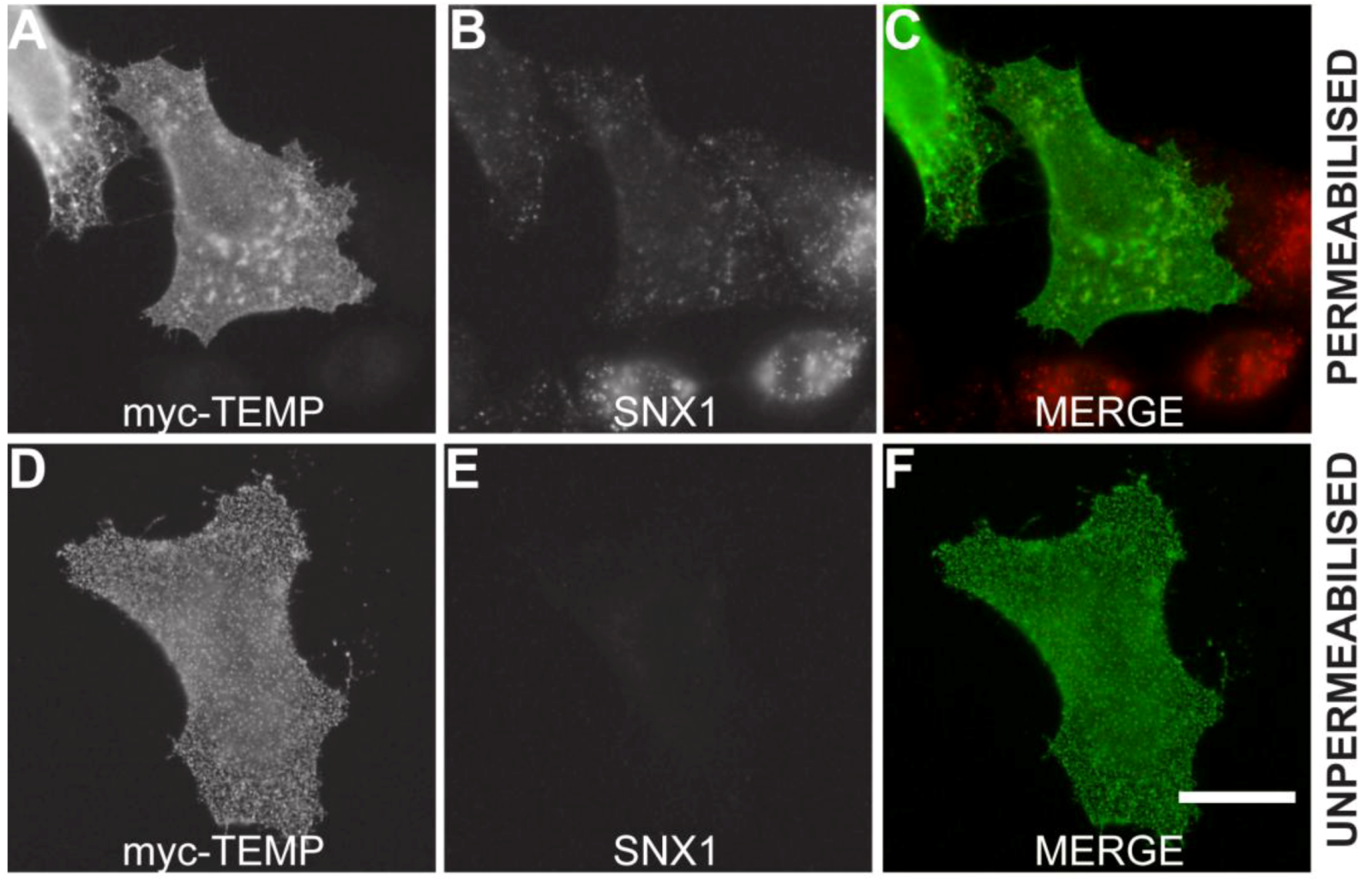
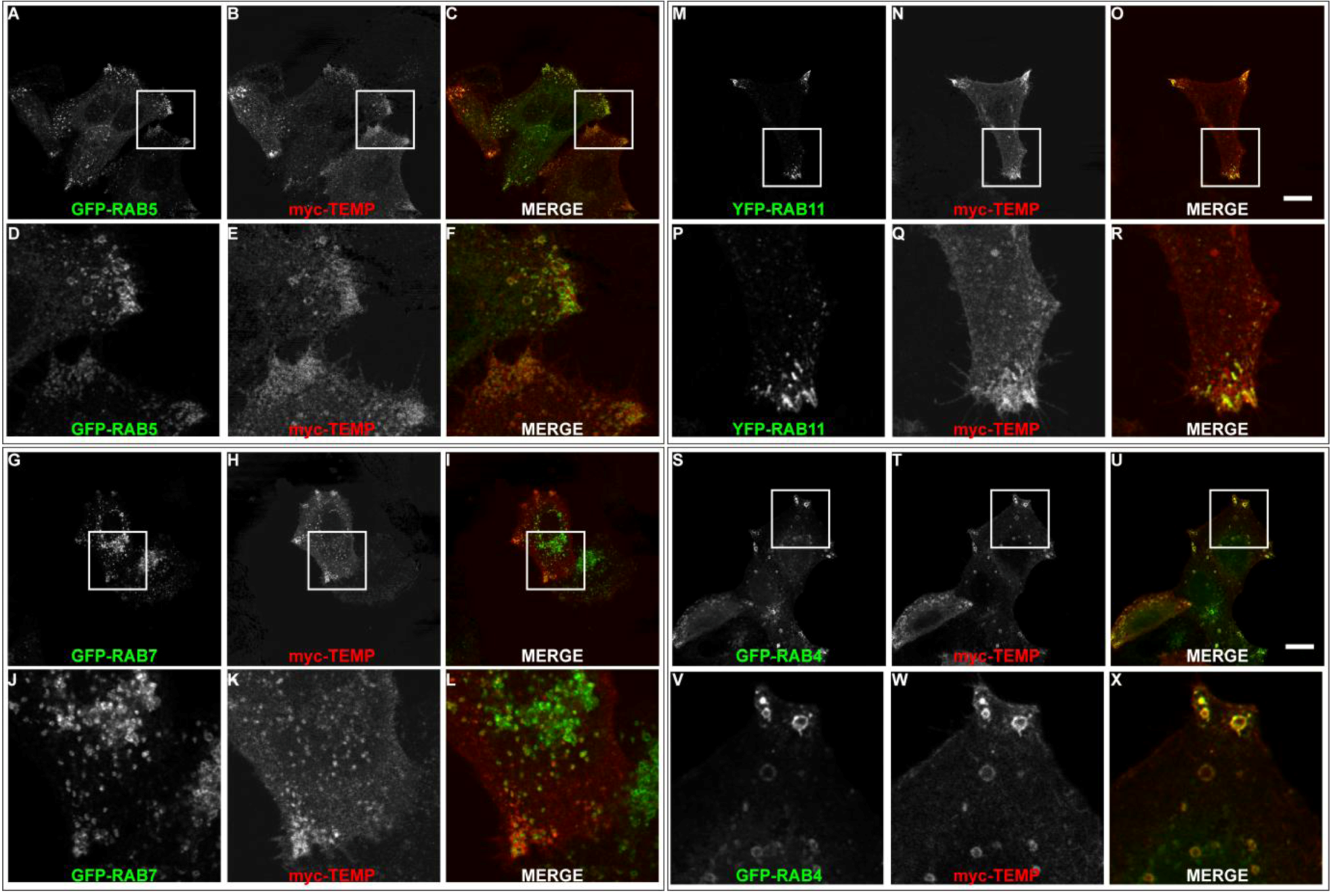
2.3. TEMP Extensively Colocalises with Early Endosomes and Recycling Endosomes
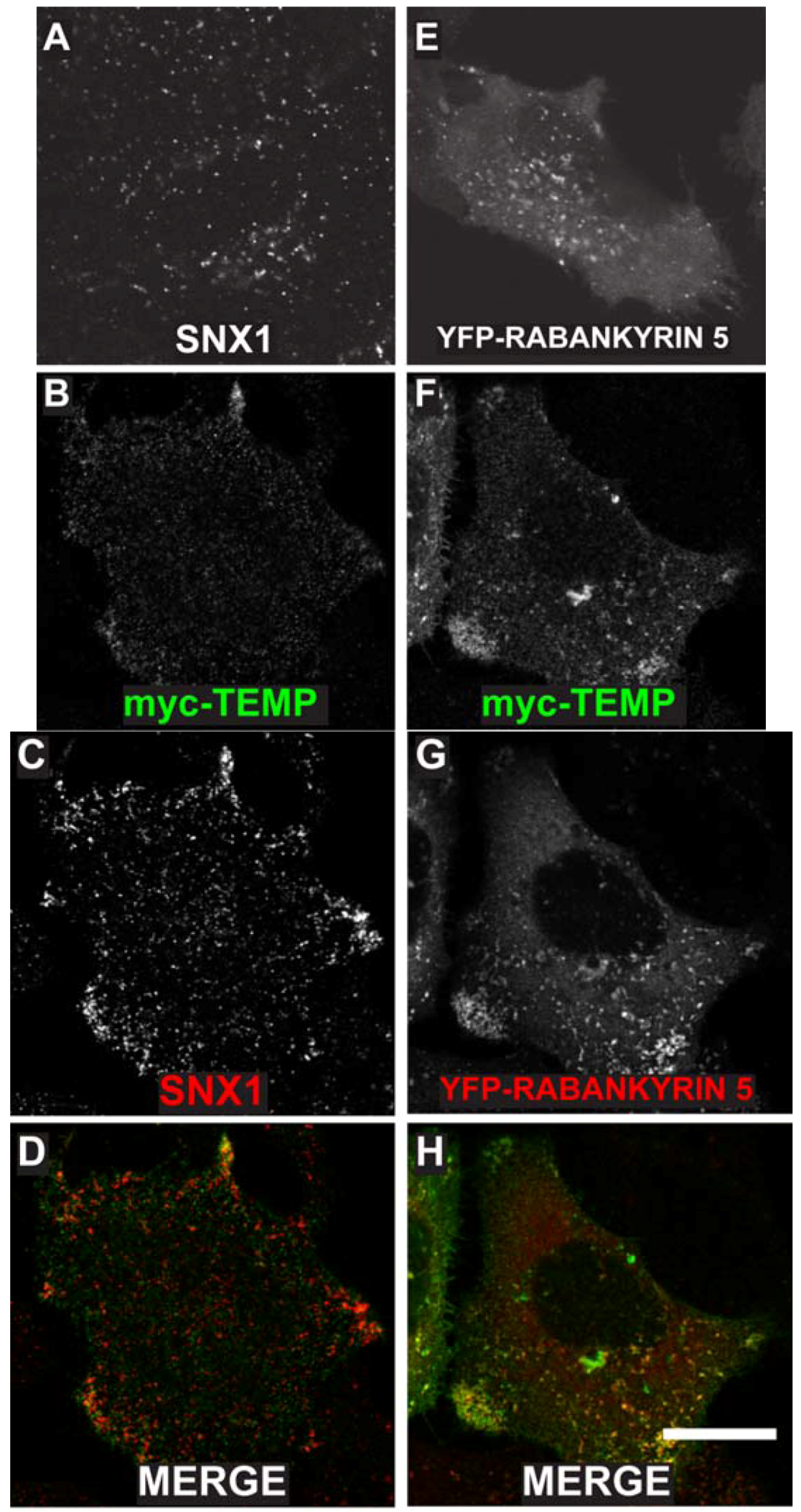
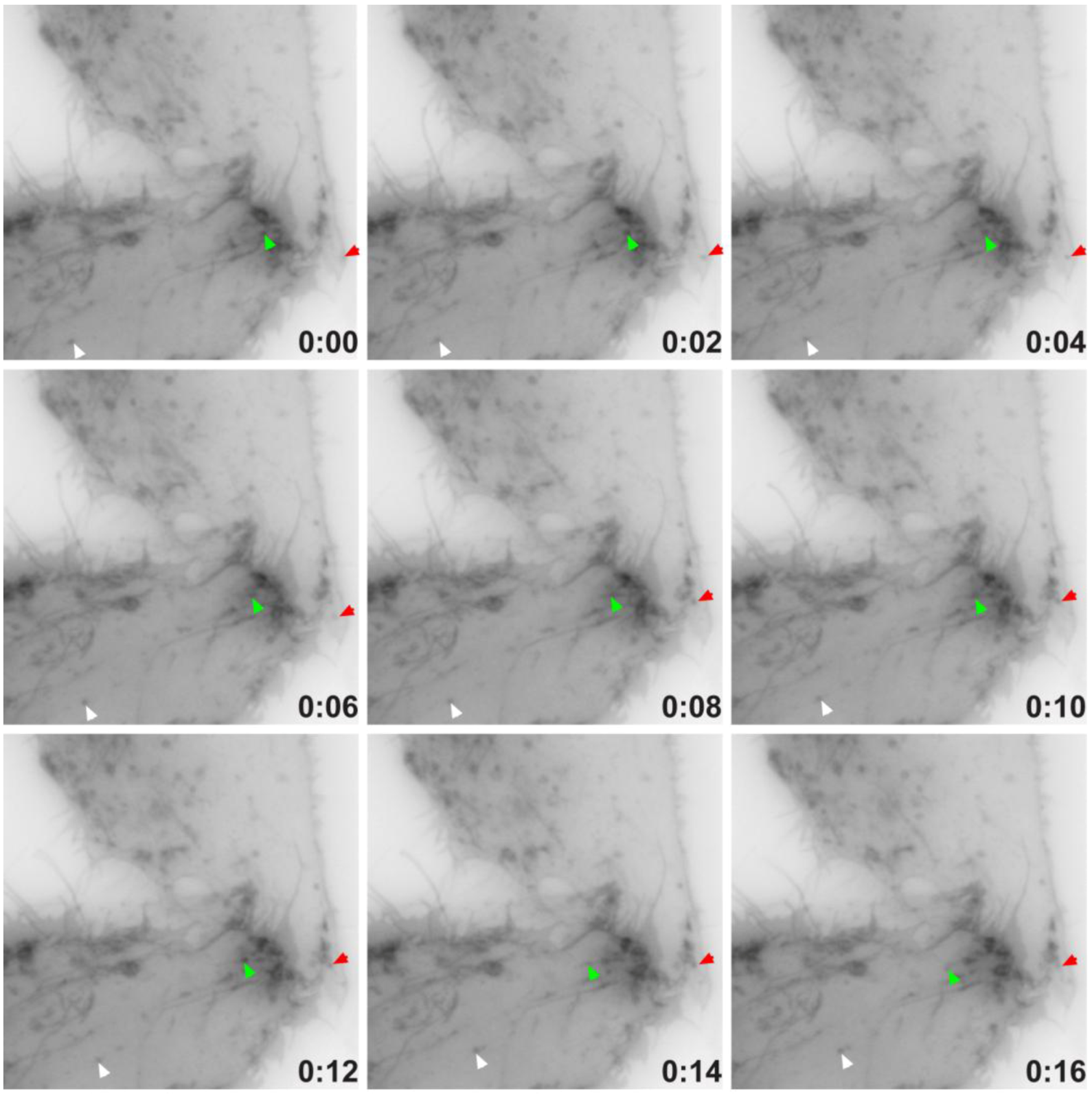
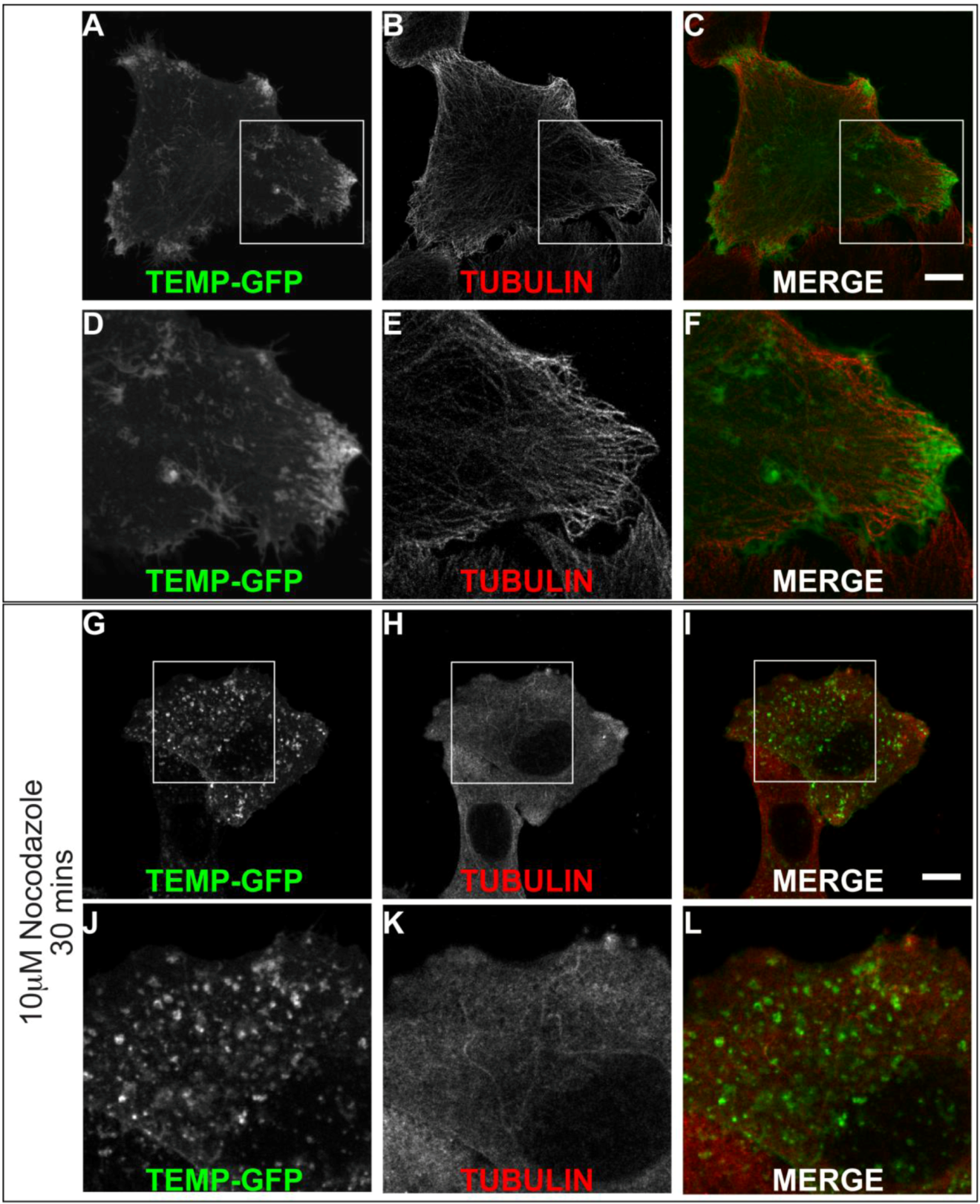
2.4. TEMP Associates with Endosomal Tubular-Vesicular Structures in Living Cells
3. Experimental
3.1. Bioinformatics
3.2. Generation of Amino-Terminal Myc-Tagged Expression Constructs
3.3. Generation of C-terminal GFP-tagged Expression Constructs
3.4. Plasmids
3.5. Antibodies
3.6. Cell Culture and Transfection
3.7. Western Immunoblotting
3.8. Surface Labelling Immunofluorescence Protocol (Unpermeabilised cells)
3.9. Indirect Immunofluorescence
3.10. Confocal Microscopy
3.11. Time-Lapse Videomicroscopy
4. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Aturaliya, R.N.; Fink, J.L.; Davis, M.J.; Teasdale, M.S.; Hanson, K.A.; Miranda, K.C.; Forrest, A.R.; Grimmond, S.M.; Suzuki, H.; Kanamori, M.; et al. Subcellular localization of mammalian type ii membrane proteins. Traffic 2006, 7, 613–625. [Google Scholar] [CrossRef]
- Fink, J.L.; Karunaratne, S.; Mittal, A.; Gardiner, D.M.; Hamilton, N.; Mahony, D.; Kai, C.; Suzuki, H.; Hayashizaki, Y.; Teasdale, R.D. Towards defining the nuclear proteome. Genome Biol 2008, 9, R15. [Google Scholar] [CrossRef]
- Forrest, A.R.; Taylor, D.F.; Fink, J.L.; Gongora, M.M.; Flegg, C.; Teasdale, R.D.; Suzuki, H.; Kanamori, M.; Kai, C.; Hayashizaki, Y.; et al. Phosphoregdb: The tissue and sub-cellular distribution of mammalian protein kinases and phosphatases. BMC bioinformatics 2006, 7, 82. [Google Scholar] [CrossRef]
- Locate—subcellular localisation database. Available online: http://www.locate.imb.uq.edu.au (accessed on 30 October 2012).
- Fink, J.L.; Aturaliya, R.N.; Davis, M.J.; Zhang, F.; Hanson, K.; Teasdale, M.S.; Kai, C.; Kawai, J.; Carninci, P.; Hayashizaki, Y.; et al. Locate: A mouse protein subcellular localization database. Nucleic. Acids Res. 2006, 34, D213–D217. [Google Scholar] [CrossRef]
- Sprenger, J.; Lynn Fink, J.; Karunaratne, S.; Hanson, K.; Hamilton, N.A.; Teasdale, R.D. Locate: A mammalian protein subcellular localization database. Nucleic. Acids Res. 2008, 36, D230–D233. [Google Scholar]
- Goder, V.; Spiess, M. Topogenesis of membrane proteins: Determinants and dynamics. FEBS Lett. 2001, 504, 87–93. [Google Scholar] [CrossRef]
- Suzuki, H.; Forrest, A.R.; van Nimwegen, E.; Daub, C.O.; Balwierz, P.J.; Irvine, K.M.; Lassmann, T.; Ravasi, T.; Hasegawa, Y.; de Hoon, M.J., et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet 2009, 41, 553–562. [Google Scholar] [CrossRef]
- Davis, M.J.; Zhang, F.; Yuan, Z.; Teasdale, R.D. Memo: A consensus approach to the annotation of a protein's membrane organization. In Silico Biol. 2006, 6, 387–399. [Google Scholar]
- Chai, L.; Dai, L.; Che, Y.; Xu, J.; Liu, G.; Zhang, Z.; Yang, R. Lrrc19, a novel member of the leucine-rich repeat protein family, activates nf-kappab and induces expression of proinflammatory cytokines. Biochem. Biophys. Res. Commun. 2009, 388, 543–548. [Google Scholar] [CrossRef]
- McMahon, H.T.; Mills, I.G. Cop and clathrin-coated vesicle budding: Different pathways, common approaches. Curr. Opin. Cell. Biol. 2004, 16, 379–391. [Google Scholar] [CrossRef]
- Su, A.I.; Cooke, M.P.; Ching, K.A.; Hakak, Y.; Walker, J.R.; Wiltshire, T.; Orth, A.P.; Vega, R.G.; Sapinoso, L.M.; Moqrich, A.; et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. USA 2002, 99, 4465–4470. [Google Scholar]
- Wu, C.; Orozco, C.; Boyer, J.; Leglise, M.; Goodale, J.; Batalov, S.; Hodge, C.L.; Haase, J.; Janes, J.; Huss, J.W., 3rd; et al. Biogps: An extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009, 10, R130. [Google Scholar] [CrossRef]
- Kurten, R.C.; Cadena, D.L.; Gill, G.N. Enhanced degradation of egf receptors by a sorting nexin, snx1. Science 1996, 272, 1008–1010. [Google Scholar]
- Nakamura, N.; Sun-Wada, G.H.; Yamamoto, A.; Wada, Y.; Futai, M. Association of mouse sorting nexin 1 with early endosomes. J. Biochem. (Tokyo) 2001, 130, 765–771. [Google Scholar]
- Haft, C.R.; de la Luz Sierra, M.; Barr, V.A.; Haft, D.H.; Taylor, S.I. Identification of a family of sorting nexin molecules and characterization of their association with receptors. Mol. Cell. Biol. 1998, 18, 7278–7287. [Google Scholar]
- Schnatwinkel, C.; Christoforidis, S.; Lindsay, M.R.; Uttenweiler-Joseph, S.; Wilm, M.; Parton, R.G.; Zerial, M. The rab5 effector rabankyrin-5 regulates and coordinates different endocytic mechanisms. PLoS Biol. 2004, 2, E261. [Google Scholar] [CrossRef]
- Mohrmann, K.; van der Sluijs, P. Regulation of membrane transport through the endocytic pathway by rabgtpases. Mol. Membr. Biol. 1999, 16, 81–87. [Google Scholar] [CrossRef]
- Stenmark, H.; Olkkonen, V.M. The rab gtpase family. Genome Biol. 2001, 2, REVIEWS3007. [Google Scholar]
- Zerial, M.; McBride, H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell. Biol. 2001, 2, 107–117. [Google Scholar] [CrossRef]
- Gorvel, J.P.; Chavrier, P.; Zerial, M.; Gruenberg, J. Rab5 controls early endosome fusion in vitro. Cell 1991, 64, 915–925. [Google Scholar] [CrossRef]
- Bucci, C.; Parton, R.G.; Mather, I.H.; Stunnenberg, H.; Simons, K.; Hoflack, B.; Zerial, M. The small gtpase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 1992, 70, 715–728. [Google Scholar] [CrossRef]
- Roberts, R.L.; Barbieri, M.A.; Pryse, K.M.; Chua, M.; Morisaki, J.H.; Stahl, P.D. Endosome fusion in living cells overexpressing gfp-rab5. J. Cell. Sci. 1999, 112 (Pt 21), 3667–3675. [Google Scholar]
- Rink, J.; Ghigo, E.; Kalaidzidis, Y.; Zerial, M. Rab conversion as a mechanism of progression from early to late endosomes. Cell 2005, 122, 735–749. [Google Scholar] [CrossRef]
- Sonnichsen, B.; De Renzis, S.; Nielsen, E.; Rietdorf, J.; Zerial, M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of rab4, rab5, and rab11. J. Cell. Biol. 2000, 149, 901–914. [Google Scholar] [CrossRef]
- Sheff, D.R.; Daro, E.A.; Hull, M.; Mellman, I. The receptor recycling pathway contains two distinct populations of early endosomes with different sorting functions. J. Cell. Biol. 1999, 145, 123–139. [Google Scholar] [CrossRef]
- Trischler, M.; Stoorvogel, W.; Ullrich, O. Biochemical analysis of distinct rab5- and rab11-positive endosomes along the transferrin pathway. J. Cell. Sci. 1999, 112 (Pt 24), 4773–4783. [Google Scholar]
- van der Sluijs, P.; Hull, M.; Webster, P.; Male, P.; Goud, B.; Mellman, I. The small gtp-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell 1992, 70, 729–740. [Google Scholar] [CrossRef]
- Ullrich, O.; Reinsch, S.; Urbe, S.; Zerial, M.; Parton, R.G. Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell. Biol. 1996, 135, 913–924. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Nielsen, H.; von Heijne, G.; Brunak, S. Improved prediction of signal peptides: Signalp 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef]
- Mulder, N.J.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Buillard, V.; Cerutti, L.; Copley, R.; et al. New developments in the interpro database. Nucleic. Acids Res. 2007, 35, D224–D228. [Google Scholar] [CrossRef]
- Mulder, N.J.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bradley, P.; Bork, P.; Bucher, P.; Cerutti, L.; et al. Interpro, progress and status in 2005. Nucleic. Acids Res. 2005, 33, D201–D205. [Google Scholar] [CrossRef]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. Interproscan: Protein domains identifier. Nuclei.c Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef]
- University of california santa cruz (ucsc) proteome browser. Available online: http://www.hgw7.cse.ucsc.edu/index.html (accessed on 30 October 2012).
- Kuhn, R.M.; Karolchik, D.; Zweig, A.S.; Trumbower, H.; Thomas, D.J.; Thakkapallayil, A.; Sugnet, C.W.; Stanke, M.; Smith, K.E.; Siepel, A.; et al. The ucsc genome browser database: Update 2007. Nucleic. Acids Res. 2007, 35, D668–D673. [Google Scholar] [CrossRef]
- Ncbi blast tool. Available online: http://www.blast.ncbi.nlm.nih.gov/ (accessed on 30 October 2012).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar]
- Martin, S.; Driessen, K.; Nixon, S.J.; Zerial, M.; Parton, R.G. Regulated localization of rab18 to lipid droplets: Effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 2005, 280, 42325–42335. [Google Scholar] [CrossRef]
- Bugarcic, A.; Zhe, Y.; Kerr, M.C.; Griffin, J.; Collins, B.M.; Teasdale, R.D. Vps26a and vps26b subunits define distinct retromer complexes. Traffic 2011, 12, 1759–1773. [Google Scholar] [CrossRef]
- Abramoff, M.D.; Magelhaes, P.J.; Ram, S.J. Image processing with imagej. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Ang, A.L.; Taguchi, T.; Francis, S.; Folsch, H.; Murrells, L.J.; Pypaert, M.; Warren, G.; Mellman, I. Recycling endosomes can serve as intermediates during transport from the golgi to the plasma membrane of mdck cells. J. Cell. Biol. 2004, 167, 531–543. [Google Scholar] [CrossRef]
- Lock, J.G.; Stow, J.L. Rab11 in recycling endosomes regulates the sorting and basolateral transport of e-cadherin. Mol. Biol. Cell. 2005, 16, 1744–1755. [Google Scholar] [CrossRef]
- Murray, R.Z.; Kay, J.G.; Sangermani, D.G.; Stow, J.L. A role for the phagosome in cytokine secretion. Science 2005, 310, 1492–1495. [Google Scholar] [CrossRef]
- Leung, S.M.; Rojas, R.; Maples, C.; Flynn, C.; Ruiz, W.G.; Jou, T.S.; Apodaca, G. Modulation of endocytic traffic in polarized madin-darby canine kidney cells by the small gtpase rhoa. Mol. Biol. Cell. 1999, 10, 4369–4384. [Google Scholar]
- D'Souza-Schorey, C.; van Donselaar, E.; Hsu, V.W.; Yang, C.; Stahl, P.D.; Peters, P.J. Arf6 targets recycling vesicles to the plasma membrane: Insights from an ultrastructural investigation. J. Cell. Biol. 1998, 140, 603–616. [Google Scholar] [CrossRef]
- Prigent, M.; Dubois, T.; Raposo, G.; Derrien, V.; Tenza, D.; Rosse, C.; Camonis, J.; Chavrier, P. Arf6 controls post-endocytic recycling through its downstream exocyst complex effector. J. Cell. Biol. 2003, 163, 1111–1121. [Google Scholar] [CrossRef]
- Radhakrishna, H.; Donaldson, J.G. Adp-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J. Cell. Biol. 1997, 139, 49–61. [Google Scholar] [CrossRef]
- D'Souza-Schorey, C.; Boshans, R.L.; McDonough, M.; Stahl, P.D.; Van Aelst, L. A role for por1, a rac1-interacting protein, in arf6-mediated cytoskeletal rearrangements. EMBO J. 1997, 16, 5445–5454. [Google Scholar] [CrossRef]
- Niedergang, F.; Colucci-Guyon, E.; Dubois, T.; Raposo, G.; Chavrier, P. Adp ribosylation factor 6 is activated and controls membrane delivery during phagocytosis in macrophages. J. Cell. Biol. 2003, 161, 1143–1150. [Google Scholar] [CrossRef]
- Radhakrishna, H.; Klausner, R.D.; Donaldson, J.G. Aluminum fluoride stimulates surface protrusions in cells overexpressing the arf6 gtpase. J. Cell. Biol. 1996, 134, 935–947. [Google Scholar] [CrossRef]
- Zhang, Q.; Cox, D.; Tseng, C.C.; Donaldson, J.G.; Greenberg, S. A requirement for arf6 in fcgamma receptor-mediated phagocytosis in macrophages. J. Biol. Chem. 1998, 273, 19977–19981. [Google Scholar] [CrossRef]
Supplementary Files
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Aturaliya, R.N.; Kerr, M.C.; Teasdale, R.D. A Novel Type III Endosome Transmembrane Protein, TEMP. Cells 2012, 1, 1029-1044. https://doi.org/10.3390/cells1041029
Aturaliya RN, Kerr MC, Teasdale RD. A Novel Type III Endosome Transmembrane Protein, TEMP. Cells. 2012; 1(4):1029-1044. https://doi.org/10.3390/cells1041029
Chicago/Turabian StyleAturaliya, Rajith N., Markus C. Kerr, and Rohan D. Teasdale. 2012. "A Novel Type III Endosome Transmembrane Protein, TEMP" Cells 1, no. 4: 1029-1044. https://doi.org/10.3390/cells1041029
APA StyleAturaliya, R. N., Kerr, M. C., & Teasdale, R. D. (2012). A Novel Type III Endosome Transmembrane Protein, TEMP. Cells, 1(4), 1029-1044. https://doi.org/10.3390/cells1041029



