Identification of Phenotypic and Physiological Markers of Salt Stress Tolerance in Durum Wheat (Triticum durum Desf.) through Integrated Analyses
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Salt Stress Sensitivity Index Determination
2.3. Metabolite Extraction and Biochemicals Analyses
2.4. Enzyme Extraction and Assay
2.5. Measurement of Carbon and Nitrogen Isotope Enrichment
2.6. Statistical Analyses
3. Results
3.1. Analysis of Variance (ANOVA) of Phenotypic and Physiological Traits
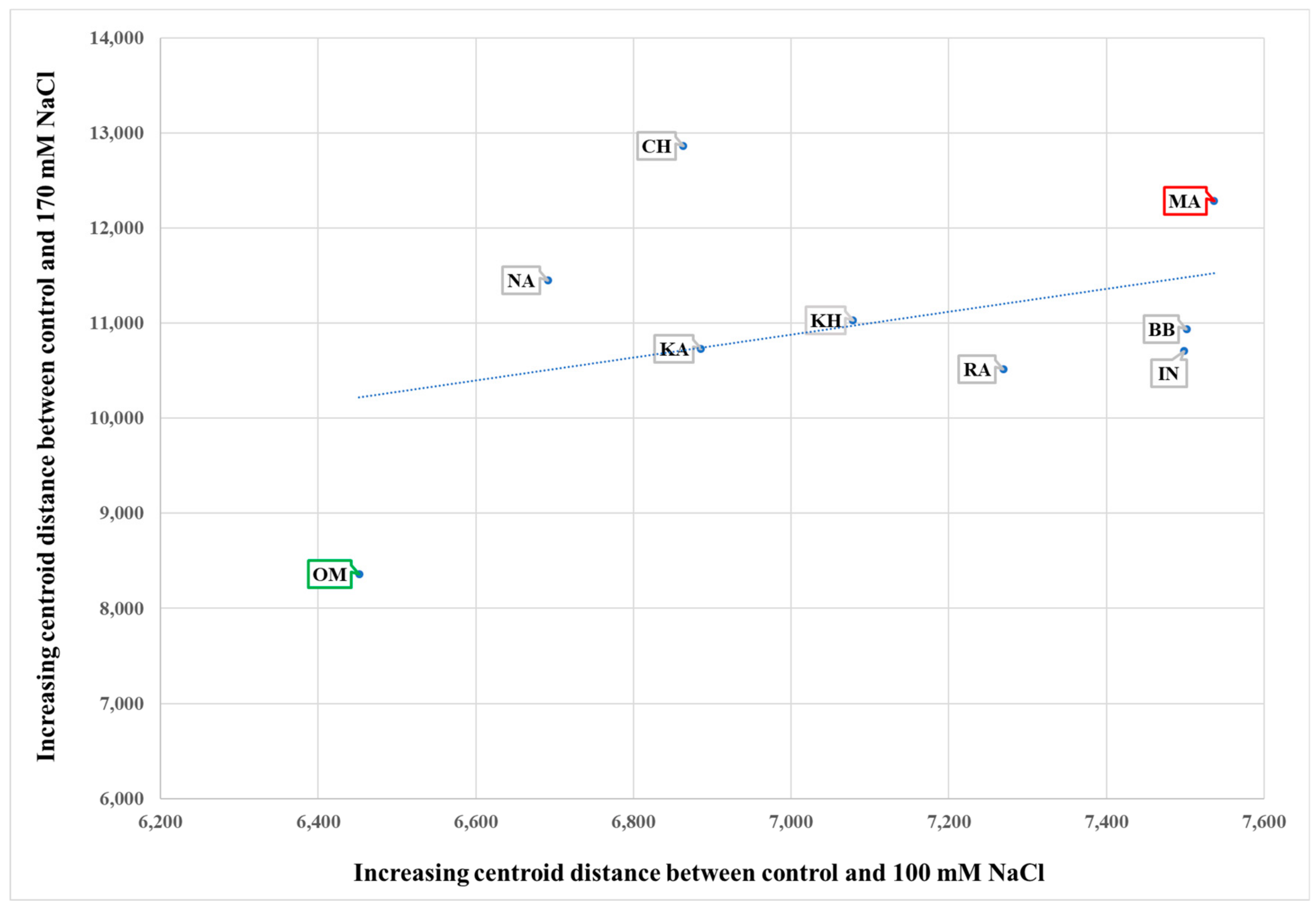
3.2. Genotype-Dependent Response to Salinity
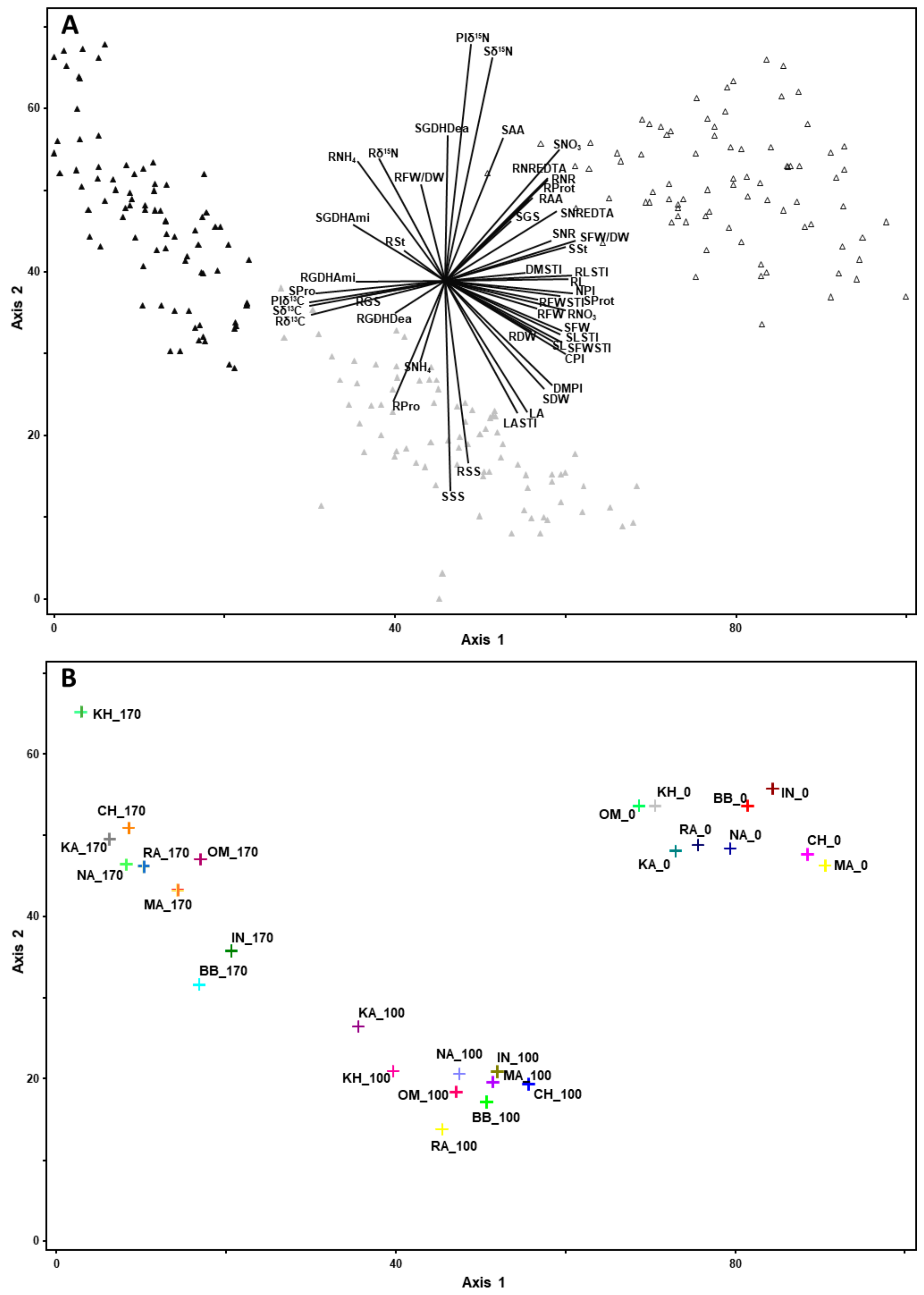
3.3. Identification of Marker Traits Representative of a Tolerance to Salinity Stress
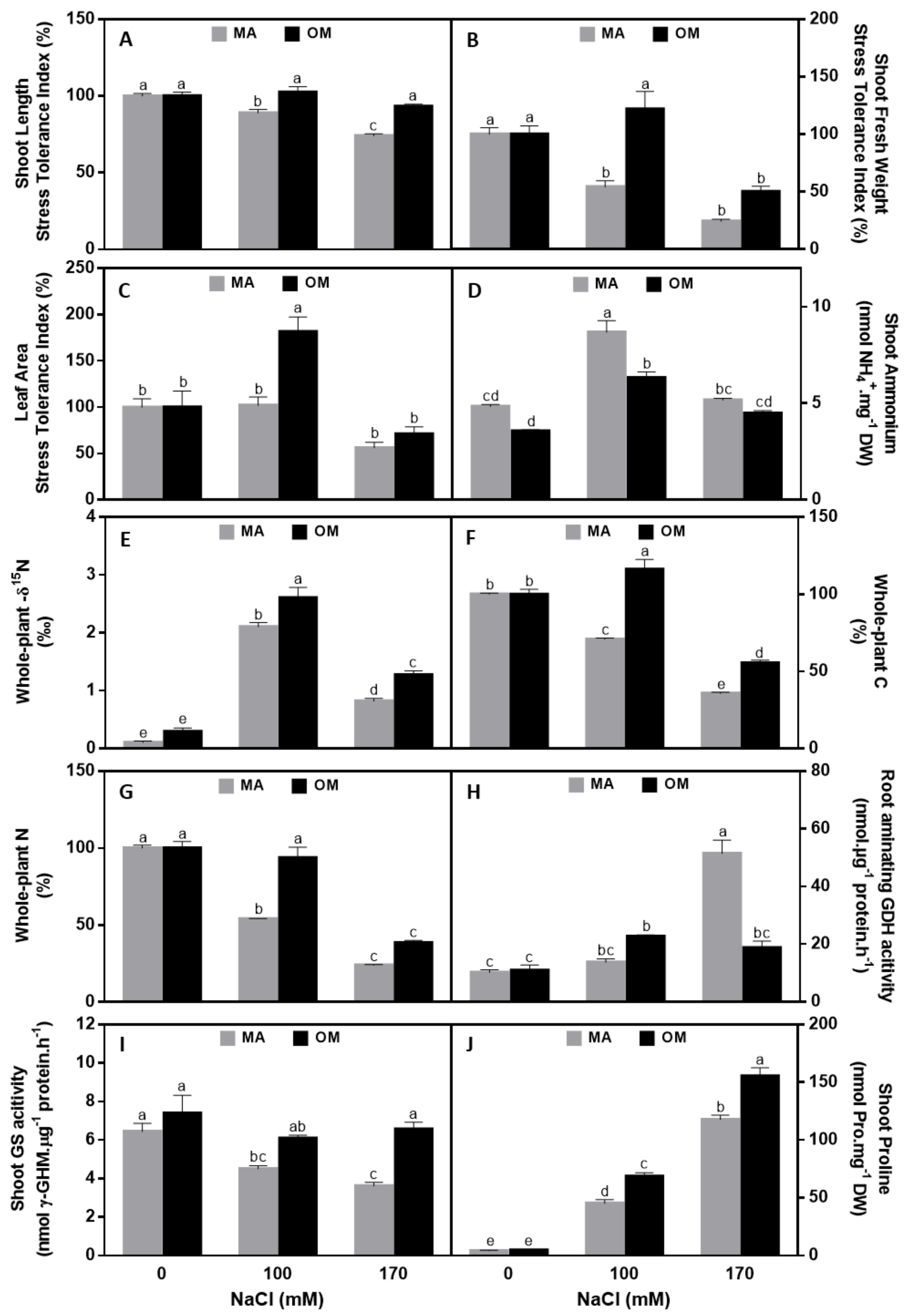
3.4. Impact of the Salt Stress on Traits Related to Growth and Development
3.5. Impact of the Salt Stress on Physiological Traits: Correlation Studies
4. Discussion
4.1. Traits Related to Growth and Development
4.2. Traits Related to Carbon Metabolism
4.3. Traits Related to Nitrogen Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Newell, N. Review: Effects of Soil Salinity on Plant Growth. Plant Physiol. 2013, 1, 1–4. [Google Scholar]
- Farooq, M.; Hussain, M.; Wakeel, A.; Siddique, K.H.M. Salt stress in maize: Effects, resistance mechanisms, and management. A review. Agron. Sustain. Dev. 2015, 35, 461–481. [Google Scholar] [CrossRef]
- Machado, R.; Serralheiro, R. Soil Salinity: Effect on Vegetable Crop Growth. Management Practices to Prevent and Mitigate Soil Salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Naliwajski, M.R.; Skłodowska, M. The relationship between carbon and nitrogen metabolism in cucumber leaves acclimated to salt stress. PeerJ 2018, 6, e6043. [Google Scholar] [CrossRef]
- Ashraf, M.; Shahzad, S.M.; Imtiaz, M.; Rizwan, M.S. Salinity effects on nitrogen metabolism in plants–focusing on the activities of nitrogen metabolizing enzymes: A review. J. Plant Nutr. 2018, 41, 1065–1081. [Google Scholar] [CrossRef]
- Jafarinia, M.; Shariati, M. Effects of salt stress on photosystem II of canola plant (Barassica napus, L.) probing by chlorophyll a fluorescence measurement. Iran. J. Sci. Technol. Trans. A Sci. 2012, 36, 73–76. [Google Scholar]
- FAO. Wheat—The Largest Primary Commodity. 2014. Available online: http://www.fao.org/assets/infographics/FAO-Infographic-wheat-en.pdf (accessed on 3 November 2019).
- Curtis, T.; Halford, N.G. Food security: The challenge of increasing wheat yield and the importance of not compromising food safety. Ann. Appl. Biol. 2014, 164, 354–372. [Google Scholar] [CrossRef]
- Kumar, P.; Yadava, R.K.; Gollen, B.; Kumar, S.; Verma, R.K.; Yadav, S. Nutritional contents and medicinal properties of wheat: A review. Life Sci. Med. Res. 2011, 22, 1–10. [Google Scholar]
- Liu, C.Y.; Shepherd, K.W.; Rathjen, A.J. Improvement of durum wheat pastamaking and breadmaking qualities. Cereal Chem. 1996, 73, 155–166. [Google Scholar]
- Nachit, M. Durum Breeding Research to Improve Dry-Land Productivity in the Mediterranean Region; SEWANA Durum Research network; Rao, S.C., Ryan, J., Eds.; ICARDA Editions: Beirut, Lebanon, 1998; pp. 1–15. [Google Scholar]
- Rana, G.; Katerji, N. Measurement and estimation of actual evapotranspiration in the field under Mediterranean climate: A review. Eur. J. Agron. 2000, 13, 125–153. [Google Scholar] [CrossRef]
- Munns, R.; Gilliham, M. Salinity tolerance of crops-what is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Zafar, S.; Ashraf, M.Y.; Niaz, M.; Kausar, A.; Hussain, J. Evaluation of wheat genotypes for salinity tolerance using physiological indices as screening tool. Pak. J. Bot. 2015, 47, 397–405. [Google Scholar]
- Wu, H.; Shabala, L.; Liu, X.; Azzarello, E.; Zhou, M.; Pandolfi, C.; Chen, Z.-H.; Bose, J.; Mancuso, S.; Shabala, S. Linking salinity stress tolerance with tissue-specific Na+ sequestration in wheat roots. Front. Plant Sci. 2015, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xia, G. The landscape of molecular mechanisms for salt tolerance in wheat. Crop J. 2018, 6, 42–47. [Google Scholar] [CrossRef]
- Kumar, S.; Beena, A.S.; Awana, M.; Singh, A. Physiological, biochemical, epigenetic and molecular analyses of wheat (Triticum aestivum) genotypes with contrasting salt tolerance. Front. Plant Sci. 2017, 8, 1151. [Google Scholar] [CrossRef]
- Yousfi, S.; Márquez, A.J.; Betti, M.; Araus, J.L.; Serret, M.D. Gene expression and physiological responses to salinity and water stress of contrasting durum wheat genotypes. J. Integr. Plant Biol. 2016, 58, 48–66. [Google Scholar] [CrossRef]
- Bartels, D.; Sunkar, R. Drought and Salt Tolerance in Plants. CRC Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef]
- Kerepesi, I.; Galiba, G. Osmotic and salt stress-induced alteration in soluble carbohydrate content in wheat seedlings. Crop Sci. 2000, 40, 482–487. [Google Scholar] [CrossRef]
- Amirjani, M.R. Effect of Salinity stress on growth, sugar content, pigments and enzyme activity of rice. Int. J. Bot. 2011, 7, 73–81. [Google Scholar] [CrossRef]
- Darko, E.; Gierczik, K.; Hudák, O.; Forgó, P.; Pál, M.; Türkösi, E.; Kovács, V.; Dulai, S.; Majláth, I.; Molnár, I.; et al. Differing metabolic responses to salt stress in wheat-barley addition lines containing different 7H chromosomal fragments. PLoS ONE 2017, 12, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Nemati, I.; Moradi, F.; Gholizadeh, S.; Esmaeili, M.A.; Bihamta, M.R. The effect of salinity stress on ions and soluble sugars distribution in leaves, leaf sheaths and roots of rice (Oryza sativa L.) seedlings. Plant Soil Environ. 2011, 57, 26–33. [Google Scholar] [CrossRef]
- Radi, A. Physiological and biochemical responses of salt-tolerant and salt-sensitive wheat and bean cultivars to salinity. J. Biol. 2013, 3, B72–B88. [Google Scholar]
- Sha Valli Khan, P.S.; Nagamallaiah, G.V.; Dhanunjay Rao, M.; Sergeant, K.; Hausman, J.F. Chapter 2—Abiotic Stress Tolerance in Plants: Insights from Proteomics. In Emerging Technologies and Management of Crop Stress Tolerance; Academic Press: Cambridge, MA, USA, 2014; pp. 23–68. [Google Scholar]
- Mahboob, W.; Khan, M.A.; Shirazi, M.U. Characterization of salt tolerant Wheat (Triticum aestivum) genotypes on the basis of physiological attributes. Int. J. Agric. Biol. 2017, 19, 726–734. [Google Scholar] [CrossRef]
- Thalmann, M.; Santelia, D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017, 214, 943–951. [Google Scholar] [CrossRef]
- Sami, F.; Yusuf, M.; Faizan, M.; Faraz, A.; Hayat, S. Role of sugars under abiotic stress. Plant Physiol. Biochem. 2016, 109, 54–61. [Google Scholar] [CrossRef]
- Yousfi, S.; Serret, M.D.; Márquez, A.J.; Voltas, J.; Araus, J.L. Combined use of δ13C, δ18O and δ15N tracks nitrogen metabolism and genotypic adaptation of durum wheat to salinity and water deficit. New Phytol. 2012, 194, 230–244. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Yuan, Y.Z.; Ou, J.Q.; Lin, Q.H.; Zhang, C.F. Glutamine synthetase and glutamate dehydrogenase contribute differentially to proline accumulation in leaves of wheat (Triticum aestivum) seedlings exposed to different salinity. J. Plant Physiol. 2007, 164, 695–701. [Google Scholar] [CrossRef]
- Dawson, T.E.; Mambelli, S.; Plamboeck, A.H.; Templer, P.H.; Tu, K.P. Dawson Stable Isotopes in Plant Ecology. Annu. Rev. Ecol. Syst. 2002, 33, 507–559. [Google Scholar] [CrossRef]
- Yousfi, S.; Serret, M.D.; Voltas, J.; Araus, J.L. Effect of salinity and water stress during the reproductive stage on growth, ion concentrations, Δ13C, and δ15N of durum wheat and related amphiploids. J. Exp. Bot. 2010, 61, 3529–3542. [Google Scholar] [CrossRef]
- Tcherkez, G.; Mahé, A.; Hodges, M. 12C/13C fractionations in plant primary metabolism. Trends Plant Sci. 2011, 16, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.; Handley, L.L.; Scrimgeour, C.M.; Gordon, D.C.; Forster, B.P.; Ellis, R.P. Using stable isotope natural abundances (δ15N and δ13C) to integrate the stress responses of wild barley (Hordeum spontaneum C. Koch.) genotypes. J. Exp. Bot. MP Spec. Issue 2000, 51, 41–50. [Google Scholar] [CrossRef]
- Robbana, C.; Kehel, Z.; Ben Naceur, M.; Sansaloni, C.; Bassi, F.; Amri, A. Genome-Wide Genetic Diversity and Population Structure of Tunisian Durum Wheat Landraces Based on DArTseq Technology. Int. J. Mol. Sci. 2019, 20, 1352. [Google Scholar] [CrossRef] [PubMed]
- Chamekh, Z.; Ayadi, S.; Karmous, C.; Trifa, Y.; Amara, H.; Boudabbous, K.; Yousfi, S.; Serret, M.D.; Araus, J.L. Comparative effect of salinity on growth, grain yield, water use efficiency, δ13C and δ15N of landraces and improved durum wheat varieties. Plant Sci. 2016, 251, 44–53. [Google Scholar] [CrossRef]
- Hoagland, D.C.; Arnon, D.I. The Water Culture Method for Growing Plant Without Soil; University of California, College of Agriculture, Agricultural Experiment Station: Berkeley, CA, USA, 1938. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671. [Google Scholar] [CrossRef]
- Takahashi, F.; Tilbrook, J.; Trittermann, C.; Berger, B.; Roy, S.J.; Seki, M.; Shinozaki, K.; Tester, M. Comparison of Leaf Sheath Transcriptome Profiles with Physiological Traits of Bread Wheat Cultivars under Salinity Stress. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Ni, Z.; Kim, E.-D.; Ha, M.; Lackey, E.; Liu, J.; Sun, Q.; Chen, Z.J. Altered circadian rhythms regulate growth vigor in hybrids and alloployploids. Nature 2009, 457, 327–331. [Google Scholar] [CrossRef]
- Smith, A.M.; Zeeman, S.C. Quantification of starch in plant tissues. Nat. Protoc. 2006, 1, 1342–1345. [Google Scholar] [CrossRef]
- Rosen, H. A modified ninhydrin colorimetric analysis for amino acids. Arch. Biochem. Biophys. 1957, 67, 10–15. [Google Scholar] [CrossRef]
- Cataldo, D.A.; Maroon, M.; Schrader, L.E.; Youngs, V.L. Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun. Soil Sci. Plant Anal. 1975, 6, 71–80. [Google Scholar] [CrossRef]
- Vega-Mas, I.; Sarasketa, A.; Marino, D. High-throughput Quantification of Ammonium Content in Arabidopsis. Bio-Protocol 2015, 5, 14–17. [Google Scholar] [CrossRef]
- Carillo, P.; Gibon, Y. PROTOCOL: Extraction and determination of proline. Prometheus Wiki. 2011. Available online: https://www.researchgate.net/publication/211353600_PROTOCOL_Extraction_and_determination_of_proline (accessed on 3 November 2019).
- Nesterenko, O.; Rashydov, N. Features of the Proline Synthesis of Pea Seedlings in Depend of Salt and Hyperthermia Treatment Coupled with Ionizing Radiation. Int. J. Second. Metab. 2018, 5, 94–108. [Google Scholar] [CrossRef][Green Version]
- Ferrario-Méry, S. Overexpression of Nitrate Reductase in Tobacco Delays Drought-Induced Decreases in Nitrate Reductase Activity and mRNA. Plant Physiol. 1998, 117, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Botrel, A.; Kaiser, W.M.; Botrel, A.; Kaiser, W.M. Nitrate reductase activation state in barley roots in relatio to the energy and carbohydrate status. Planta 1997, 201, 496–501. [Google Scholar] [CrossRef]
- O’Neal, D.; Joy, K.W. Glutamine synthetase of pea leaves. I. Purification, stabilization, and pH optima. Arch. Biochem. Biophys. 1973, 159, 113–122. [Google Scholar] [CrossRef]
- Turano, F.; Dashner, R.; Upadhyaya, A.R.; Caldwell, C. Purification of Mitochondrial Glutamate Dehydrogenase from Dark-Grown Soybean Seedlings. Plant Physiol. 1996, 112, 1357–1364. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis, 2nd ed.; Springer Series in Statistics; Springer: Berlin/Heidelberg, Germany, 2002; Volume 98, p. 487. [Google Scholar]
- McCune, B.; Mefford, M.J. PC-ORD: Multivariate Analysis of Ecological Data [v7]; MjM Softw: Gleneden Beach, OR, USA, 2016. [Google Scholar]
- Sadia, S.; Zhang, J.; Sheayi, A.A.; Tariq, A.; Cao, K. Tools and Techniques in Plant Ecology-A review. J. Environ. Agric. Sci. 2016, 7, 35–41. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant. Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Acosta-Motos, J.; Ortuño, M.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.; Hernandez, J. Plant Responses to Salt Stress: Adaptive Mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Vijayalakshmi, T.; Vijayakumar, A.S.; Kiranmai, K.; Nareshkumar, A.; Sudhakar, C. Salt Stress Induced Modulations in Growth, Compatible Solutes and Antioxidant Enzymes Response in Two Cultivars of Safflower (Carthamus tinctorius L. Cultivar TSF1 and Cultivar SM) Differing in Salt Tolerance. Am. J. Plant Sci. 2016, 07, 1802–1819. [Google Scholar] [CrossRef]
- Sayar, R.; Bchini, H.; Mosbahi, M.; Ezzine, M. Effects of salt and drought stresses on germination, emergence and seedling growth of Durum wheat (Triticum durum Desf.). J. Agric. Res. 2010, 5, 2008–2016. [Google Scholar]
- Ayed, S.; Rassaa, N.; Chamekh, Z.; Beji, S.; Karoui, F.; Bouzaien, T.; Mrabit, M.; Ben, Y.M. Effect of salt stress (sodium chloride) on germination and seedling growth of durum wheat (Triticum durum Desf.) genotypes. Int. J. Biodivers. Conserv. 2014, 6, 320–325. [Google Scholar] [CrossRef]
- Houshmand, S.; Arzani, A.; Maibody, S.A.M.; Feizi, M. Evaluation of salt-tolerant genotypes of durum wheat derived from in vitro and field experiments. Field Crop. Res. 2005, 91, 345–354. [Google Scholar] [CrossRef]
- Płazek, A.; Tatrzańska, M.; Maciejewski, M.; Kościelniak, J.; Gondek, K.; Bojarczuk, J.; Dubert, F. Investigation of the salt tolerance of new Polish bread and durum wheat cultivars. Acta Physiol. Plant. 2013, 35, 2513–2523. [Google Scholar] [CrossRef]
- Ouerghi, Z.; Zid, E.; Hajji, M.; Soltani, A. Comportement physiologique du blé dur (Triticum durum L.) en milieu salé. In Durum Wheat Improvement in the Mediterranean Region: New Challenges; Royo, C., Nachit, M., Di Fonzo, N., Araus, J.L., Eds.; CIHEAM: Zaragoza, Spain, 2000; pp. 309–313. [Google Scholar]
- Majoul, T.; Chahed, K.; Zamiti, E.; Ouelhazi, L.; Ghrir, R. Analysis by two-dimensional electrophoresis of the effect of salt stress on the polypeptide patterns in roots of a salt-tolerant and a salt-sensitive cultivar of wheat. Electrophoresis 2000, 21, 2562–2565. [Google Scholar] [CrossRef]
- Mallek-Maalej, E.; Boulasnem, F.; Salem, M. Effet de la salinité sur la germination de graines de céréales cultivées en Tunisie. Cah. Agric. 1998, 7, 153–156. [Google Scholar]
- Chaabane, R.; Bchini, H.; Ouji, H.; Ben Salah, H.; Khamassi, K.; Khoufi, S.; Babay, E.; Naceur, M. Ben Behaviour of tunisian durum wheat {Triticum turgidum L.) varieties under saline stress. Pak. J. Nutr. 2011, 10, 539–542. [Google Scholar]
- Bouaouina, S.; Zid, E.; Hajji, M. Tolérance à la salinité, transports ioniques et fluorescence chlorophyllienne chez le blé dur (Triticum turgidum L.). In Durum wheat Improvement In the Mediterranean Region New Challenges; Instituto Agronómico Mediterráneo: Zaragoza, Spain, 2000; Volume 40, pp. 239–243. [Google Scholar]
- Brini, F.; Amara, I.; Feki, K.; Hanin, M.; Khoudi, H.; Masmoudi, K. Physiological and molecular analyses of seedlings of two Tunisian durum wheat (Triticum turgidum L. subsp. Durum [Desf.]) varieties showing contrasting tolerance to salt stress. Acta Physiol. Plant. 2009, 31, 145–154. [Google Scholar] [CrossRef]
- Asl, R.G.; Arbat, H.K.; Yarnia, M.; Aminzade, G.; Asl, L.G.; Ghanifathi, T. Evaluation of drought tolerance indices and grain yield in wheat genotypes using principal components analysis. Adv. Environ. Biol. 2011, 5, 2153–2157. [Google Scholar]
- Oyiga, B.C.; Sharma, R.C.; Shen, J.; Baum, M.; Ogbonnaya, F.C.; Léon, J.; Ballvora, A. Identification and Characterization of Salt Tolerance of Wheat Germplasm Using a Multivariable Screening Approach. J. Agron. Crop Sci. 2016, 202, 472–485. [Google Scholar] [CrossRef]
- Hameed, A.; Naseer, S.; Iqbal, T.; Syed, H.; Haq, M.A. Effects of nacl salinity on seedling growth, senescence, catalase and protease activities in two wheat genotypes differing in salt tolerance. Pak. J. Bot. 2008, 40, 1043–1051. [Google Scholar]
- Hamam, K.A.; Negim, O. Evaluation of wheat genotypes and some soil properties under saline water irrigation. Ann. Agric. Sci. 2014, 59, 165–176. [Google Scholar] [CrossRef]
- Yousfi, S.; Serret, M.D.; Araus, J.L. Shoot δ15N gives a better indication than ion concentration or Δ13C of genotypic differences in the response of durum wheat to salinity. Funct. Plant Biol. 2009, 36, 144–155. [Google Scholar] [CrossRef]
- Sadras, V.O.; Milroy, S.P. Soil-water thresholds for the responses of leaf expansion and gas exchange: A review. Field Crop. Res. 1996, 47, 253–266. [Google Scholar] [CrossRef]
- Pattanagul, W.; Thitisaksakul, M. Effect of salinity stress on growth and carbohydrate metabolism in three rice (Oryza sativa L.) cultivars differing in salinity tolerance. Indian J. Exp. Biol. 2008, 46, 736–742. [Google Scholar]
- Lee, B.-R.; Jin, Y.-L.; Jung, W.-J.; Avice, J.-C.; Morvan-Bertrand, A.; Ourry, A.; Park, C.-W.; Kim, T.-H. Water-deficit accumulates sugars by starch degradation—Not by de novo synthesis—In white clover leaves (Trifolium repens). Physiol. Plant 2008, 134, 403–411. [Google Scholar] [CrossRef]
- Hummel, I.; Pantin, F.; Sulpice, R.; Piques, M.; Rolland, G.; Dauzat, M.; Christophe, A.; Pervent, M.; Bouteillé, M.; Stitt, M.; et al. Arabidopsis Plants Acclimate to Water Deficit at Low Cost through Changes of Carbon Usage: An Integrated Perspective Using Growth, Metabolite, Enzyme, and Gene Expression Analysis. Plant Physiol. 2010, 154, 357–372. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Hartmann, H.; Gleixner, G.; Thoma, M.; Schwab, V.F. Carbon isotope fractionation including photosynthetic and post-photosynthetic processes in C3 plants: Low [CO2] matters. Geochim. Cosmochim. Acta 2019, 245, 1–15. [Google Scholar] [CrossRef]
- Yousfi, S.; Serret, M.D.; Araus, J.L. Comparative response of δ13C, δ18O and δ15N in durum wheat exposed to salinity at the vegetative and reproductive stages. Plant Cell Environ. 2013, 36, 1214–1227. [Google Scholar] [CrossRef]
- Farquhar, G.D.D.; Ehleringer, J.R.R.; Hubick, K.T.T. Carbon isotope discrimination and photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989, 40, 503–537. [Google Scholar] [CrossRef]
- Rivelli, A.R.; James, R.A.; Munns, R.; Condon, A.G. Effect of salinity on water relations and growth of wheat genotypes with contrasting sodium uptake. Funct. Plant Biol. 2002, 29, 1065–1074. [Google Scholar] [CrossRef]
- Hirel, B.; Lea, P.J. Ammonia assimilation. In Plant Nitrogen; Springer: Berlin/Heidelberg, Germany, 2001; pp. 79–99. [Google Scholar]
- Kwinta, J.; Cal, K. Effects of salinity stress on the activity of glutamine synthetase and glutamate dehydrogenase in triticale seedlings. Pol. J. Environ. Stud. 2005, 14, 125–130. [Google Scholar]
- Teixeira, J.; Fidalgo, F. Salt stress affects glutamine synthetase activity and mRNA accumulation on potato plants in an organ-dependent manner. Plant. Physiol. Biochem. 2009, 47, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, Z.; Zhou, Y.; Han, J.; Shi, D. Effects of salt stress on ion balance and nitrogen metabolism in rice. Plant Soil Environ. 2012, 58, 62–67. [Google Scholar] [CrossRef]
- Skopelitis, D.S.; Paranychianakis, N.V.; Paschalidis, K.A.; Pliakonis, E.D.; Delis, I.D.; Yakoumakis, D.I.; Kouvarakis, A.; Papadakis, A.K.; Stephanou, E.G.; Roubelakis-Angelakis, K.A. Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 2006, 18, 2767–2781. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Shen, Q.; Xu, G.; Guo, S. New insight into the strategy for nitrogen metabolism in plant cells. Int. Rev. Cell Mol. Biol. 2014, 310, 1–37. [Google Scholar]
- Majumdar, R.; Barchi, B.; Turlapati, S.A.; Gagne, M.; Minocha, R.; Long, S.; Minocha, S.C. Glutamate, Ornithine, Arginine, Proline, and Polyamine Metabolic Interactions: The Pathway Is Regulated at the Post-Transcriptional Level. Front. Plant Sci. 2016, 7, 1–17. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef]
- Udawat, P.; Jha, R.K.; Sinha, D.; Mishra, A.; Jha, B. Overexpression of a Cytosolic Abiotic Stress Responsive Universal Stress Protein (SbUSP) Mitigates Salt and Osmotic Stress in Transgenic Tobacco Plants. Front. Plant Sci. 2016, 7, 1–21. [Google Scholar] [CrossRef]
- Pageau, K.; Reisdorf-Cren, M.; Morot-Gaudry, J.-F.; Masclaux-Daubresse, C. The two senescence-related markers, GS1 (cytosolic glutamine synthetase) and GDH (glutamate dehydrogenase), involved in nitrogen mobilization, are differentially regulated during pathogen attack and by stress hormones and reactive oxygen species in Nicoti. J. Exp. Bot. 2006, 57, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Sairam, R.K.; Srivastava, G.; Agarwal, S.; Meena, R.C. Differences in antioxidant activity in response to salinity stress in tolerant and susceptible wheat genotypes. Biol. Plant. 2005, 49, 85–91. [Google Scholar] [CrossRef]
- Velicevici, G.; Madosa, E.; Sumalan, R.; Ciulca, S.; Bitea, N.; Petrescu, I.; Cretescu, I. Proline accumulation in some barley genotypes exposed to drought. J. Hortic. For. Biotechnol. 2011, 15, 48–54. [Google Scholar]
- Surabhi, G.K.; Reddy, A.; Sudhakar, C. NaCl effects on proline metabolism in two high yielding genotypes of mulberry (Morus alba L.) with contrasting salt tolerance. Plant Sci. 2003, 165, 1245–1251. [Google Scholar]
- Pakniyat, H.; Armion, M. Sodium and proline accumulation as osmoregulators in tolerance of sugar beet genotypes to salinity. Pak. J. Biol. Sci. 2007, 10, 4081–4086. [Google Scholar]
- Misra, N.; Gupta, A.K. Interactive Effects of Sodium and Calcium on Proline Metabolism in Salt Tolerant Green Gram Cultivar. Am. J. Plant Physiol. 2006, 1. [Google Scholar] [CrossRef][Green Version]
- Silva-Ortega, C.O.; Ochoa-Alfaro, A.E.; Reyes-Agüero, J.A.; Aguado-Santacruz, G.A.; Jiménez-Bremont, J.F. Salt stress increases the expression of P5CS gene and induces proline accumulation in cactus pear. Plant. Physiol. Biochem. 2008, 46, 82–92. [Google Scholar] [CrossRef]
- Huang, Z.; Zhao, L.; Chen, D.; Liang, M.; Liu, Z.; Shao, H.; Long, X. Salt Stress Encourages Proline Accumulation by Regulating Proline Biosynthesis and Degradation in Jerusalem Artichoke Plantlets. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Brugière, N.; Dubois, F.; Limami, A.M.; Lelandais, M.; Roux, Y.; Sangwan, R.S.; Hirel, B. Glutamine synthetase in the phloem plays a major role in controlling proline production. Plant Cell 1999, 11, 1995–2011. [Google Scholar] [CrossRef]
- Handley, L.L.; Robinson, D.; Forster, B.P.; Ellis, R.P.; Scrimgeour, C.M.; Gordon, D.C.; Nevo, E.; Raven, J.A. Shoot δ15N correlates with genotype and salt stress in barley. Planta 1997, 201, 100–102. [Google Scholar] [CrossRef]
| Value | F | Df | Df Error | P-Value | |
|---|---|---|---|---|---|
| Model Constant | 19,009.85 | 66,930.51 | 48 | 169 | <0.001 |
| Treatment | 341.29 | 597.26 | 96 | 336 | <0.001 |
| Genotype | 625.74 | 272.54 | 384 | 1338 | <0.001 |
| Treatment × Genotype | 56.02 | 12.19 | 768 | 2674 | <0.001 |
| Plant Traits | Abbreviations | P-Value | ||
|---|---|---|---|---|
| Treatment a | Genotype b | Treatment × Genotype c | ||
| Root Amino Acids (nmol amino acids.mg−1 dry weight) | RAA | <0.001 | <0.001 | <0.001 |
| Shoot Amino Acids (nmol amino acids.mg−1 dry weight) | SAA | <0.001 | <0.001 | <0.001 |
| Whole-plant total Carbon (mg) | CPl | <0.001 | <0.001 | <0.001 |
| Whole-Plant δ13C (‰) | Plδ13C | <0.001 | <0.001 | <0.001 |
| Root δ13C (‰) | Rδ13C | <0.001 | <0.001 | <0.001 |
| Shoot δ13C (‰) | Sδ13C | <0.001 | <0.001 | <0.001 |
| Dry Matter Whole Plant (g) | DMPl | <0.001 | <0.001 | 0.014 |
| Dry Matter Stress Tolerance Index (%) | DMSTI | <0.001 | 0.993 | 1.000 |
| Whole-Plant δ15N (‰) | Plδ15N | <0.001 | <0.001 | <0.001 |
| Root δ15N (‰) | Rδ15N | <0.001 | <0.001 | <0.001 |
| Shoot δ15N (‰) | Sδ15N | <0.001 | <0.001 | <0.001 |
| Root Dry Weight (g) | RDW | <0.001 | 0.002 | 0.985 |
| Shoot Dry Weight (g) | SDW | <0.001 | <0.001 | 0.001 |
| Root Fresh Weight/Root Dry Weight Ratio | RFW/DW | <0.001 | <0.001 | 0.564 |
| Shoot Fresh Weight/Shoot Dry Weight Ratio | SFW/DW | <0.001 | <0.001 | <0.001 |
| Root Fresh Weight (g) | RFW | <0.001 | <0.001 | 0.347 |
| Shoot Fresh Weight (g) | SFW | <0.001 | <0.001 | <0.001 |
| Root Fresh Weight Stress Tolerance Index (%) | RFWSTI | <0.001 | 0.533 | 0.983 |
| Shoot Fresh Weight Stress Tolerance Index (%) | SFWSTI | <0.001 | <0.001 | 0.006 |
| Root Aminating GDH Activity (nmol. µg−1 protein.h−1) | RGDHAmi | <0.001 | <0.001 | <0.001 |
| Shoot Aminating GDH Activity (nmol. µg−1 protein.h−1) | SGDHAmi | <0.001 | <0.001 | <0.001 |
| Root Deaminating GDH Activity (nmol. µg−1 protein.h−1) | RGDHDea | <0.001 | <0.001 | <0.001 |
| Shoot Deaminating GDH Activity (nmol. µg−1 protein.h−1) | SGDHDea | <0.001 | <0.001 | <0.001 |
| Root Soluble Sugar (nmol D-glucose.mg−1 dry weight) | RSS | <0.001 | <0.001 | 0.000 |
| Shoot Soluble Sugar (nmol D-glucose.mg−1 dry weight) | SSS | <0.001 | <0.001 | <0.001 |
| Root GS Activity (nmol γ-glutamyl hydroxamate. µg−1 protein.h−1) | RGS | <0.001 | <0.001 | <0.001 |
| Shoot GS Activity (nmol γ-glutamyl hydroxamate µg−1 protein.h−1) | SGS | <0.001 | <0.001 | 0.008 |
| Leaf Area (mm2) | LA | <0.001 | <0.001 | 0.013 |
| Leaf Area Stress Tolerance Index (%) | LASTI | <0.001 | <0.001 | <0.001 |
| Root Length (cm) | RL | <0.001 | <0.001 | <0.001 |
| Shoot Length (cm) | SL | <0.001 | <0.001 | <0.001 |
| Root Length Stress Tolerance Index (%) | RLSTI | <0.001 | <0.001 | 0.141 |
| Shoot Length Stress Tolerance Index (%) | SLSTI | <0.001 | <0.001 | 0.007 |
| Root Ammonium (nmol ammonium.mg−1 dry weight) | RNH4 | <0.001 | 0.002 | <0.001 |
| Shoot Ammonium (nmol ammonium.mg−1 dry weight) | SNH4 | <0.001 | <0.001 | <0.001 |
| Root Nitrate (µg nitrate.mg−1 dry weight) | RNO3 | <0.001 | 0.001 | <0.001 |
| Shoot Nitrate (µg nitrate.mg−1 dry weight) | SNO3 | <0.001 | <0.001 | <0.001 |
| Whole-Plant Total Nitrogen (mg) | NPl | <0.001 | <0.001 | <0.001 |
| Root NR-EDTA Activity (nmol nitrite. µg−1 protein.h−1) | RNREDTA | <0.001 | <0.001 | <0.001 |
| Shoot NR-EDTA Activity (nmol nitrite. µg−1 protein.h−1) | SNREDTA | <0.001 | <0.001 | <0.001 |
| Root NR activity (nmol nitrite. µg−1 protein.h−1) | RNR | <0.001 | <0.001 | <0.001 |
| Shoot NR activity (nmol nitrite. µg−1 protein.h−1) | SNR | <0.001 | <0.001 | <0.001 |
| Root Proline (nmol proline.mg−1 dry weight) | RPro | <0.001 | <0.001 | <0.001 |
| Shoot Proline (nmol proline.mg−1 dry weight) | SPro | <0.001 | <0.001 | <0.001 |
| Root Proteins (µg proteins.mg−1 dry weight) | RProt | <0.001 | <0.001 | <0.001 |
| Shoot Proteins (µg proteins.mg−1 dry weight) | SProt | <0.001 | <0.001 | <0.001 |
| Root Starch (nmol equivalent D-glucose.mg−1 dry weight) | RSt | <0.001 | 0.006 | 0.116 |
| Shoot Starch (nmol equivalent D-glucose.mg−1 dry weight) | SSt | <0.001 | <0.001 | <0.001 |
| Plant Traits | Abbreviations | P-value | |
|---|---|---|---|
| 100 mM NaCl | 170 mM NaCl | ||
| Whole-Plant Total Carbon (mg) | CPl | <0.0001 | 0.0377 |
| Whole-Plant δ13C (‰) | Plδ13C | 0.9986 | 0.0001 |
| Whole-Plant δ15N (‰) | Plδ15N | 0.0134 | 0.0404 |
| Shoot Fresh Weight Stress Tolerance Index (%) | SFWSTI | <0.0001 | 0.204 |
| Root Aminating GDH Activity (nmol µg−1 protein h−1) | RGDHAmi | 0.1105 | <0.0001 |
| Shoot Aminating GDH Activity (nmol µg−1 protein h−1) | SGDHAmi | 0.9996 | 0.0028 |
| Root Deaminating GDH Activity (nmol µg−1 protein h−1) | RGDHDea | <0.0001 | <0.0001 |
| Root GS Activity (nmol γ-glutamyl hydroxamate µg−1 protein h−1) | RGS | 0.1229 | <0.0001 |
| Shoot GS Activity (nmol γ-glutamyl hydroxamate µg−1 protein h−1) | SGS | 0.4581 | 0.0035 |
| Leaf Area Stress Tolerance Index (%) | LASTI | <0.0001 | 0.988 |
| Shoot Length Stress Tolerance Index (%) | SLSTI | 0.0037 | <0.0001 |
| Shoot Ammonium (nmol ammonium.mg−1 dry weight) | SNH4 | <0.0001 | 0.8938 |
| Whole-Plant Total Nitrogen (mg) | NPl | <0.0001 | 0.0699 |
| Shoot Proline (nmol proline. mg−1 dry weight) | SPro | 0.0008 | <0.0001 |
| Shoot Starch (nmol equivalent D-glucose.mg−1 dry weight) | SSt | <0.0001 | 0.996 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guellim, A.; Catterou, M.; Chabrerie, O.; Tetu, T.; Hirel, B.; Dubois, F.; Ben Ahmed, H.; Kichey, T. Identification of Phenotypic and Physiological Markers of Salt Stress Tolerance in Durum Wheat (Triticum durum Desf.) through Integrated Analyses. Agronomy 2019, 9, 844. https://doi.org/10.3390/agronomy9120844
Guellim A, Catterou M, Chabrerie O, Tetu T, Hirel B, Dubois F, Ben Ahmed H, Kichey T. Identification of Phenotypic and Physiological Markers of Salt Stress Tolerance in Durum Wheat (Triticum durum Desf.) through Integrated Analyses. Agronomy. 2019; 9(12):844. https://doi.org/10.3390/agronomy9120844
Chicago/Turabian StyleGuellim, Amira, Manuella Catterou, Olivier Chabrerie, Thierry Tetu, Bertrand Hirel, Frédéric Dubois, Hela Ben Ahmed, and Thomas Kichey. 2019. "Identification of Phenotypic and Physiological Markers of Salt Stress Tolerance in Durum Wheat (Triticum durum Desf.) through Integrated Analyses" Agronomy 9, no. 12: 844. https://doi.org/10.3390/agronomy9120844
APA StyleGuellim, A., Catterou, M., Chabrerie, O., Tetu, T., Hirel, B., Dubois, F., Ben Ahmed, H., & Kichey, T. (2019). Identification of Phenotypic and Physiological Markers of Salt Stress Tolerance in Durum Wheat (Triticum durum Desf.) through Integrated Analyses. Agronomy, 9(12), 844. https://doi.org/10.3390/agronomy9120844






