Integrative Transcriptomic and Metabolomic Analysis Reveals the Molecular Mechanisms Underlying Flowering Time Variation in Camellia Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Plants and Sample Preparation
2.2. RNA Extraction, Library Construction, and Transcriptome Sequencing
2.3. Identification of DEGs and Enrichment Analysis
2.4. Metabolite Extraction and UPLC-MS/MS Analysis
2.5. Extraction and Detection of Plant Hormones
2.6. Combined Transcriptome and Metabolome Analysis
2.7. Validation of DEGs by RT-qPCR Analysis
3. Results
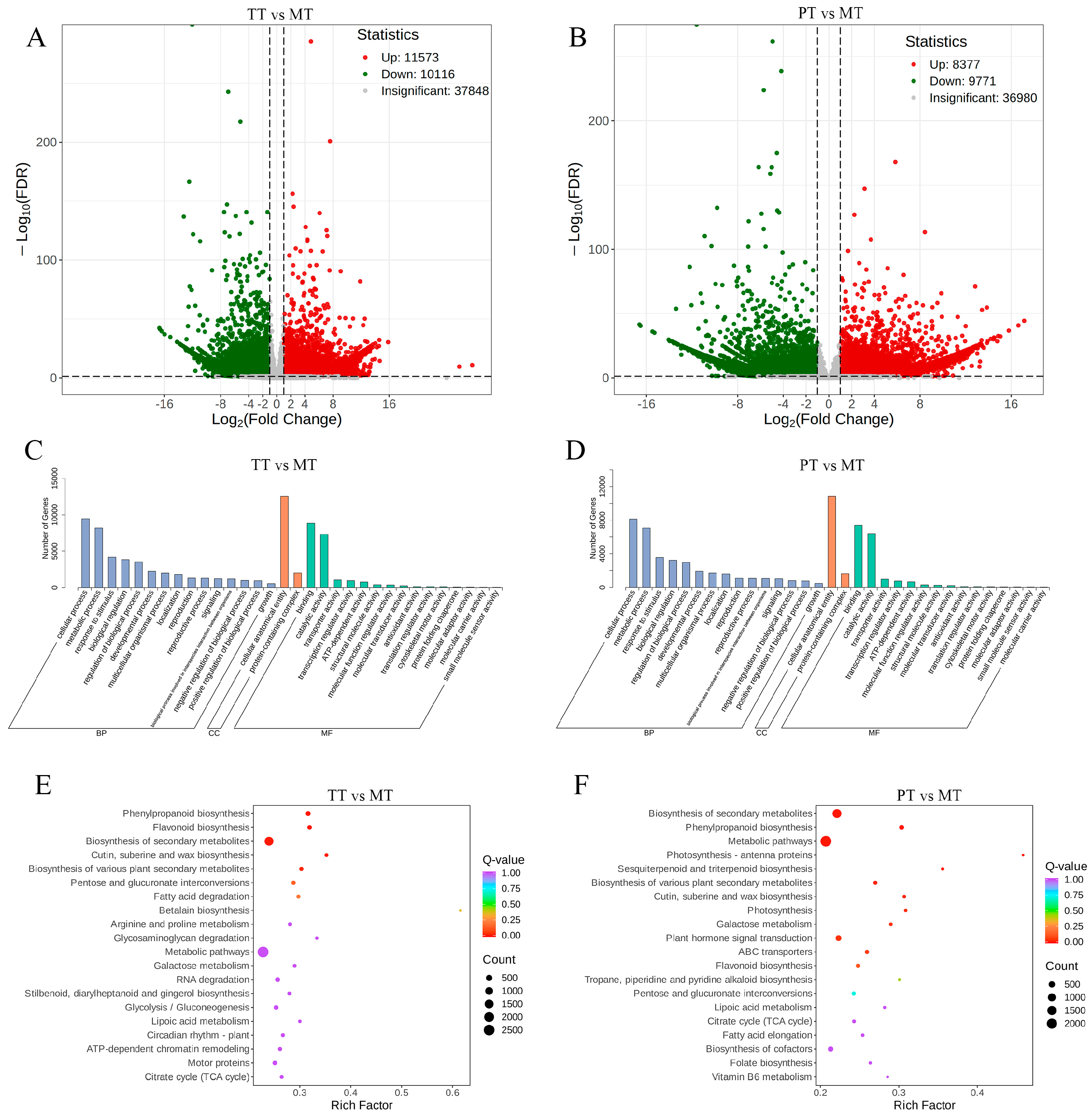
3.1. Screening of Differentially Expressed Genes from Transcriptome
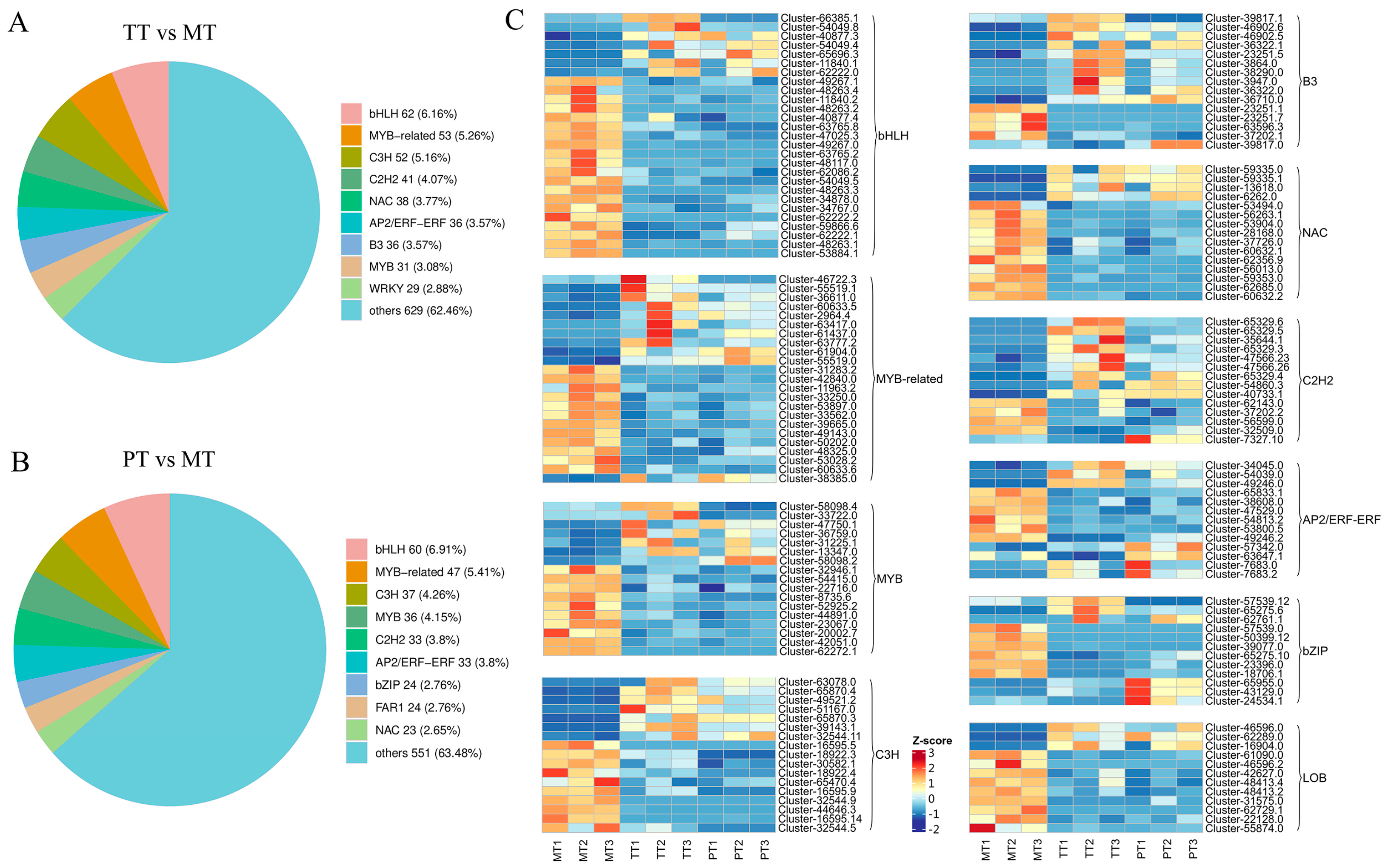
3.2. Analysis of Differentially Expressed Transcription Factors
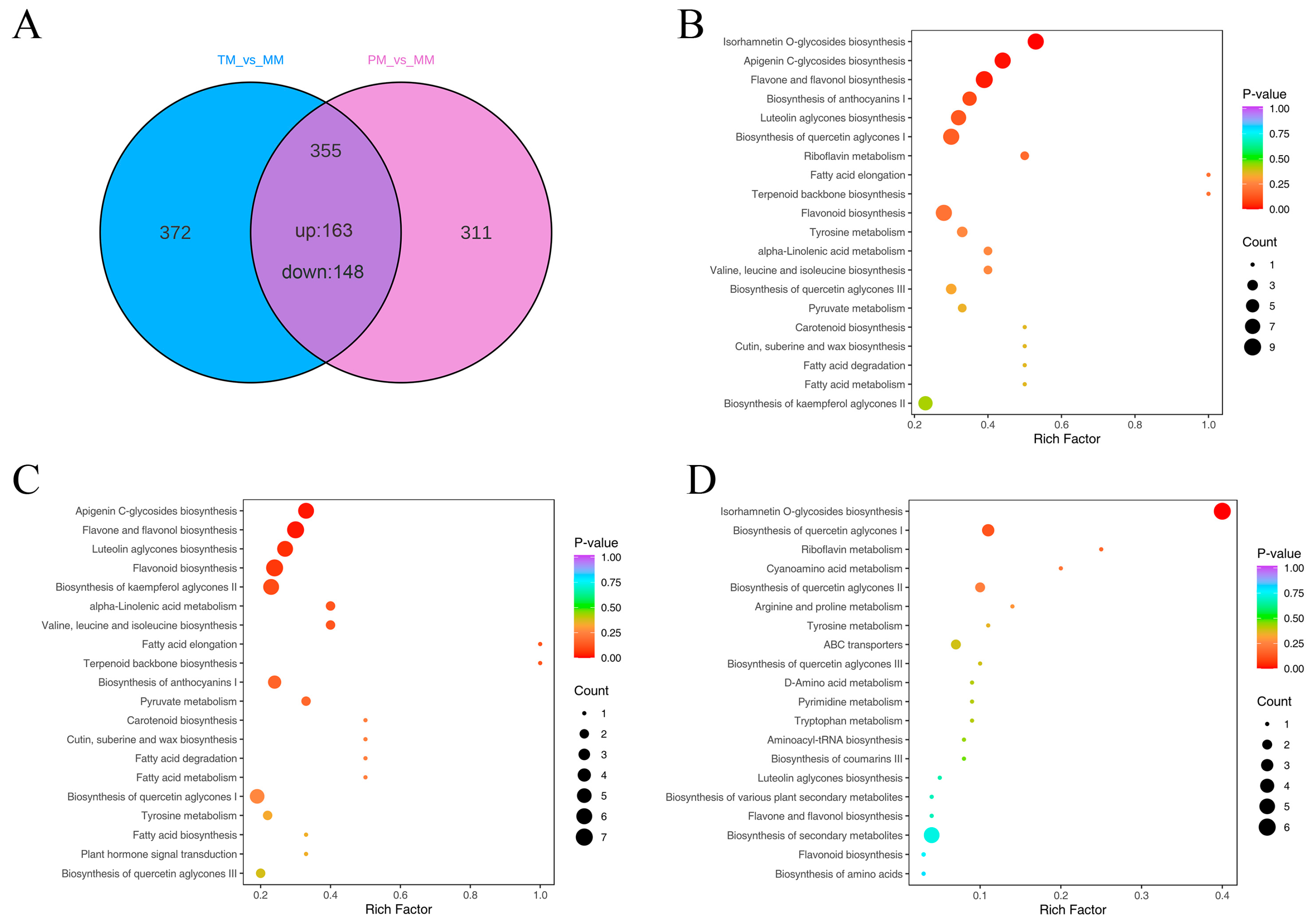
3.3. Screening of Differential Accumulated Metabolites from Metabolome
3.4. KEGG Functional Annotation and Enrichment Analysis of DAMs
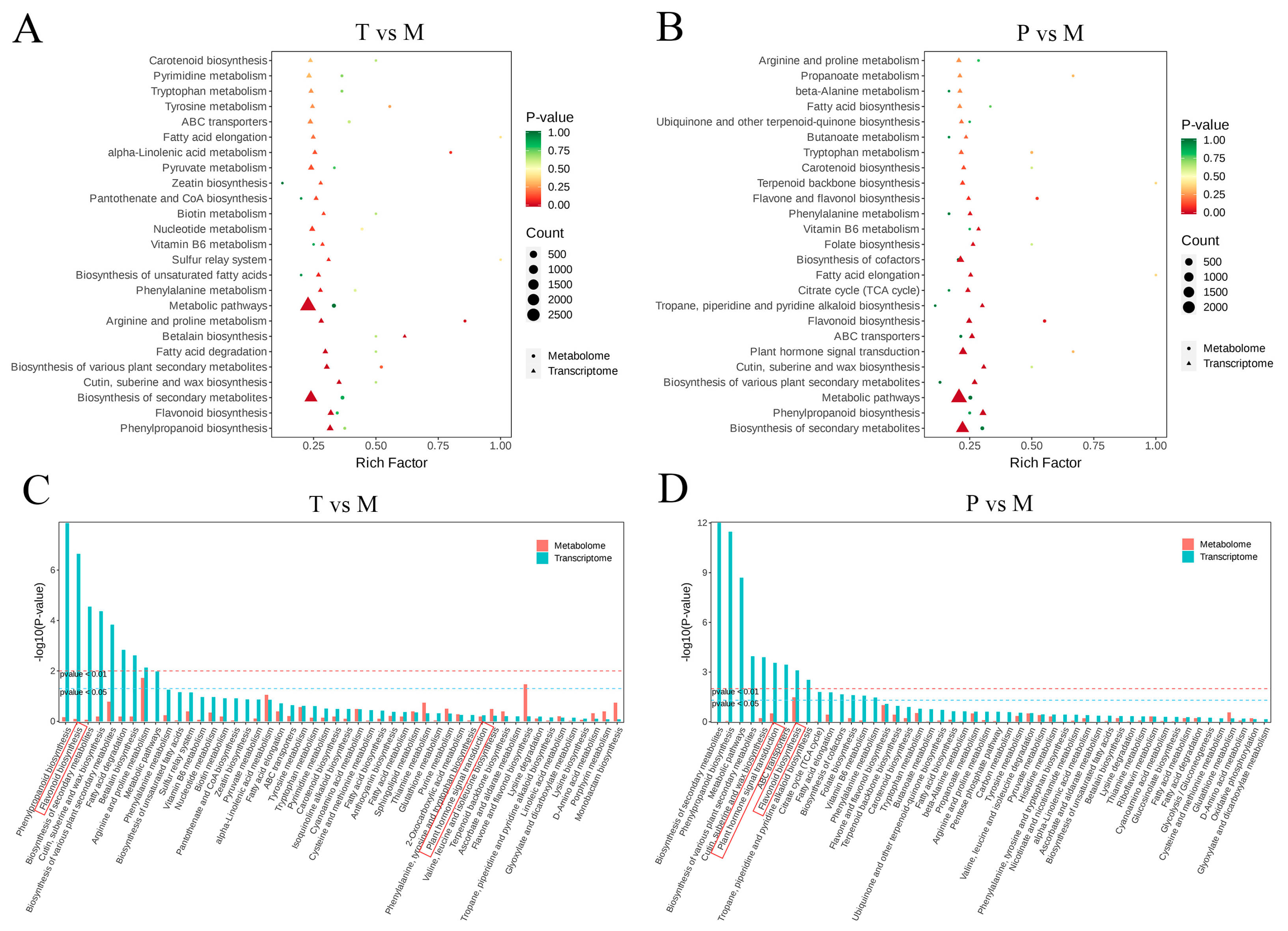
3.5. Integrated Analysis of the Transcriptome and Metabolome
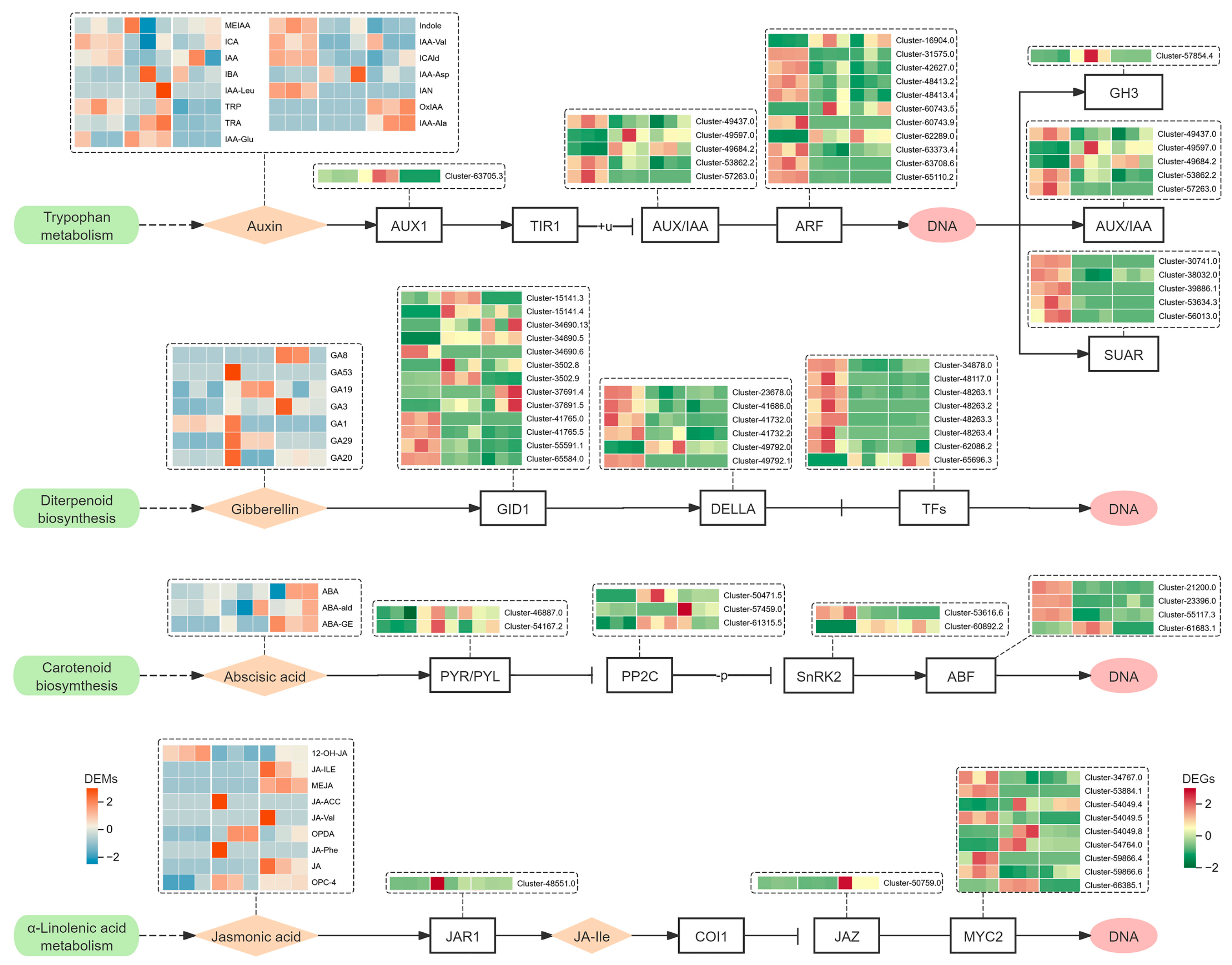
3.6. Analysis of Plant Hormone Signal Transduction Pathway
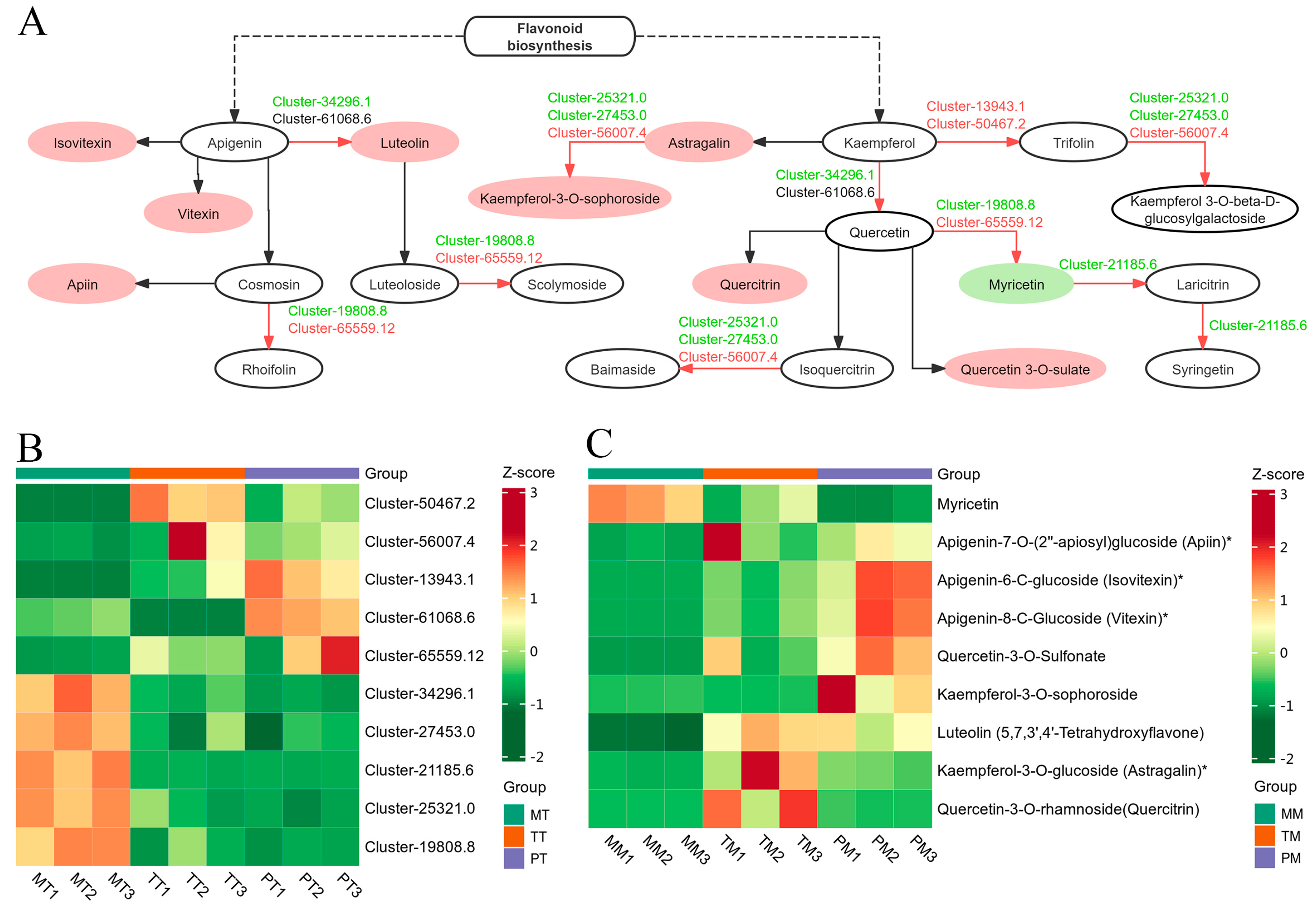
3.7. Analysis of Flavonoid Biosynthesis Pathway
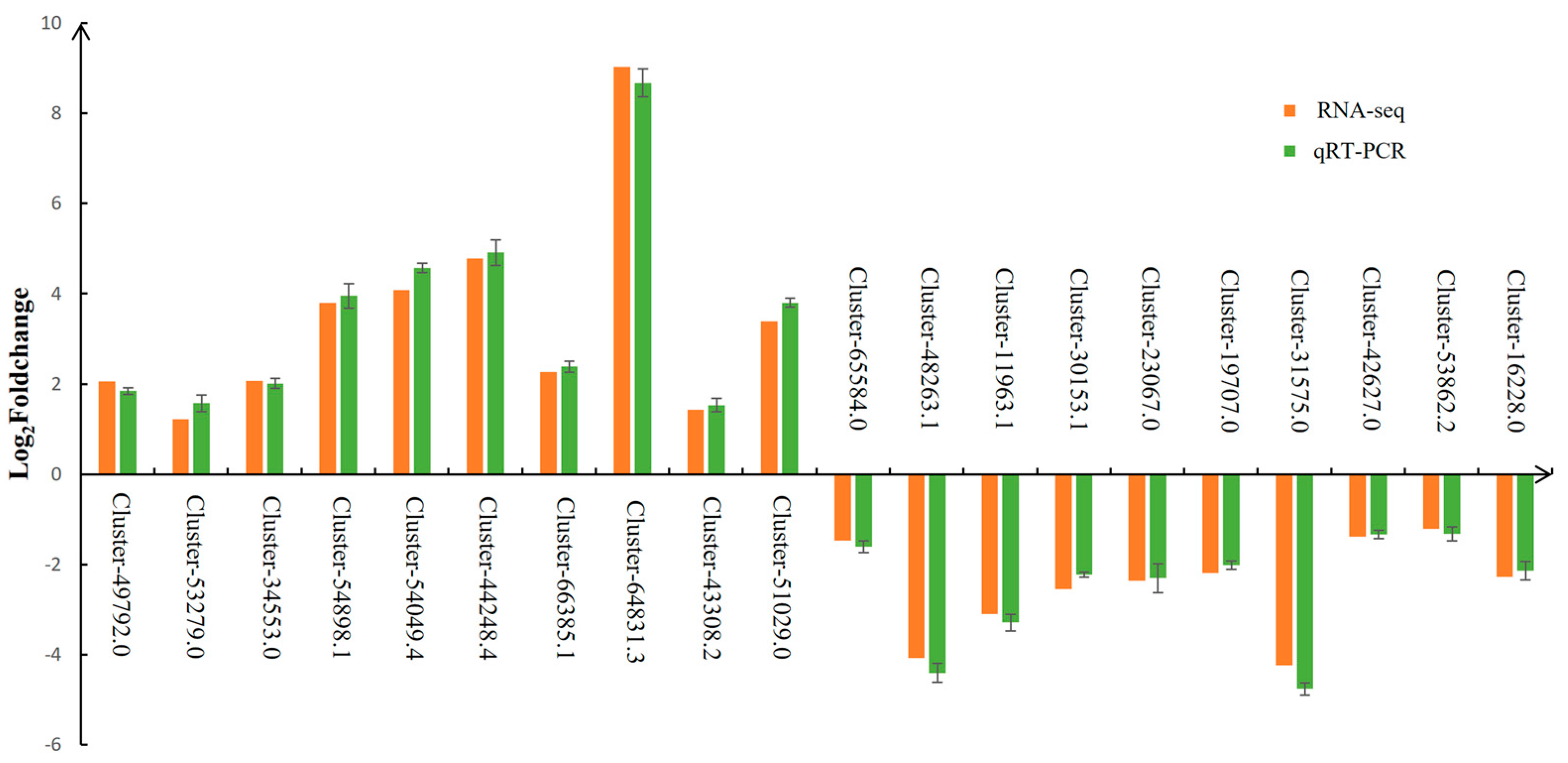
3.8. Expression Validation of RNA-Seq Data by RT-qPCR
4. Discussion
4.1. Transcriptional Regulation of Flowering Time in Camellia
4.2. Photoperiodic Regulation and Circadian Adaptations
4.3. Hormonal Crosstalk in Floral Initiation
4.4. Flavonoid Dynamics and Bud Development
4.5. Future Directions and Breeding Applications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Vijayan, K.; Zhang, W.-J.; Tsou, C.-H. Molecular taxonomy of Camellia (Theaceae) inferred from nrITS sequences. Am. J. Bot. 2009, 96, 1348–1360. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Hyun, C.C.; Misuk, Y.; Kim, S.-C. The complete chloroplast genome sequence of the Japanese Camellia (Camellia japonica L.). Mitochondrial DNA Part B 2017, 2, 583–584. [Google Scholar] [CrossRef][Green Version]
- Vela, P.; Salinero, C.; Sainz, M.J. Phenological growth stages of Camellia japonica. Ann. Appl. Biol. 2013, 162, 182–190. [Google Scholar] [CrossRef]
- Wang, Y.; Zhuang, H.; Shen, Y.; Wang, Y.; Wang, Z. The Dataset of Camellia Cultivars Names in the World. Biodivers. Data J. 2021, 9, 61646. [Google Scholar] [CrossRef]
- Ren, H.; Jian, S.; Chen, Y.; Liu, H.; Zhang, Q.; Liu, N.; Xu, Y.; Luo, J. Distribution, status, and conservation of Camellia changii Ye (Theaceae), a Critically Endangered plant endemic to southern China. Oryx 2014, 48, 358–360. [Google Scholar] [CrossRef]
- Fan, Z.; Li, J.; Li, X.; Wu, B.; Wang, J.; Liu, Z.; Yin, H. Genome-wide transcriptome profiling provides insights into floral bud development of summer-flowering Camellia azale. Sci. Rep. 2015, 5, 9729. [Google Scholar] [CrossRef]
- Niu, F.; Rehmani, M.S.; Yan, J. Multilayered regulation and implication of flowering time in plants. Plant Physiol. Biochem. 2024, 213, 108842. [Google Scholar] [CrossRef] [PubMed]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Shen, Y.; Chang, H.C.; Hou, Y.; Harris, A.; Ma, S.F.; McPartland, M.; Hymus, G.J.; Adam, L.; Marion, C.; et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010, 187, 57–66. [Google Scholar] [CrossRef]
- Sharma, N.; Geuten, K.; Giri, B.S.; Varma, A. The molecular mechanism of vernalization in Arabidopsis and cereals: Role of Flowering Locus C and its homologs. Physiol. Plant 2020, 170, 373–383. [Google Scholar] [CrossRef]
- Sharma, N.; Ruelens, P.; D’hauw, M.; Maggen, T.; Dochy, N.; Torfs, S.; Kaufmann, K.; Rohde, A.; Geuten, K. A Flowering Locus C Homolog Is a Vernalization-Regulated Repressor in Brachypodium and Is Cold Regulated in Wheat. Plant Physiol. 2016, 173, 1301–1315. [Google Scholar] [CrossRef]
- Ferrándiz, C.; Gu, Q.; Martienssen, R.; Yanofsky, M.F. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 2000, 127, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Rehman, S.; Bahadur, S.; Xia, W. An overview of floral regulatory genes in annual and perennial plants. Gene 2023, 885, 147699. [Google Scholar] [CrossRef]
- Davière, J.-M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Estelle, M. Recent advances and emerging trends in plant hormone signalling. Nature 2009, 459, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Conti, L. Hormonal control of the floral transition: Can one catch them all? Dev. Biol. 2017, 430, 288–301. [Google Scholar] [CrossRef]
- Bao, S.; Hua, C.; Shen, L.; Yu, H. New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 118–131. [Google Scholar] [CrossRef]
- Zhang, C.; Jian, M.; Li, W.; Yao, X.; Tan, C.; Qian, Q.; Hu, Y.; Liu, X.; Hou, X. Gibberellin signaling modulates flowering via the DELLA-BRAHMA-NF-YC module in Arabidopsis. Plant Cell 2023, 35, 3470–3484. [Google Scholar] [CrossRef]
- Riboni, M.; Robustelli Test, A.; Galbiati, M.; Tonelli, C.; Conti, L. ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6309–6322. [Google Scholar] [CrossRef]
- Zhai, Q.; Zhang, X.; Wu, F.; Feng, H.; Deng, L.; Xu, L.; Zhang, M.; Wang, Q.; Li, C. Transcriptional Mechanism of Jasmonate Receptor COI1-Mediated Delay of Flowering Time in Arabidopsis. Plant Cell 2015, 27, 2814–2828. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Li, Y.; Guan, Y.; Wang, S.; Luo, H.; Li, X.; Li, H.; Zhang, Z. Auxin-induced AUXIN RESPONSE FACTOR4 activates APETALA1 and FRUITFULL to promote flowering in woodland strawberry. Hortic. Res. 2021, 8, 115. [Google Scholar] [CrossRef]
- D’Aloia, M.; Bonhomme, D.; Bouché, F.; Tamseddak, K.; Ormenese, S.; Torti, S.; Coupland, G.; Périlleux, C. Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant J. 2011, 65, 972–979. [Google Scholar] [CrossRef] [PubMed]
- Segarra, S.; Mir, R.; Martínez, C.; León, J. Genome-wide analyses of the transcriptomes of salicylic acid-deficient versus wild-type plants uncover Pathogen and Circadian Controlled 1 (PCC1) as a regulator of flowering time in Arabidopsis. Plant Cell Environ. 2010, 33, 11–22. [Google Scholar] [CrossRef]
- Achard, P.; Baghour, M.; Chapple, A.; Hedden, P.; Van Der Straeten, D.; Genschik, P.; Moritz, T.; Harberd, N.P. The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. USA 2007, 104, 6484–6489. [Google Scholar] [CrossRef]
- Lee, Z.; Kim, S.; Choi, S.J.; Joung, E.; Kwon, M.; Park, H.J.; Shim, J.S. Regulation of Flowering Time by Environmental Factors in Plants. Plants 2023, 12, 3680. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Fan, Z.; Li, S.; Wang, M.; Huang, M.; Ma, X.; Liu, W.; Wang, Y.; Yu, Y.; Li, Y.; et al. Genomics insights into flowering and floral pattern formation: Regional duplication and seasonal pattern of gene expression in Camellia. BMC Biol. 2024, 22, 50. [Google Scholar] [CrossRef]
- Lin, M.; Wang, S.; Liu, Y.; Li, J.; Zhong, H.; Zou, F.; Yuan, D. Hydrogen cyanamide enhances flowering time in tea oil camellia (Camellia oleifera Abel.). Ind. Crops Prod. 2022, 176, 114313. [Google Scholar] [CrossRef]
- Pan, X.; Welti, R.; Wang, X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 2010, 5, 986–992. [Google Scholar] [CrossRef]
- Šimura, J.; Antoniadi, I.; Široká, J.; Tarkowská, D.; Strnad, M.; Ljung, K.; Novák, O. Plant Hormonomics: Multiple Phytohormone Profiling by Targeted Metabolomics. Plant Physiol. 2018, 177, 476–489. [Google Scholar] [CrossRef]
- Cui, K.; Lin, Y.; Zhou, X.; Li, S.; Liu, H.; Zeng, F.; Zhu, F.; Ouyang, G.; Zeng, Z. Comparison of sample pretreatment methods for the determination of multiple phytohormones in plant samples by liquid chromatography–electrospray ionization-tandem mass spectrometry. Microchem. J. 2015, 121, 25–31. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Li, X.; Liang, G.; Yu, D. Two DELLA-interacting proteins bHLH48 and bHLH60 regulate flowering under long-day conditions in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 2757–2767. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Ye, L.; Zhai, T.; Wang, S.; Pan, Y.; Qu, K.; Gu, M.; Wang, Y.; Zhang, J.; Li, X.; et al. CELL DIVISION CYCLE 5 controls floral transition by regulating flowering gene transcription and splicing in Arabidopsis. Plant Physiol. 2024, 197, 616. [Google Scholar] [CrossRef] [PubMed]
- Lyu, T.; Cao, J. Cys2/His2 Zinc-Finger Proteins in Transcriptional Regulation of Flower Development. Int. J. Mol. Sci. 2018, 19, 2589. [Google Scholar] [CrossRef]
- Casal, J.J.; Qüesta, J.I. Light and temperature cues: Multitasking receptors and transcriptional integrators. New Phytol. 2018, 217, 1029–1034. [Google Scholar] [CrossRef]
- Shim, J.S.; Kubota, A.; Imaizumi, T. Circadian Clock and Photoperiodic Flowering in Arabidopsis: CONSTANS Is a Hub for Signal Integration. Plant Physiol. 2017, 173, 5–15. [Google Scholar] [CrossRef]
- Li, X.; Liang, T.; Liu, H. How plants coordinate their development in response to light and temperature signals. Plant Cell 2021, 34, 955–966. [Google Scholar] [CrossRef]
- Zhao, Z.; Dent, C.; Liang, H.; Lv, J.; Shang, G.; Liu, Y.; Feng, F.; Wang, F.; Pang, J.; Li, X.; et al. CRY2 interacts with CIS1 to regulate thermosensory flowering via FLM alternative splicing. Nat. Commun. 2022, 13, 7045. [Google Scholar] [CrossRef]
- Guo, H.; Yang, H.; Mockler, T.C.; Lin, C. Regulation of Flowering Time by Arabidopsis Photoreceptors. Science 1998, 279, 1360–1363. [Google Scholar] [CrossRef]
- Soy, J.; Leivar, P.; González-Schain, N.; Martín, G.; Diaz, C.; Sentandreu, M.; Al-Sady, B.; Quail, P.H.; Monte, E. Molecular convergence of clock and photosensory pathways through PIF3–TOC1 interaction and co-occupancy of target promoters. Proc. Natl. Acad. Sci. USA 2016, 113, 4870–4875. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kita, M.; Niinuma, K.; Ito, S.; Yamashino, T.; Mizoguchi, T.; Mizuno, T. Arabidopsis Clock-Associated Pseudo-Response Regulators PRR9, PRR7 and PRR5 Coordinately and Positively Regulate Flowering Time Through the Canonical CONSTANS-Dependent Photoperiodic Pathway. Plant Cell Physiol. 2007, 48, 822–832. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Yu, R.; Fan, L.-M.; Wei, N.; Chen, H.; Deng, X.W. DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 2016, 7, 11868. [Google Scholar] [CrossRef]
- Li, X.; Lin, C.; Lan, C.; Tao, Z. Genetic and epigenetic basis of phytohormonal control of floral transition in plants. J. Exp. Bot. 2024, 75, 4180–4194. [Google Scholar] [CrossRef] [PubMed]
- Campos-Rivero, G.; Osorio-Montalvo, P.; Sánchez-Borges, R.; Us-Camas, R.; Duarte-Aké, F.; De-la-Peña, C. Plant hormone signaling in flowering: An epigenetic point of view. J. Plant Physiol. 2017, 214, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Calderon-Villalobos, L.I.A.; Estelle, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 2009, 5, 301–307. [Google Scholar] [CrossRef]
- Harberd, N.P. Relieving DELLA Restraint. Science 2003, 299, 1853–1854. [Google Scholar] [CrossRef]
- Hirano, K.; Asano, K.; Tsuji, H.; Kawamura, M.; Mori, H.; Kitano, H.; Ueguchi-Tanaka, M.; Matsuoka, M. Characterization of the Molecular Mechanism Underlying Gibberellin Perception Complex Formation in Rice. Plant Cell 2010, 22, 2680–2696. [Google Scholar] [CrossRef]
- Dill, A.; Thomas, S.G.; Hu, J.; Steber, C.M.; Sun, T.-P. The Arabidopsis F-Box Protein SLEEPY1 Targets Gibberellin Signaling Repressors for Gibberellin-Induced Degradation. Plant Cell 2004, 16, 1392–1405. [Google Scholar] [CrossRef]
- Silverstone, A.L.; Ciampaglio, C.N.; Sun, T.-P. The Arabidopsis RGA Gene Encodes a Transcriptional Regulator Repressing the Gibberellin Signal Transduction Pathway. Plant Cell 1998, 10, 155–169. [Google Scholar] [CrossRef]
- Cheng, H.; Qin, L.; Lee, S.; Fu, X.; Richards, D.E.; Cao, D.; Luo, D.; Harberd, N.P.; Peng, J. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 2004, 131, 1055–1064. [Google Scholar] [CrossRef]
- Davière, J.-M.; Achard, P. A Pivotal Role of DELLAs in Regulating Multiple Hormone Signals. Mol. Plant 2016, 9, 10–20. [Google Scholar] [CrossRef]
- Hou, X.; Lee, L.Y.C.; Xia, K.; Yan, Y.; Yu, H. DELLAs Modulate Jasmonate Signaling via Competitive Binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef]
- Riboni, M.; Galbiati, M.; Tonelli, C.; Conti, L. GIGANTEA Enables Drought Escape Response via Abscisic Acid-Dependent Activation of the Florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1. Plant Physiol. 2013, 162, 1706–1719. [Google Scholar] [CrossRef]
- Shu, K.; Chen, Q.; Wu, Y.; Liu, R.; Zhang, H.; Wang, S.; Tang, S.; Yang, W.; Xie, Q. ABSCISIC ACID-INSENSITIVE 4 negatively regulates flowering through directly promoting Arabidopsis FLOWERING LOCUS C transcription. J. Exp. Bot. 2015, 67, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, L.; Ye, T.; Lu, Y.; Chen, X.; Wu, Y. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 2013, 64, 675–684. [Google Scholar] [CrossRef]
- Achard, P.; Cheng, H.; De Grauwe, L.; Decat, J.; Schoutteten, H.; Moritz, T.; Van Der Straeten, D.; Peng, J.; Harberd, N.P. Integration of Plant Responses to Environmentally Activated Phytohormonal Signals. Science 2006, 311, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Guo, Y.; Ali, I.; Lin, J.; Xu, Y.; Yang, M. Accumulation characteristics of plant flavonoids and effects of cultivation measures on their biosynthesis: A review. Plant Physiol. Biochem. 2024, 215, 108960. [Google Scholar] [CrossRef] [PubMed]
- Zuk, M.; Szperlik, J.; Hnitecka, A.; Szopa, J. Temporal biosynthesis of flavone constituents in flax growth stages. Plant Physiol. Biochem. 2019, 142, 234–245. [Google Scholar] [CrossRef]
- Solar, A.; Colarič, M.; Usenik, V.; Stampar, F. Seasonal variations of selected flavonoids, phenolic acids and quinones in annual shoots of common walnut (Juglans regia L.). Plant Sci. 2006, 170, 453–461. [Google Scholar] [CrossRef]
- Liu, H.; Cheng, Z.; Luo, M.; Xie, J. The dynamic variations of flavonoid metabolites in flower buds for Zingiber mioga at different developmental stages. J. Food Compos. Anal. 2023, 123, 105537. [Google Scholar] [CrossRef]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, J.; Chen, Y.; Tang, H.; Wang, Y.; He, Y.; Ou, Y.; Sun, X.; Wang, S.; Yao, Y. Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins. Hortic. Res. 2018, 5, 27. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Guo, T.; Zou, S.; Li, L.; Li, X.; Wang, J.; Zhu, Z.; Ai, L. Integrative Transcriptomic and Metabolomic Analysis Reveals the Molecular Mechanisms Underlying Flowering Time Variation in Camellia Species. Agronomy 2025, 15, 1288. https://doi.org/10.3390/agronomy15061288
Zhou L, Guo T, Zou S, Li L, Li X, Wang J, Zhu Z, Ai L. Integrative Transcriptomic and Metabolomic Analysis Reveals the Molecular Mechanisms Underlying Flowering Time Variation in Camellia Species. Agronomy. 2025; 15(6):1288. https://doi.org/10.3390/agronomy15061288
Chicago/Turabian StyleZhou, Ling, Tao Guo, Shihui Zou, Lingli Li, Xuemei Li, Jiao Wang, Zilin Zhu, and Lijiao Ai. 2025. "Integrative Transcriptomic and Metabolomic Analysis Reveals the Molecular Mechanisms Underlying Flowering Time Variation in Camellia Species" Agronomy 15, no. 6: 1288. https://doi.org/10.3390/agronomy15061288
APA StyleZhou, L., Guo, T., Zou, S., Li, L., Li, X., Wang, J., Zhu, Z., & Ai, L. (2025). Integrative Transcriptomic and Metabolomic Analysis Reveals the Molecular Mechanisms Underlying Flowering Time Variation in Camellia Species. Agronomy, 15(6), 1288. https://doi.org/10.3390/agronomy15061288



