Identification of Elite Agronomic Traits Using Chromosome Arm Substitution Lines of Triticum dicoccoides in the Background of Common Wheat
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Field Experimental Design
2.3. Investigation of Agronomic Traits at the Seedling Stage
2.4. Agronomic Trait Survey at Heading Stage
2.5. CASLs Propagation
2.6. Yield Trait Survey
2.7. Spike Trait Survey
3. Results
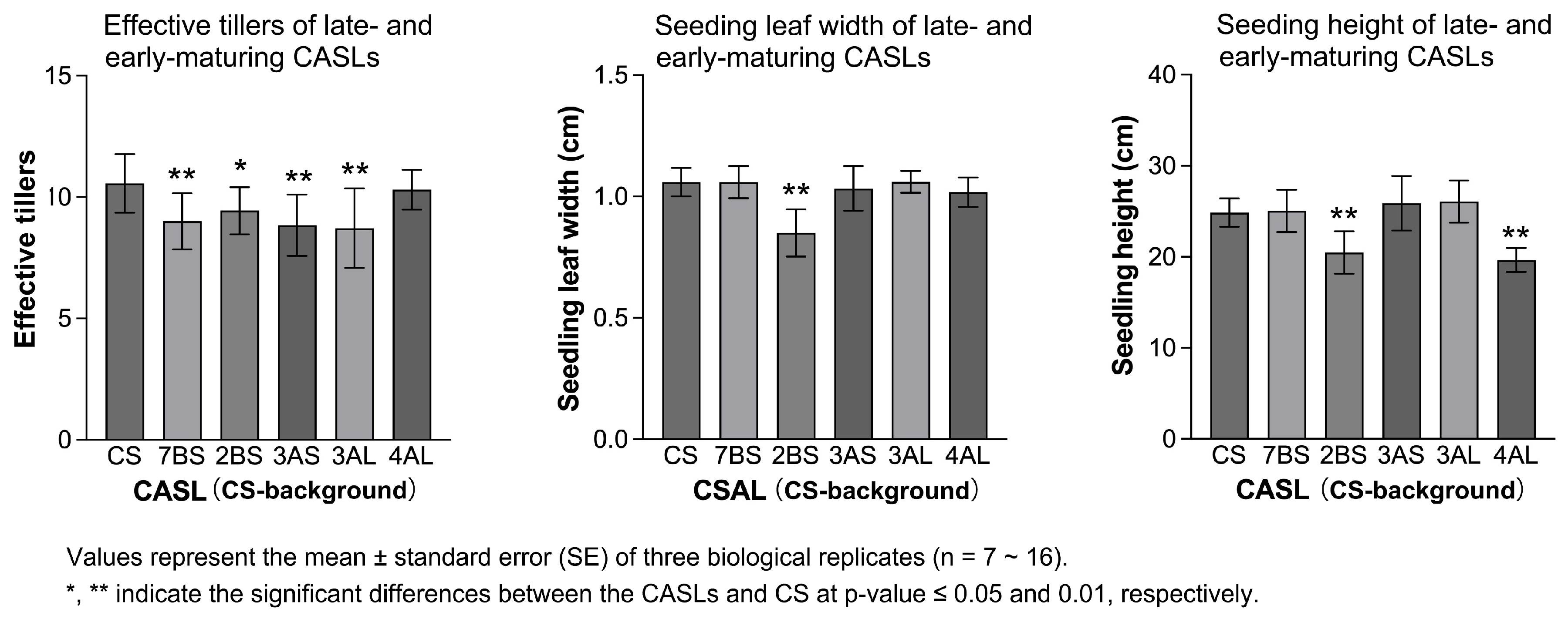
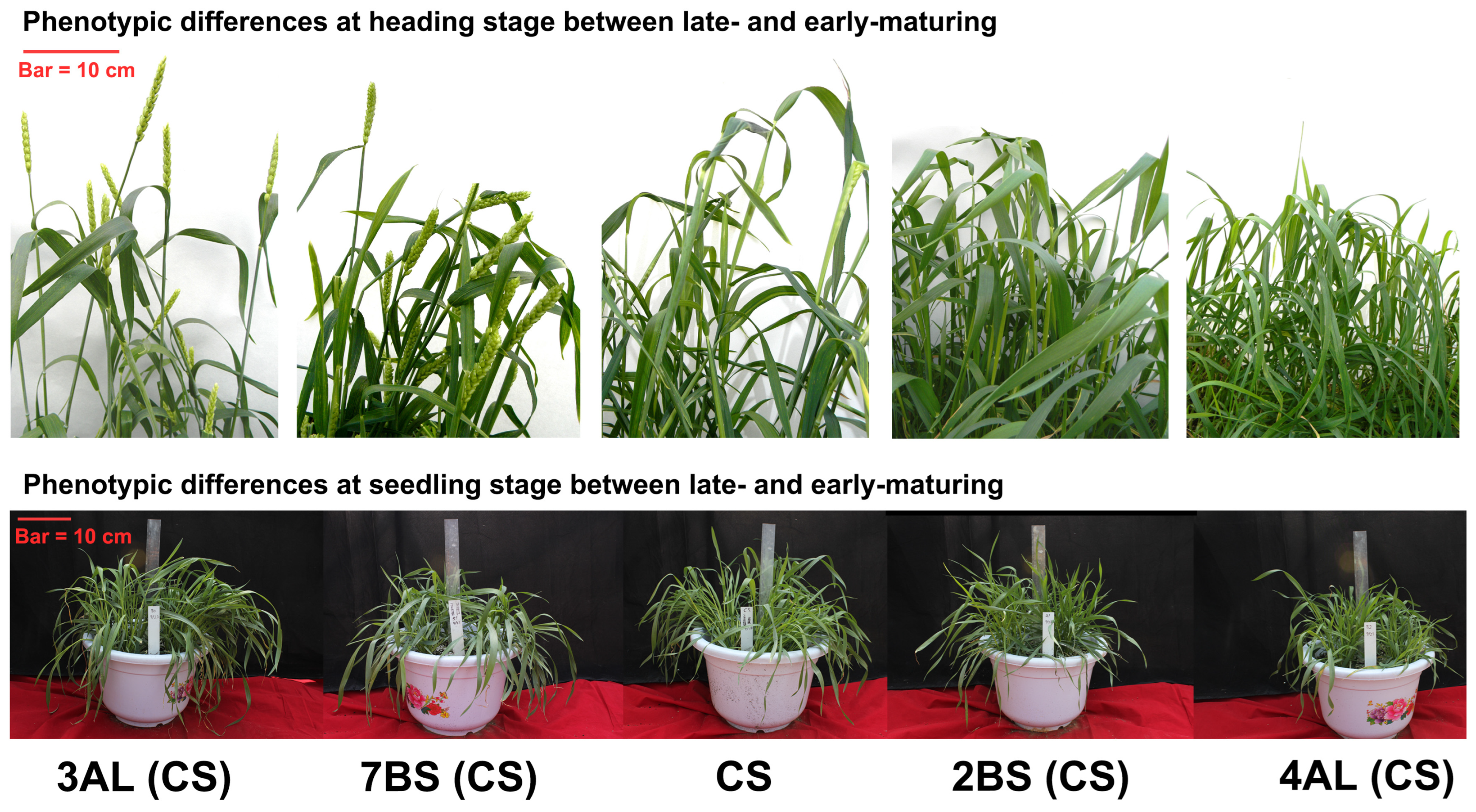
3.1. Statistical Analysis of Agronomic Traits During the Seedling Stage
3.2. Agronomic Trait Statistical Analysis at Heading Stage
3.3. Investigation and Data Analysis of Grain Traits
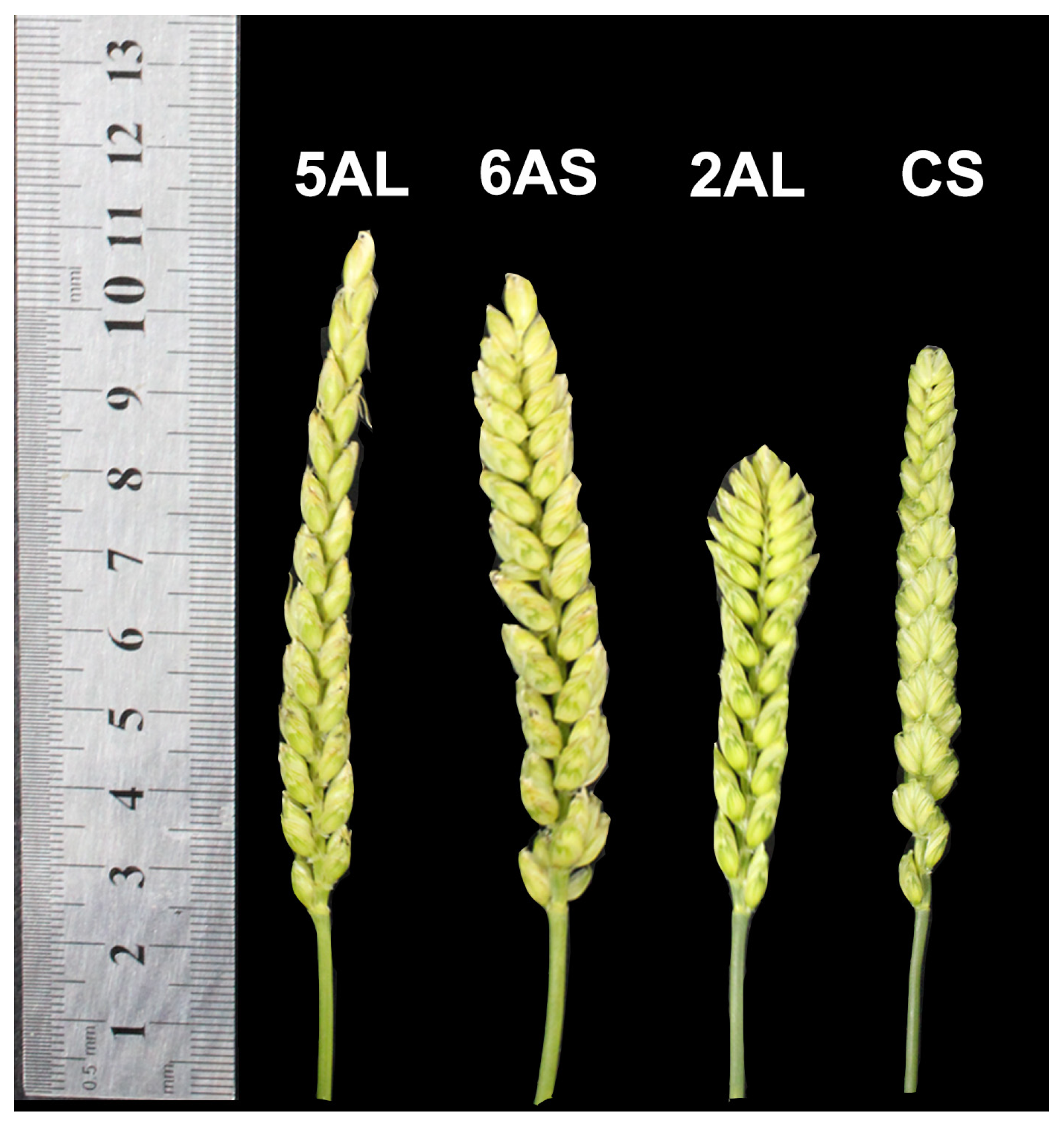
3.4. Statistical Analysis of Spike Traits
3.5. Statistical Analysis of Other Field Agronomic Traits
4. Discussion
4.1. The Utilization of Wild Emmer Wheat in Breeding
4.2. The Important Agronomic Traits and Their Controlling Candidate Chromosomal Regions Analyses in CASLs
4.3. CASLs Offer a Powerful Tool for Dissecting the Genetic Control of These Complex Traits and Understanding Their Interactions
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, A.; Mantovani, E.; Simsek, S.; Jain, S.; Elias, E.; Mergoum, M. Genome wide genetic dissection of wheat quality and yield related traits and their relationship with grain shape and size traits in an elite × non-adapted bread wheat cross. PLoS ONE 2019, 14, e0221826. [Google Scholar] [CrossRef] [PubMed]
- Yuli, J.; Tiantian, G.; Hong, L.; Diaoguo, A. Research progress on the wheat powdery mildew resistance gene Pm2. Chin. J. Eco-Agric. 2022, 30, 779–786. [Google Scholar]
- Ye, X.; Lin, Z.; Wang, K.; Tang, H.; Han, Z. Annual progress on new gene characterization and novel technology development for wheat genetic improvement in 2023. Sci. Technol. Rev. 2024, 42, 174–187. [Google Scholar]
- Millet, E.; Rong, J.; Qualset, C.; McGuire, P.; Bernard, M.; Sourdille, P.; Feldman, M. Production of chromosome-arm substitution lines of wild emmer in common wheat. Euphytica 2013, 190, 1–17. [Google Scholar] [CrossRef]
- Feldman, M. Historical aspects and significance of the discovery of wild wheats. Stadler Symp. 1977, 9, 121–146. [Google Scholar]
- Feldman, M.; Sears, E. The wild gene resources of wheat. Sci. Am. 1981, 244, 102–112. [Google Scholar] [CrossRef]
- Feldman, M.; Millet, E. Methodologies for identification, allocation and transfer of quantitative genes from wild emmer into cultivated wheat. In Proceedings of the 8th International Wheat Genetics Symposium, Beijing, China, 20–25 July 1993; pp. 19–27. [Google Scholar]
- Millet, E.; Rong, J.; Qualset, C.; Mcguire, P.; Bernard, M.; Sourdille, P.; Feldman, M. Grain yield and grain protein percentage of common wheat lines with wild emmer chromo some-arm substitutions. Euphytica 2014, 195, 69–81. [Google Scholar] [CrossRef]
- Orlovskaya, O.; Leonova, I.; Solovey, L.; Dubovets, N. Molecular cytological analysis of alien introgressions in common wheat lines created by crossing of Triticum aestivum with T. dicoccoides and T. dicoccum. Vavilov J. Genet. Breeding 2023, 27, 553–564. [Google Scholar] [CrossRef]
- Almadi, H.; Nazarian, F. The inheritance and chromosomal location of morphological traits in wildwheat, Triticum turgidum L. ssp. dicoccoides. Euphytica 2007, 158, 103–108. [Google Scholar]
- Zheng, Q.; Zhao, L.; Li, B.; Li, H.; Wan, J.; Zhang, X. Wheat wide hybridization and chromosome engineering breeding in China. Hereditas 2025, 47, 289–299. [Google Scholar] [CrossRef]
- Xie, W.; Nevo, E. Wild emmer: Genetic resources, gene mapping and potential for wheat improvement. Euphytica 2008, 164, 603–614. [Google Scholar] [CrossRef]
- Simmonds, J.; Fish, L.; Leverington-Waite, M.; Wang, Y.; Howell, P.; Snape, J. Mapping of a gene (Vir) for a non-glaucous, viridescent phenotype in bread wheat derived from Triticum dicoccoides, and its association with yield variation. Euphytica 2008, 159, 333–341. [Google Scholar] [CrossRef]
- Joppa, L.; Cantrell, R. Chromosome location of genes for grain protein content of wildtetraploid wheat. Crop Sci. 1990, 30, 1059–1064. [Google Scholar]
- Kumar, N.; Kulwal, P.; Balyan, H.; Gupta, P. QTL mapping for yield and yield contributing traits in two mapping populations of bread wheat. Mol. Breed. 2007, 19, 163–177. [Google Scholar] [CrossRef]
- Feldman, M.; Millet, E.; Abbo, S. Exploitation of wild emmer wheat to increase yield and protein content in durum and common wheat. In Proceedings of the EUCARPIA Meeting of the Genetic Resources Section, Clermont-Ferrand, France, 15–18 March 1994. [Google Scholar]
- Zhou, W.; Jiang, Y.; Zhang, W.; Xu, G.; Rong, J. Characterization of Large Chromosome Segment Introgressions from Triticum turgidum subsp. dicoccoides into Bread Wheat with Simple Sequence Repeat Markers. Crop Sci. 2013, 53, 1555–1565. [Google Scholar]
- Rong, J.; Millet, E.; Manisterski, J.; Feldman, M. A new powdery mildew resistance gene: Introgression from wild emmer into common wheat and rflp-based mapping. Euphytica 2000, 115, 121–126. [Google Scholar]
- Yu, J.; Zhao, Y.; Ding, M.; Yu, Z.; Jiang, Y.; Ma, W.; Rong, J. Wild emmer chromosome arm substitution lines: Useful resources for wheat genetic study and breeding. Crop Sci. 2020, 60, 1761–1769. [Google Scholar]
- Zhao, Y.; Miao, Y.; Cao, Y.; Rong, J. Investigation on the Seed Germination and Seedling Growth of Chromosome Arm Substitution Lines of Triticum dicoccoidesin the Background of T. aestivum. J. Triticeae Crops 2017, 37, 287–294. [Google Scholar]
- Merchuk-Ovnat, L.; Fahima, T.; Ephrath, J.; Krugman, T.; Saranga, Y. Ancestral QTL alleles from wild emmer wheat enhance root development under drought in modern wheat. Front. Plant Sci. 2017, 8, 703. [Google Scholar] [CrossRef]
- Kunert, A.; Naz, A.; Dedeck, O.; Pillen, K.; Leon, J. AB-QTL, analysis imn winter wheat: I. Synthetichexaploid wheat (T. tugidum ssp., dicoccoides × T. tauschii) as a source of favourable alleles for millingand baking quality traits. Theor. Appl. Genet. 2007, 115, 683–695. [Google Scholar] [CrossRef]
- Naz, A.; Kunert, A.; Lind, V.; Pillen, K.; Leon, J. AB-QTL, analysis in winter wheat: Il. Genetic analysisof seedling and field resistance against leafrust in a wheat advanced backcross, population. Theor. Appl. Genet. 2008, 116, 1095–1104. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.; Gadaleta, A.; Cenci, A.; Carluccio, A.; Abdelbacki, A.; Simeone, R. Molecular mapping of the novel powdery mildew resistance gene Pm36 introgressed from Triticum turgidumvar. dicoccoides in durum wheat. Theor. Appl. Genet. 2008, 117, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Uauy, C.; Distelfeld, A.; Blechl, A.; Epstein, L.; Chen, X.; Dubcovsky, J. A kinase-start gene confers temperaturedependent resistance to wheat stripe rust. Science 2009, 323, 1357–1360. [Google Scholar] [CrossRef] [PubMed]
- Klymiuk, V.; Yaniv, E.; Huang, L.; Raats, D.; Fatiukha, A.; Chen, S.; Fahima, T. Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat. Commun. 2018, 9, 3735. [Google Scholar]
- Qie, Y.; Liu, Y.; Wang, M.; Li, X.; See, D.; An, D.; Chen, X. Development, validation, and re-selection of wheat Lines with pyramided genes Yr64 and Yr15 linked on the short arm of chromosome 1B for resistance to stripe rust. Plant Dis. 2019, 103, 51–58. [Google Scholar]
- Yaniv, E.; Raats, D.; Ronin, Y.; Korol, A.; Grama, A.; Bariana, H.; Fahima, T. Evaluation of marker-assisted selection for the stripe rust resistance gene Yr15, introgressed from wild emmer wheat. Mol. Breed. 2015, 35, 43. [Google Scholar] [CrossRef]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J.A. NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar]
- Khan, M.; Anwar, S.; Khan, M. Genetic variability for seedling traits in wheat (Triticum aestivum L.) under moisture stress conditions. Asian J. Plant Sci. 2002, 1, 588–590. [Google Scholar] [CrossRef]
- Salih, A. SNP Markers for Seedling Traits Revealed by Association Analysis Andgenetic Variation in Durum Wheat Accessions (Triticum turgidum L. ssp. durum Desf.). Ph.D. Dissertation, Huazhong Agricultural University, Wuhan, China, 2016. [Google Scholar]
- Meles, B.; Genetic, V. Correlation and path analysis of yield and grain quality traits in bread wheat (Tritium aestivum L.) genotypes at Axum, Northern Ethiopia. J. Plant Breed. Crop Sci. 2017, 9, 175–185. [Google Scholar]
- Bavec, M.; Bavec, F.; Varga, B.; Kovačević, V. Relationships among yield, it’s quality and yield components in winter wheat (Triticum aestivum L.) cultivars affected by seeding rates. Die Bodenkult. 2002, 53, 143–151. [Google Scholar]
- Botwright, T.; Condon, A.; Rebetzke, G.; Richards, R. Field evaluation of early vigour for genetic improvement of grain yield in wheat. Aust. J. Agric. Res. 2002, 53, 1137–1145. [Google Scholar]
- Gegas, V.; Nazari, A.; Griffiths, S.; Simmonds, J.; Fish, L.; Orford, S.; Sayers, L.; Doonan, J.; Snape, J. A genetic framework for grain size and shape variation in wheat. Plant Cell 2010, 22, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Farahani, H.; Moaveni, P.; Maroufi, K. Effect of seed size on seedling production in wheat (Triticum aestivum L.). Adv. Environ. Biol. 2011, 5, 1711–1715. [Google Scholar]
- Russo, M.; Ficco, D.; Laido, G.; Marone, D.; Papa, R.; Blanco, A.; Gadaleta, A.; Vita, P.; Mastrangelo, A. A dense durum wheat × T. dicoccum linkage map based on SNP markers for the study of seed morphology. Mol Breed 2014, 34, 1579–1597. [Google Scholar] [CrossRef]
- Kumar, A.; Mantovani, E.; Seetan, R.; Soltani, A.; Echeverry-Solarte, M.; Jain, S.; Simsek, S.; Doehlertm, D.; Alamri, M.; Elias, E. Dissection of Genetic Factors underlying Wheat Kernel Shape and Size in an Elite × Nonadapted Cross using a High Density SNP Linkage Map. Plant Genome 2016, 9, 1–22. [Google Scholar] [CrossRef]
- Feng, F.; Han, Y.; Wang, S.; Yin, S.; Peng, Z.; Min, Z.; Gao, W.; Wen, X.; Qin, X.; Siddique, K. The Effect of Grain Position on Genetic Improvement of Grain Number and Thousand Grain Weight in Winter Wheat in North China. Front. Plant Sci. 2018, 9, 129. [Google Scholar] [CrossRef]
- Guy, G.; Idan, A.; Aviad, P.; Zimran, G.; Peleg, Z. GNI-A1 mediates trade-off between grain number and grain weight in tetraploid wheat. TAG Theor. Appl. Genet. 2019, 132, 2353–2365. [Google Scholar]
- Slafer, G.; Calderini, D.; Miralles, G. Yield components and compensation in wheat: Opportunities for future increasing yield potential. In Increasing Yield Potential in Wheat: Breaking the Barriers; Reynolds, M.P., Rajaram, S., Eds.; CIMMYT: Ciudad Obregón, Mexico, 1996; pp. 101–133. [Google Scholar]
- Wu, Q.; Chen, Y.; Zhou, S.; Fu, L.; Chen, J.; Xiao, Y.; Zhang, D.; Ouyang, S.; Zhao, X.; Cui, Y. High-Density Genetic Linkage Map Construction and QTL Mapping of Grain Shape and Size in the Wheat Population Yanda1817×Beinong6. PLoS ONE 2015, 12, e0118144. [Google Scholar]
- Xu, Y. Response of Seedling Biomass and Agronomic Traitsin Maturity in Stage of Wheat Nitrogen, Phosphorus and Potassium Deficiency and It’s Genetican Alysis. Master’s Thesis, Shandong Agricultural University, Tai’an, China, 2022. [Google Scholar]
- Sun, N.; Zhou, Q.; Zhi, L.; Qiao, P.; Mou, L.; Ni, S.; Chen, L. Identification of Drought Resistance at Seedling Stage and Correlation Analysisbetween Drought Resistance Indexes and lmportant Agronomic Characters of 240 Wheat Materials. Acta Agric. Boreali-Occident. Sin. 2022, 2, 147–156. [Google Scholar]
- Jiang, Y.; Schmidt, R.; Zhao, Y.; Reif, J. A quantitative genetic framework highlights the role of epistatic effects for grain-yield heterosis in bread wheat. Nat. Genet. 2017, 49, 1741–1746. [Google Scholar] [CrossRef]
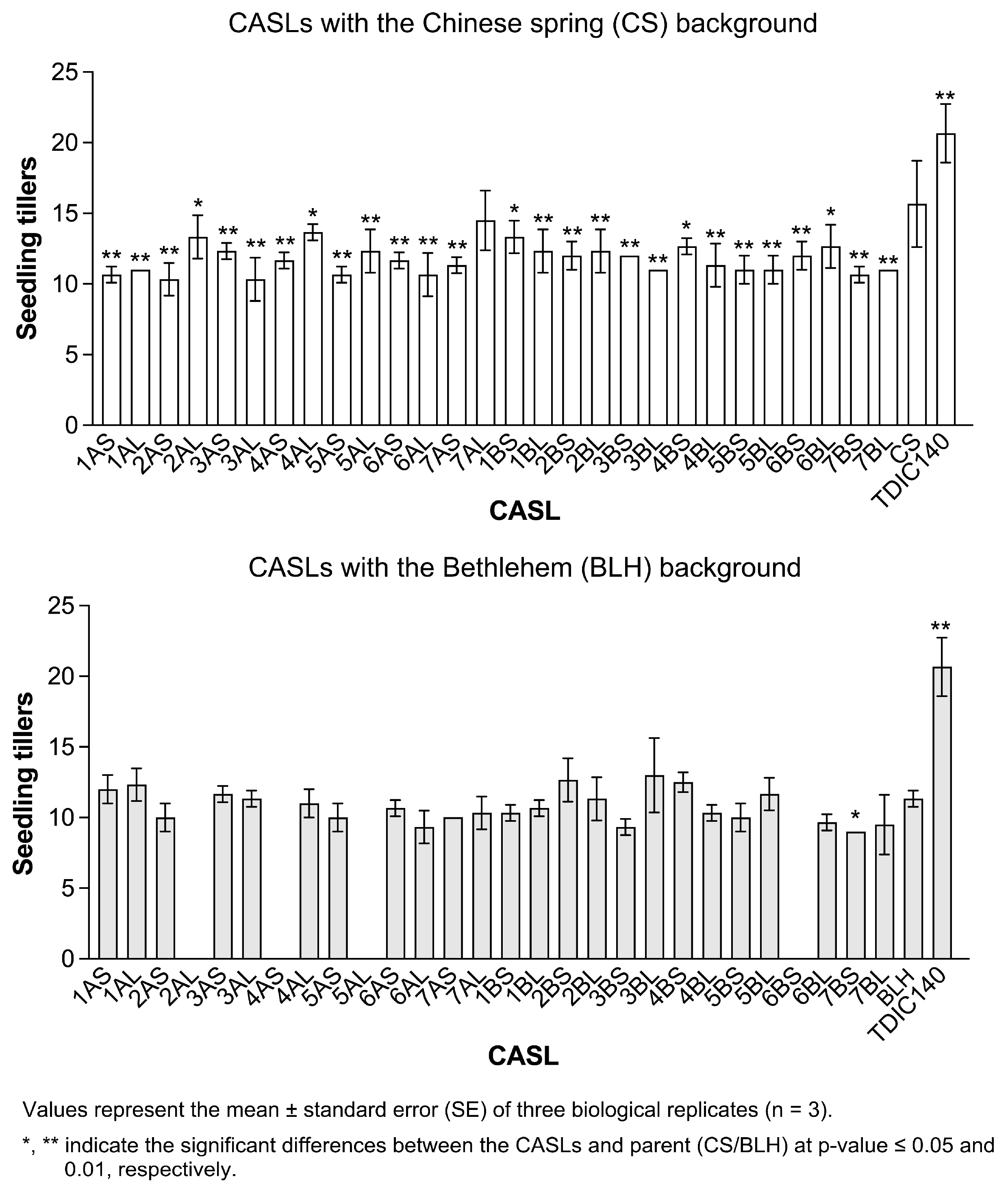
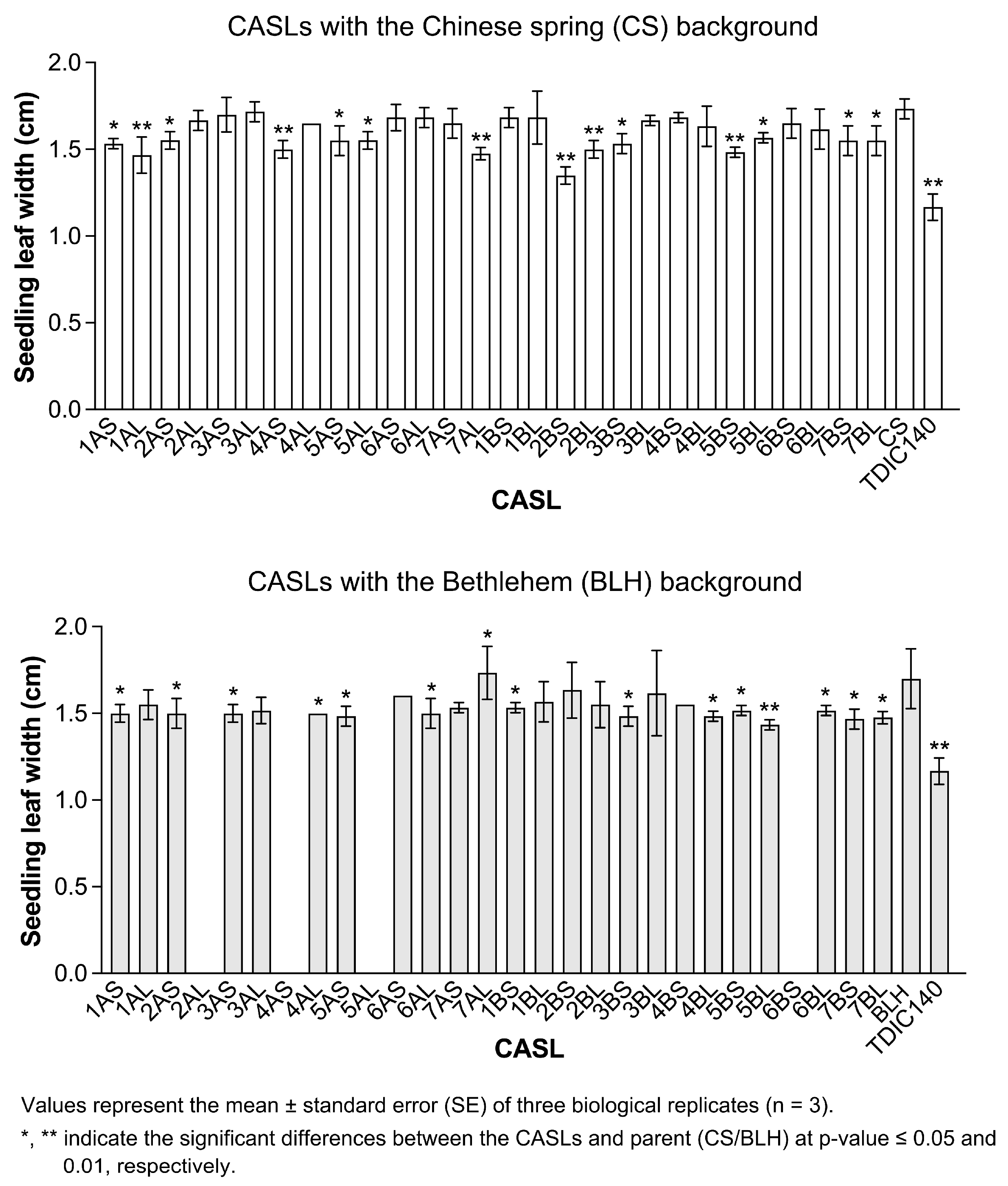
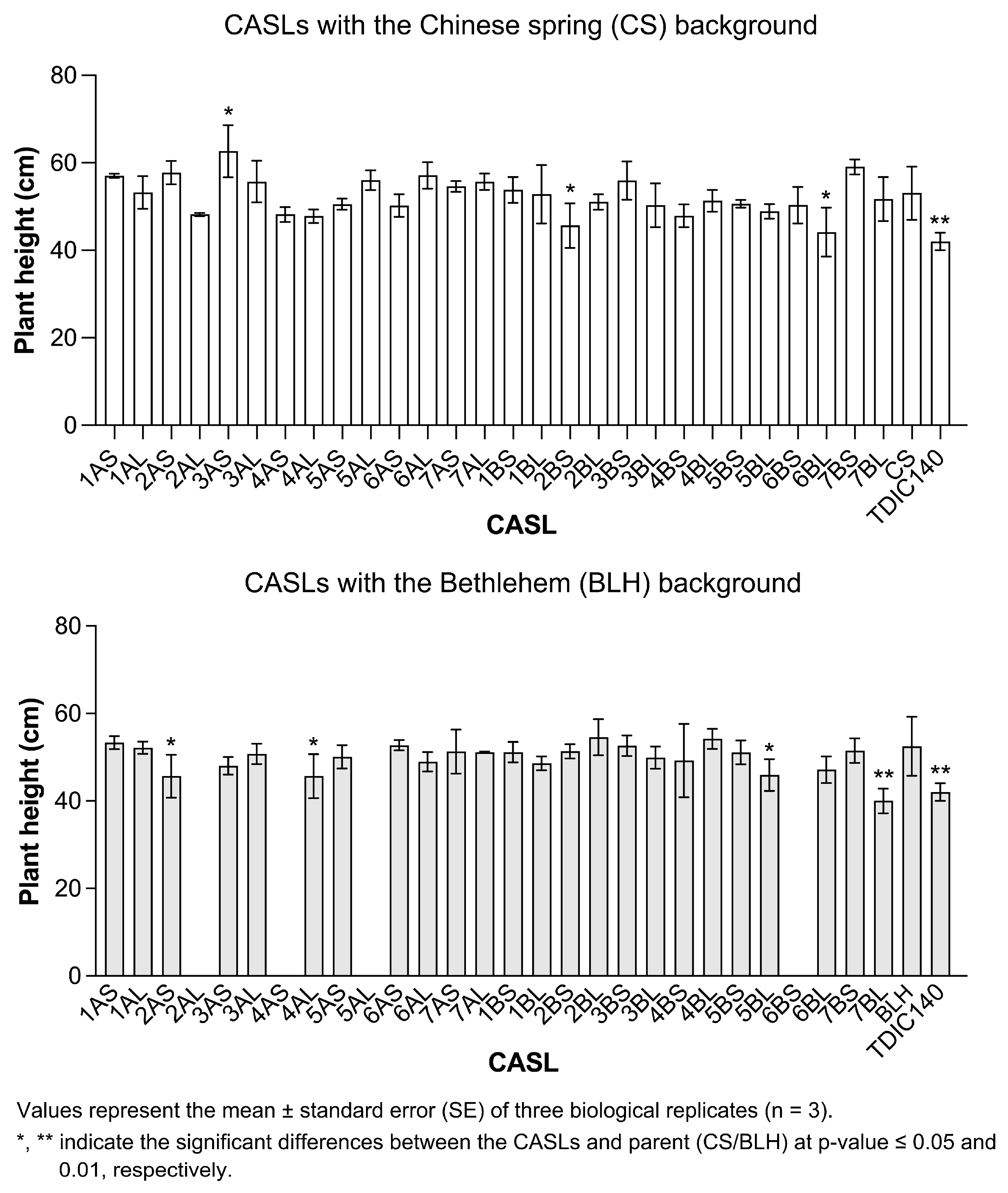
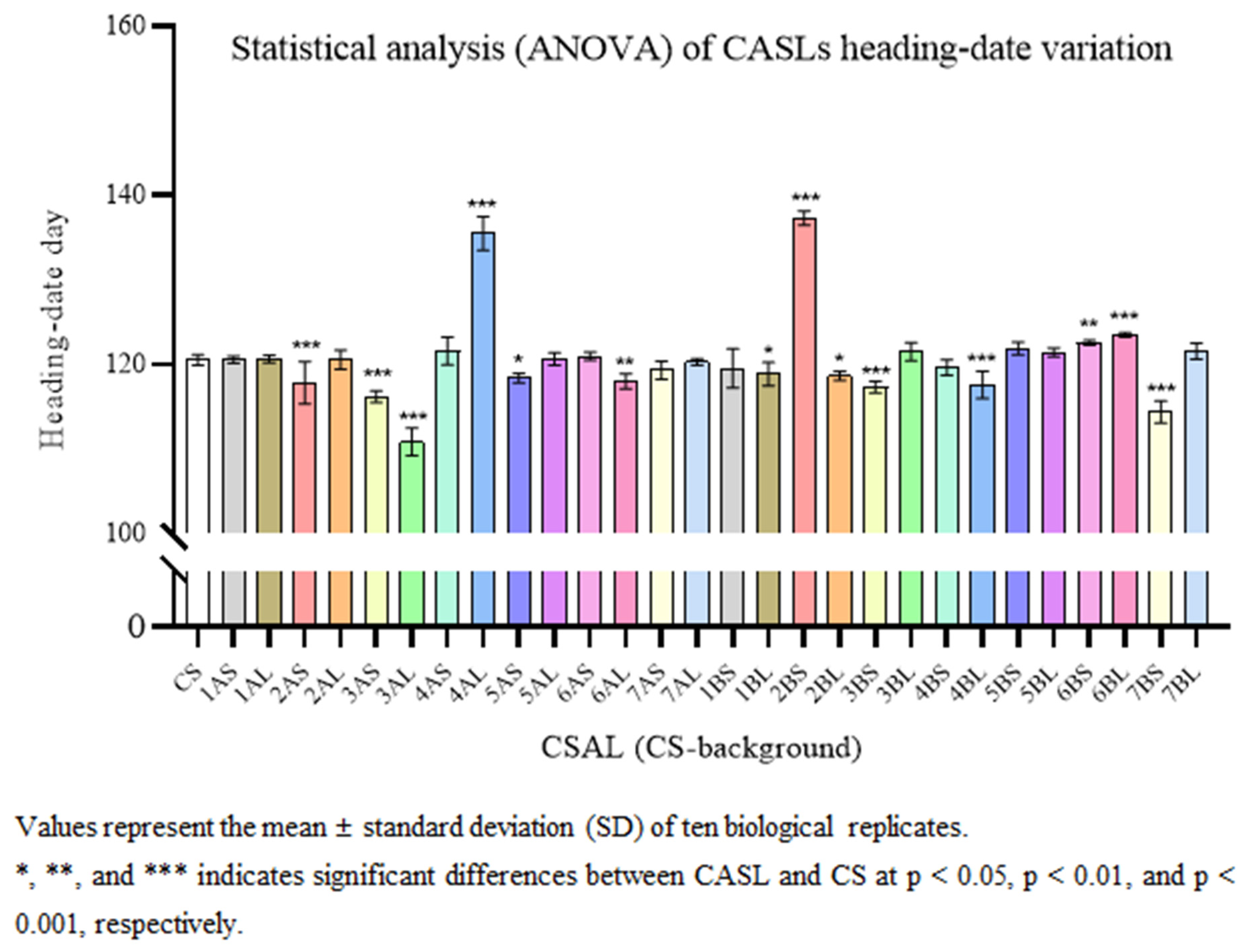

| BLH Background | CS Background | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Homologous Group | A(L) | A(S) | B(L) | B(S) | Homologous Group | A(L) | A(S) | B(L) | B(S) |
| 1 | + | + | + | + | 1 | + | + | + | + |
| 2 | + | + | + | 2 | + | + | + | + | |
| 3 | + | + | + | + | 3 | + | + | + | + |
| 4 | + | − | + | + | 4 | + | + | + | + |
| 5 | − | + | + | + | 5 | + | + | + | + |
| 6 | + | + | + | − | 6 | + | + | + | + |
| 7 | + | + | + | + | 7 | + | + | + | + |
| Awn Type | Awn Length and Position |
|---|---|
| No Awn | Completely without awns or awns are extremely short. |
| Top Awn | Awns present at the top of the spike, with a length of <5 mm, no awns at the lower part of the spike. |
| Curved Awn | The base of the awn is swollen and curved. |
| Short Awn | Awns present both at the top and bottom of the spike, with a length < 40 mm. |
| Long Awn | Awn length > 40 mm. |
| Spike Type | Phenotype Description |
|---|---|
| Spindle-shaped | The ear is pointed at both ends, with the middle slightly larger. |
| Oval-shaped | The middle is wide, with both ends slightly smaller, resembling an ellipse. |
| Rectangular-shaped | The ear is relatively short, and the shape is consistent in the upper, middle, and lower parts, appearing column-like. |
| Club-shaped | The ear is smaller at the bottom and larger at the top, with tightly clustered spikelets at the upper part, resembling a big head. |
| Conical-shaped (Pagoda-shaped) | The ear is larger at the bottom and smaller at the top, resembling a cone. |
| CASL | Grayscale Value | CASL | R Value | CASL | G Value | CASL | B Value |
|---|---|---|---|---|---|---|---|
| 4AL | 142.28 ± 2.73 * | 2BS | 180.8 ± 4.24 ** | 4AL | 138.78 ± 3.96 ** | 2BL | 82.9 ± 6.93 |
| 2BS | 141.08 ± 1.61 * | 4AS | 179.37 ± 8.16 ** | 2BS | 137.98 ± 0.35 ** | 2AL | 79.62 ± 2.11 |
| 2BL | 140.75 ± 2.07 | 2BL | 177.75 ± 3.44 ** | 2BL | 135.07 ± 3.94 | 5BL | 78.37 ± 1.1 |
| 2AL | 139.72 ± 0.68 | 1AL | 176.92 ± 0.79 ** | 2AL | 133.8 ± 0.48 | 1BS | 78.12 ± 4.77 |
| 1AL | 138.73 ± 2.87 | 2AL | 175.9 ± 3.38 | 1AL | 132.38 ± 3.61 | 4AL | 77.88 ± 4.54 |
| 5BL | 136.97 ± 1.46 | 1BS | 172.58 ± 0.8 | 3AS | 131.63 ± 3.46 | CS | 77.87 ± 2.92 |
| 3AS | 136.83 ± 3.6 | 3AS | 172.35 ± 5.03 | 5BL | 131.48 ± 1.23 | 6BS | 77.13 ± 4.92 |
| 6BS | 136.07 ± 1.84 | 5BL | 171.77 ± 2.2 | CS | 131.33 ± 2.16 | 2BS | 76.22 ± 4.53 |
| 1AS | 135.8 ± 2.16 | 2AS | 171.75 ± 2.39 | 6BS | 130.75 ± 4.29 | 5AL | 75.53 ± 6.76 |
| CS | 135.4 ± 2.43 | 1BL | 171.7 ± 0.88 | 1AS | 130.72 ± 2.96 | 4AS | 75.5 ± 2.05 |
| 1BS | 135.38 ± 2.27 | 1AS | 171.47 ± 0.78 | 3AL | 130.12 ± 0.42 | 1AL | 75.47 ± 5.39 |
| 4AS | 135.28 ± 0.9 | 4BS | 171.42 ± 3.05 | 4AS | 130.08 ± 1.63 | 7BL | 75.05 ± 1.48 |
| 7AS | 135.17 ± 0.28 | 6BS | 170.7 ± 2.59 | 7AS | 130.02 ± 0.46 | 3AS | 74.9 ± 1.73 |
| 3AL | 134.65 ± 0.73 | 3BS | 170.58 ± 6.72 | 1BS | 128.4 ± 2.42 | 3AL | 73.93 ± 2.28 |
| 4BS | 133.32 ± 1.58 | 7BS | 170.5 ± 1.98 | 3BL | 127.17 ± 3.05 | 7AS | 73.38 ± 4.87 |
| 3BL | 133.22 ± 2.24 | 7AS | 170.32 ± 1.43 | 5AL | 126.93 ± 0.96 | 1AS | 73 ± 3.55 |
| 5AL | 132.95 ± 0.51 | 3BL | 169.65 ± 0.59 | 4BS | 126.92 ± 1.34 | 3BL | 72.88 ± 3.01 |
| 3BS | 132.38 ± 1.31 | 6AS | 169.28 ± 0.46 | 6BL | 125.62 ± 4.2 | 4BL | 72.82 ± 3.27 |
| 2AS | 132.25 ± 2.69 | 6BL | 168.72 ± 6.18 | 3BS | 125.6 ± 4.73 | 6AL | 72.72 ± 1.99 |
| 1BL | 131.72 ± 1.01 | 3AL | 168.37 ± 2.49 | 2AS | 125.48 ± 3.08 | 3BS | 70.8 ± 5.55 |
| 6BL | 131.7 ± 2.79 | 5AL | 168.22 ± 2.05 | 6AS | 125.35 ± 2.43 | 6BL | 70.55 ± 4.67 |
| 6AS | 131.45 ± 2.3 | 4AL | 167.78 ± 3.19 | 6AL | 125.1 ± 3.89 | 4BS | 70.33 ± 1.38 |
| 6AL | 131.33 ± 3.58 | 5AS | 167.78 ± 3.19 | 7BL | 124.69 ± 2.67 | 5BS | 69.97 ± 2.55 |
| 7BS | 131.08 ± 0.44 | 6AL | 167.47 ± 3.74 | 5AS | 124.5 ± 3.56 | 1BL | 69.38 ± 2.42 |
| 7BL | 130.83 ± 2.77 | CS | 167.12 ± 4.74 | 5BS | 124.33 ± 4 | 7AL | 69.17 ± 5.59 |
| 5AS | 130.37 ± 2.51 | 5BS | 166.68 ± 1.97 | 1BL | 124.3 ± 0.94 | 6AS | 67.97 ± 6.75 |
| 5BS | 130.33 ± 2.13 | 4BL | 166.63 ± 3.04 | 7BS | 124.3 ± 0.79 | 2AS | 67.62 ± 2.34 |
| 4BL | 130.07 ± 2.57 | 7BL | 165.77 ± 3.71 | 7AL | 123.77 ± 1.03 ** | 5AS | 67 ± 6.22 |
| 7AL | 129.53 ± 0.34 * | 7AL | 165.68 ± 4.31 | 4BL | 123.35 ± 2.19 ** | 7BS | 66.83 ± 3.73 ** |
| CASL | Length/mm | CASL | Width/mm | CASL | Thickness/mm | CASL | Projected Area/mm2 | CASL | TGW |
|---|---|---|---|---|---|---|---|---|---|
| 6AS | 8.34 ± 0.16 * | 2AS | 3.73 ± 0.01 * | 4BL | 2.82 ± 0.08 * | 2AS | 23.41 ± 0.1 * | 7AL | 35.48 ± 1.16 * |
| 2AS | 8.34 ± 0.21 * | 1BL | 3.7 ± 0.1 * | 3BL | 2.78 ± 0.19 * | 1BL | 22.65 ± 0.65 * | 7AS | 34.89 ± 1.01 * |
| 4BL | 8.28 ± 0.07 * | 3BS | 3.67 ± 0 | 2BL | 2.75 ± 0.07 * | 6AS | 22.27 ± 0.38 * | 4BS | 32.99 ± 1.36 |
| 1BL | 8.14 ± 0.13 * | 7AL | 3.66 ± 0.07 | 7AS | 2.74 ± 0.09 * | 7AL | 22.04 ± 0.81 * | 6AL | 32.65 ± 0.79 |
| 7AL | 8.03 ± 0.1 | 3AS | 3.66 ± 0.04 | 4AS | 2.7 ± 0.13 * | 4BL | 21.96 ± 0.99 * | 6AS | 32.56 ± 1.47 |
| 5AS | 7.91 ± 0.16 | 5AS | 3.65 ± 0 | 7BS | 2.7 ± 0.05 * | 5AS | 21.74 ± 0.59 * | 1BL | 32.54 ± 0.98 |
| 3AS | 7.82 ± 0.14 | 2AL | 3.64 ± 0.01 | 2AS | 2.65 ± 0.05 | 3AS | 21.56 ± 0.56 * | 7BL | 32.3 ± 1.15 |
| 5AL | 7.82 ± 0.25 | 5BS | 3.63 ± 0.01 | 3AS | 2.65 ± 0.12 | 7BL | 21.56 ± 0.26 * | 2AS | 32.29 ± 1.49 |
| 2BS | 7.81 ± 0.06 | 4BS | 3.63 ± 0.07 | 7AL | 2.65 ± 0.02 | 2AL | 21.51 ± 0.42 | 1AL | 32.28 ± 1.36 |
| 7AS | 7.8 ± 0.22 | 4BL | 3.62 ± 0.11 | 3AL | 2.64 ± 0 | 3BS | 21.32 ± 0.16 | 3AL | 32.25 ± 1.51 |
| 6AL | 7.7 ± 0.15 | 1AL | 3.61 ± 0.01 | 5AS | 2.64 ± 0.03 | 5BS | 21.27 ± 0.17 | 3AS | 31.97 ± 1.91 |
| 3AL | 7.68 ± 0.15 | 7BS | 3.59 ± 0.02 | 3BS | 2.57 ± 0 | 3AL | 21.1 ± 0.54 | 5BS | 31.78 ± 1.38 |
| 7BL | 7.68 ± 0.42 | 3AL | 3.57 ± 0.09 | 2BS | 2.56 ± 0.04 | 7AS | 21.09 ± 0.34 | 4BL | 31.61 ± 1.57 |
| 3BS | 7.66 ± 0.16 | 6AS | 3.57 ± 0.02 | CS | 2.55 ± 0.04 | 5AL | 21.07 ± 0.72 | 5AS | 31.56 ± 1.67 |
| 4BS | 7.64 ± 0.22 | 7BL | 3.57 ± 0.05 | 1BL | 2.55 ± 0.02 | 4BS | 21.06 ± 0.38 | 3BL | 31.44 ± 1.19 |
| 2AL | 7.62 ± 0.26 | 2BL | 3.56 ± 0.09 | 5AL | 2.55 ± 0.03 | 2BS | 20.74 ± 0.17 | CS | 31.28 ± 1.13 |
| 5BL | 7.62 ± 0.26 | 5BL | 3.54 ± 0.03 | 7BL | 2.54 ± 0.02 | 7BS | 20.65 ± 0.42 | 4AS | 30.95 ± 1.6 |
| 1BS | 7.61 ± 0.21 | 5AL | 3.53 ± 0.04 | 6BL | 2.53 ± 0.07 | 5BL | 20.63 ± 0.3 | 1AS | 30.78 ± 1.13 |
| 1AL | 7.61 ± 0 | 7AS | 3.53 ± 0.02 | 6AL | 2.53 ± 0.01 | 6AL | 20.46 ± 0.61 | 1BS | 30.76 ± 2.02 |
| 1AS | 7.59 ± 0.06 | 6AS | 3.52 ± 0.04 | 5BL | 2.52 ± 0.01 | 1AL | 20.42 ± 0.36 | 5AL | 30.45 ± 1.15 |
| 5BS | 7.59 ± 0.27 | CS | 3.47 ± 0.02 | 6BS | 2.51 ± 0.01 | CS | 20.42 ± 0.27 | 7BS | 30.29 ± 1.37 |
| 6BL | 7.58 ± 0.26 | 1BS | 3.47 ± 0.07 | 2AL | 2.48 ± 0.02 | 1BS | 19.54 ± 1.11 | 3BS | 30.23 ± 1.37 |
| CS | 7.52 ± 0.11 | 4AS | 3.46 ± 0.04 | 5BS | 2.47 ± 0.03 | 4AS | 19.47 ± 1.17 | 2BS | 29.22 ± 0.95 |
| 4AS | 7.42 ± 0.22 | 2BS | 3.45 ± 0.13 | 1AL | 2.46 ± 0.05 | 4AL | 18.96 ± 0.8 * | 2BL | 29.22 ± 0.79 |
| 4AL | 7.28 ± 0.34 | 3BL | 3.42 ± 0.09 | 4BS | 2.45 ± 0.05 | 2BL | 18.95 ± 0.73 * | 5BL | 29.18 ± 1 |
| 7BS | 7.08 ± 0.76 | 4AL | 3.34 ± 0 | 1BS | 2.45 ± 0.07 | 3BL | 18.87 ± 0.34 * | 6BL | 28.83 ± 1.81 |
| 3BL | 6.96 ± 0.59 * | 1AS | 3.28 ± 0 | 1AS | 2.43 ± 0.04 * | 1AS | 18.19 ± 0.31 * | 2AL | 28.65 ± 0.96 |
| 6BS | 6.74 ± 0.67 * | 6BS | 3.17 ± 0.06 * | 6AS | 2.39 ± 0.04 * | 6BS | 17.73 ± 0.1 * | 6BS | 27.05 ± 1.15 * |
| 2BL | 6.57 ± 0.67 * | 6BL | 2.98 ± 0.03 * | 4AL | 2.12 ± 0.02 * | 6BL | 17.2 ± 0.14 * | 4AL | 26.63 ± 1.66 * |
| CASLs | Heading Period | R Value | G Value | B Value | Grayscale Value | Width/mm | Length/mm | Grain Thickness | Projected Area/mm2 | TGW |
|---|---|---|---|---|---|---|---|---|---|---|
| Heading Period | 1 | |||||||||
| R Value | 0.56 ** | 1 | ||||||||
| G Value | 0.47 * | 0.78 ** | 1 | |||||||
| B Value | 0.29 | 0.49 ** | 0.79 ** | 1 | ||||||
| Grayscale Value | 0.51 ** | 0.88 ** | 0.98 ** | 0.78 ** | 1 | |||||
| Width/mm | −0.35 | 0.02 | −0.28 | −0.34 | −0.21 | 1 | ||||
| Length/mm | −0.14 | −0.28 | −0.49 ** | −0.58 ** | −0.47 ** | 0.54 ** | 1 | |||
| Grain Thickness | −0.51 ** | −0.34 | −0.32 | −0.14 | −0.33 | 0.24 | 0.07 | 1 | ||
| Projected Area/mm2 | −0.29 | −0.12 | −0.40 * | −0.51 ** | −0.35 | 0.90 ** | 0.84 ** | 0.19 | 1 | |
| TGW | −0.48 ** | −0.54 ** | −0.54 ** | −0.51 ** | −0.58 ** | 0.48 ** | 0.55 ** | 0.38 * | 0.57 ** | 1 |
| CASL | Spike Length (cm) | CASL | Spikelet Number | CASL | Number of Effective Tillers | CASL | Grain Number per Panicle |
|---|---|---|---|---|---|---|---|
| 6AS | 8.04 ± 0.41 * | 6BL | 22.16 ± 0.89 * | 5AL | 17.00 ± 1.73 * | 7AL | 51.18 ± 5.18 * |
| 5AL | 8.68 ± 1.56 * | 4BS | 21.83 ± 1.6 | 2BL | 16.33 ± 1.53 * | 3AS | 49.77 ± 4.05 * |
| 4AL | 8.04 ± 0.38 | 4AS | 20.86 ± 0.93 | 3BS | 15.33 ± 0.58 | 2AS | 47.12 ± 3.64 |
| 4BS | 7.77 ± 0.79 | 5BL | 20.52 ± 1.29 | 7AS | 15.00 ± 2.58 | 5BL | 46.07 ± 4.67 |
| 2AS | 8.03 ± 0.31 | 7BS | 20.48 ± 2.01 | 5BL | 14.33 ± 2.21 | 1BS | 45.8 ± 5.53 |
| 3AS | 7.77 ± 0.34 | 1BL | 19.72 ± 2.21 | 1BS | 14.00 ± 1.36 | 3BL | 45.73 ± 2.69 |
| 3BL | 8.02 ± 0.61 | CS | 19.49 ± 1.07 | 2AS | 13.67 ± 2.08 | 1BL | 45.13 ± 5.74 |
| 7AL | 7.91 ± 0.48 | 3BS | 19.43 ± 1.3 | 7AL | 13.67 ± 2.69 | 6BL | 44.54 ± 1.47 |
| 4AS | 7.49 ± 0.02 | 5AL | 19.39 ± 1.97 | 3AS | 13.33 ± 2.52 | 3AL | 43.11 ± 3.29 |
| 3AL | 7.43 ± 0.05 | 7AL | 19.38 ± 1.37 | 3AL | 13.33 ± 2.52 | 7BS | 42.98 ± 1.75 |
| 6AL | 7.62 ± 0.29 | 7BL | 19.09 ± 0.66 | 4AL | 13.33 ± 2.52 | 4BS | 42.81 ± 1.11 |
| 1BL | 7.62 ± 0.37 | 5AS | 18.95 ± 2.05 | 4BS | 13.33 ± 1.08 | 6AL | 42.03 ± 3.21 |
| 5AS | 7.45 ± 0.1 | 6BS | 18.84 ± 0.99 | 6BS | 13.33 ± 0.58 | 7BL | 41.47 ± 1.93 |
| 5BL | 7.7 ± 0.28 | 1BS | 18.83 ± 1 | 5AS | 12.33 ± 1.53 | 5AL | 41.43 ± 5.63 |
| 4BL | 7.22 ± 0.19 | 4AL | 18.82 ± 0.49 | 1BL | 12.00 ± 1.61 | 5BS | 40.74 ± 3.88 |
| 1AL | 6.71 ± 0.26 | 6AS | 18.61 ± 2.76 | 3BL | 12.00 ± 1 | 4AL | 40.49 ± 5.93 |
| 1BS | 7.42 ± 0.55 | 7AS | 18.51 ± 1.86 | 4BL | 12.00 ± 1.61 | CS | 39.42 ± 2.86 |
| 7BL | 6.91 ± 0.7 | 2BL | 17.75 ± 1.41 | 6AS | 11.67 ± 2.06 | 2AL | 38.58 ± 2.73 |
| CS | 7.03 ± 0.6 | 5BS | 17.62 ± 2.04 | 7BS | 11.67 ± 1.53 | 2BL | 38.48 ± 3.84 |
| 7AS | 7.06 ± 0.38 | 2AS | 17.36 ± 1.01 | 6AL | 11.33 ± 0.58 | 4BL | 38.46 ± 2.63 |
| 3BS | 6.62 ± 0.43 | 6AL | 17.26 ± 1.13 | CS | 11.00 ± 1.00 | 6BS | 37.55 ± 3.65 |
| 7BS | 6.7 ± 0.55 | 3BL | 17.19 ± 0.44 | 2AL | 10.67 ± 1.79 | 6AS | 36.36 ± 3.6 |
| 6BL | 7.02 ± 0.16 | 4BL | 17.11 ± 1.08 | 2BS | 10.00 ± 1.00 | 7AS | 36.3 ± 4.86 |
| 1AS | 6.57 ± 0.14 | 1AL | 17.08 ± 0.63 | 7BL | 10.00 ± 1.04 | 2BS | 34.29 ± 3.28 |
| 2BL | 6.62 ± 0.76 | 3AL | 16.78 ± 0.54 | 4AS | 9.67 ± 1.43 | 4AS | 33.97 ± 2.19 |
| 5BS | 6.87 ± 0.32 | 3AS | 16.7 ± 1.74 | 5BS | 9.00 ± 0.73 | 1AL | 32.81 ± 2.76 |
| 2BS | 6.74 ± 0.35 | 2BS | 16.37 ± 1.03 * | 6BL | 9.00 ± 1.65 | 5AS | 31.28 ± 3.04 |
| 6BS | 6.38 ± 0.03 | 1AS | 16.2 ± 0.9 * | 1AL | 6.33 ± 1.53 * | 1AS | 30.57 ± 2.41 * |
| 2AL | 6.04 ± 0.38 | 2AL | 15.71 ± 1.52 * | 1AS | 6.00 ± 1.00 * | 3BS | 30.13 ± 2.75 * |
| CASL | Spike Length | Effective Grains | Effective Tiller | Spikelet Number | TGW |
|---|---|---|---|---|---|
| Spike Length | 1 | ||||
| Effective Grains | 0.54 ** | 1 | |||
| Effective Tiller | 0.41 * | 0.33 * | 1 | ||
| Spikelet Number | 0.23 | 0.17 | 0.19 | 1 | |
| TGW | 0.23 | 0.21 | 0.00 | −0.03 | 1 |
| BLH Background | CS Background | ||||||
|---|---|---|---|---|---|---|---|
| CASL | Plant Height (cm) | Panicle Type | Type of Spike Awn | CASL | Plant Height (cm) | Panicle Type | Type of Spike Awn |
| 5BS | 91.92 ± 2.44 ** | Elliptic-type | Long awn | 2BS | 136.19 ± 3.81 ** | Elliptic-type | No awn |
| 7AL | 91.74 ± 2.83 ** | Elliptic-type | Long awn | 4AL | 126.52 ± 3.48 ** | Elliptic-type | Base short awn Top long awn |
| 1AL | 91.10 ± 2.03 ** | Elliptic-type | Long awn | 7AL | 124.47 ± 3.53 ** | Elliptic-type | No awn |
| 5BL | 90.89 ± 2.50 ** | Elliptic-type | Long awn | 1BL | 122.67 ± 4.33 ** | Elliptic-type | No awn |
| 1BS | 90.87 ± 2.55 ** | Elliptic-type | Long awn | 4BL | 122.98 ± 4.02 ** | Elliptic-type | No awn |
| 6AS | 90.70 ± 2.93 ** | Elliptic-type | Long awn | 1AS | 122.08 ± 4.92 ** | Rectangle | No awn |
| 1AS | 89.73 ± 2.86 ** | Elliptic-type | Long awn | 1AL | 121.53 ± 3.47 ** | Clavate-type | No awn |
| 7AS | 89.73 ± 2.86 ** | Elliptic-type | Long awn | 1BS | 121.37 ± 3.63 ** | Elliptic-type | No awn |
| 2AS | 87.93 ± 2.4 | Elliptic-type | Long awn | 4AS | 120.13 ± 3.87 | Elliptic-type | Base short awn Top long awn |
| 2BS | 87.87 ± 2.55 | Elliptic-type | Long awn | 3AS | 117.62 ± 5.38 | Rectangle | No awn |
| BLH | 86.26 ± 1.67 | Elliptic-type | Long awn | 7BS | 117.02 ± 2.98 | Elliptic-type | No awn |
| 3AL | 86.07 ± 2.1 | Elliptic-type | Long awn | 6AS | 116.01 ± 3.99 | Elliptic-type | No awn |
| 2BL | 85.74 ± 2.84 | Elliptic-type | Long awn | 5AS | 115.24 ± 4.76 | Elliptic-type | No awn |
| 3BL | 84.52 ± 3.33 | Elliptic-type | Long awn | 7BL | 114.04 ± 3.96 | Elliptic-type | No awn |
| 3AS | 84.13 ± 1.95 | Elliptic-type | Long awn | 5AL | 113.82 ± 4.18 | cone model | No awn |
| 5AS | 84.02 ± 2.2 | Elliptic-type | Long awn | 6BS | 113.13 ± 3.87 | Elliptic-type | Base short awn Top long awn |
| 3BS | 83.96 ± 2.33 | Elliptic-type | Long awn | CS | 112.61 ± 2.39 | Elliptic-type | No awn |
| 4BL | 83.87 ± 2.54 | Elliptic-type | Long awn | 2AS | 111.97 ± 3.03 | Elliptic-type | No awn |
| 4BS | 82.12 ± 1.98 ** | Elliptic-type | Long awn | 4BS | 111.82 ± 3.18 | Elliptic-type | No awn |
| 4AL | 81.92 ± 2.42 ** | Elliptic-type | Long awn | 5BL | 110.59 ± 4.41 | Elliptic-type | No awn |
| 7BS | 81.71 ± 2.9 ** | Elliptic-type | Long awn | 6AL | 109.83 ± 5.17 | Elliptic-type | No awn |
| 6BL | 80.26 ± 1.67 ** | Elliptic-type | Long awn | 3AL | 109.16 ± 5.84 | Elliptic-type | No awn |
| 6AL | 80.02 ± 2.2 ** | Elliptic-type | Long awn | 6BL | 109.25 ± 3.75 | Elliptic-type | Base short awn Top long awn |
| 1BL | 78.03 ± 2.18 ** | Elliptic-type | Long awn | 3BS | 108.46 ± 4.54 | Rectangle | No awn |
| 7BL | 76.87 ± 2.55 ** | Elliptic-type | Long awn | 5BS | 108.74 ± 3.26 | Elliptic-type | No awn |
| TTD | 73.46 ± 1.54 ** | Spindle-type | Long awn | 3BL | 105.08 ± 2.92 | Elliptic-type | No awn |
| 7AS | 103.69 ± 3.31 | Elliptic-type | No awn | ||||
| 2AL | 102.99 ± 4.01 | Clavate-type | No awn | ||||
| 2BL | 101.37 ± 3.63 | Elliptic-type | No awn | ||||
| TTD | 73.46 ± 1.54 ** | Spindle-type | Long awn | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Wei, X.; Jiang, Y.; Hu, X.; Rong, J. Identification of Elite Agronomic Traits Using Chromosome Arm Substitution Lines of Triticum dicoccoides in the Background of Common Wheat. Agronomy 2025, 15, 752. https://doi.org/10.3390/agronomy15030752
Zhao Y, Wei X, Jiang Y, Hu X, Rong J. Identification of Elite Agronomic Traits Using Chromosome Arm Substitution Lines of Triticum dicoccoides in the Background of Common Wheat. Agronomy. 2025; 15(3):752. https://doi.org/10.3390/agronomy15030752
Chicago/Turabian StyleZhao, Yanhao, Xiaofen Wei, Yurong Jiang, Xin Hu, and Junkang Rong. 2025. "Identification of Elite Agronomic Traits Using Chromosome Arm Substitution Lines of Triticum dicoccoides in the Background of Common Wheat" Agronomy 15, no. 3: 752. https://doi.org/10.3390/agronomy15030752
APA StyleZhao, Y., Wei, X., Jiang, Y., Hu, X., & Rong, J. (2025). Identification of Elite Agronomic Traits Using Chromosome Arm Substitution Lines of Triticum dicoccoides in the Background of Common Wheat. Agronomy, 15(3), 752. https://doi.org/10.3390/agronomy15030752






