A Corn Point Cloud Stem-Leaf Segmentation Method Based on Octree Voxelization and Region Growing
Abstract
1. Introduction
2. Related Works
3. Materials and Methods
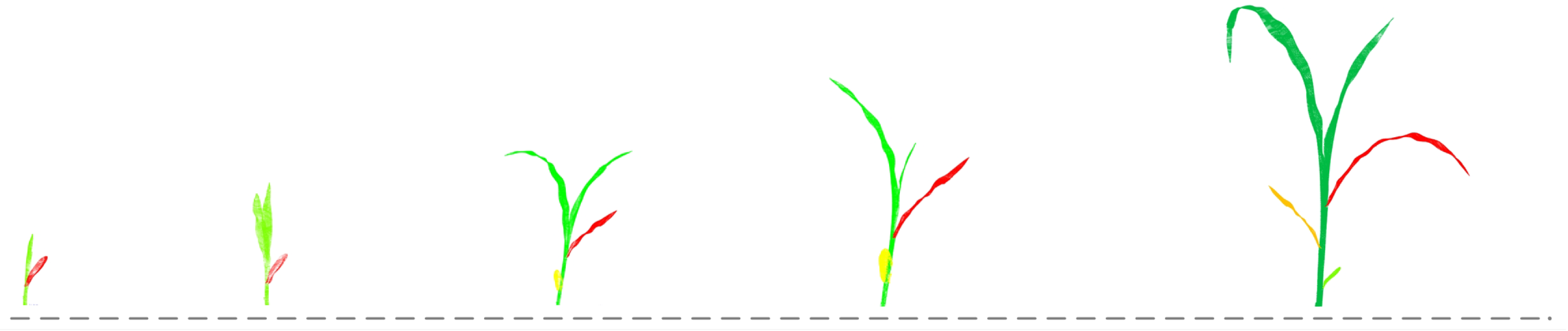
3.1. Data Preparation
3.2. Point Cloud Sampling
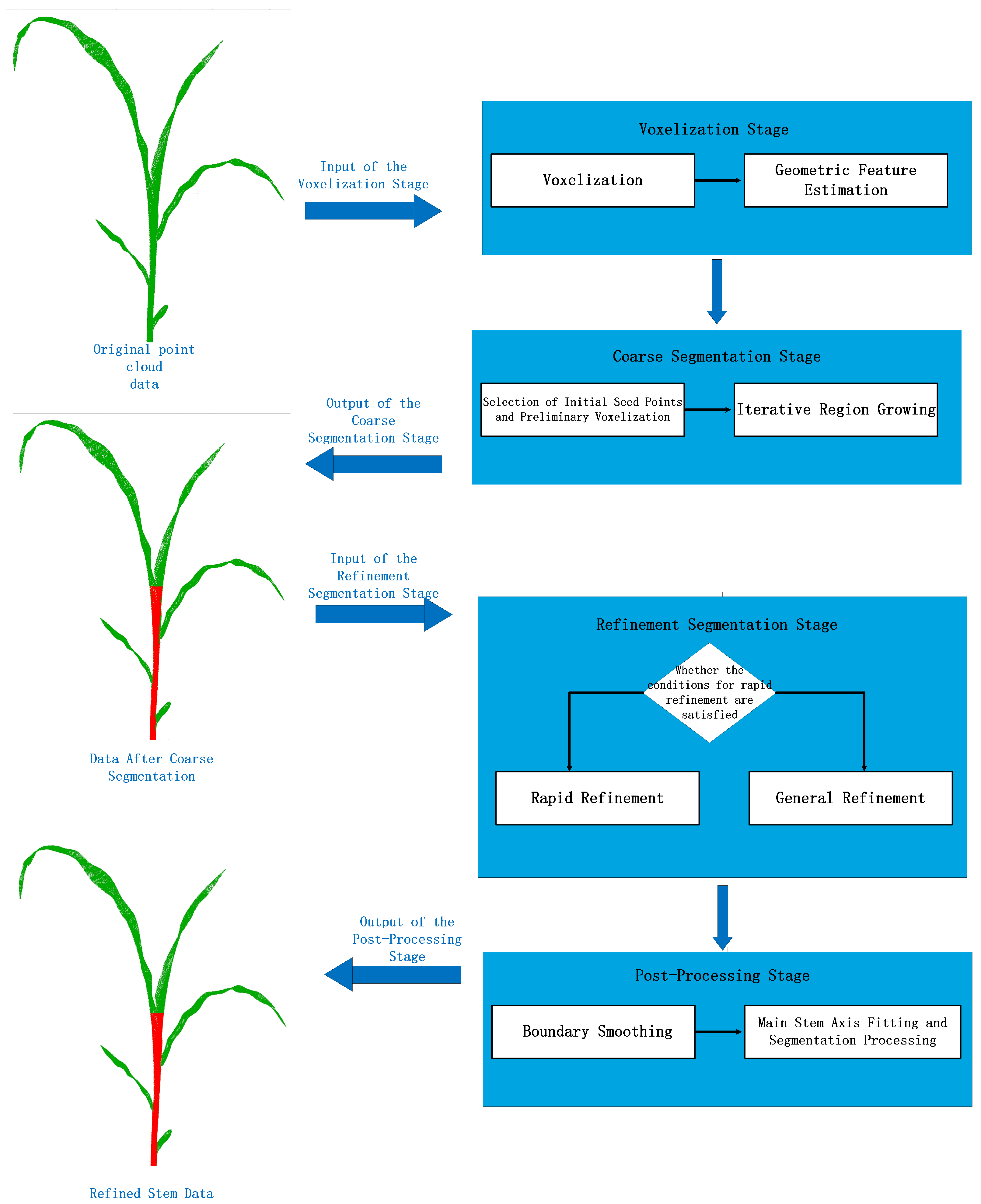
3.3. Stem Segmentation
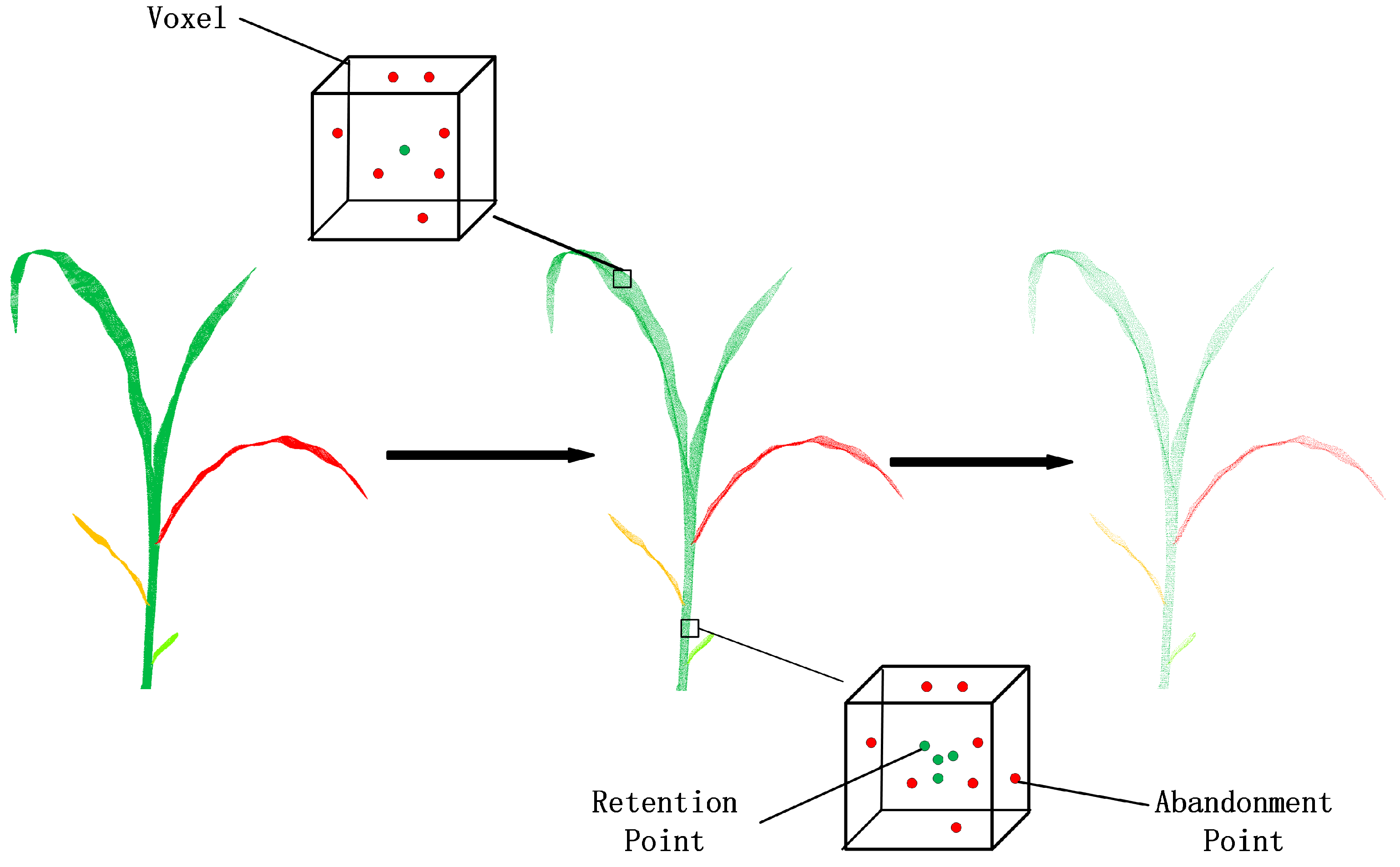
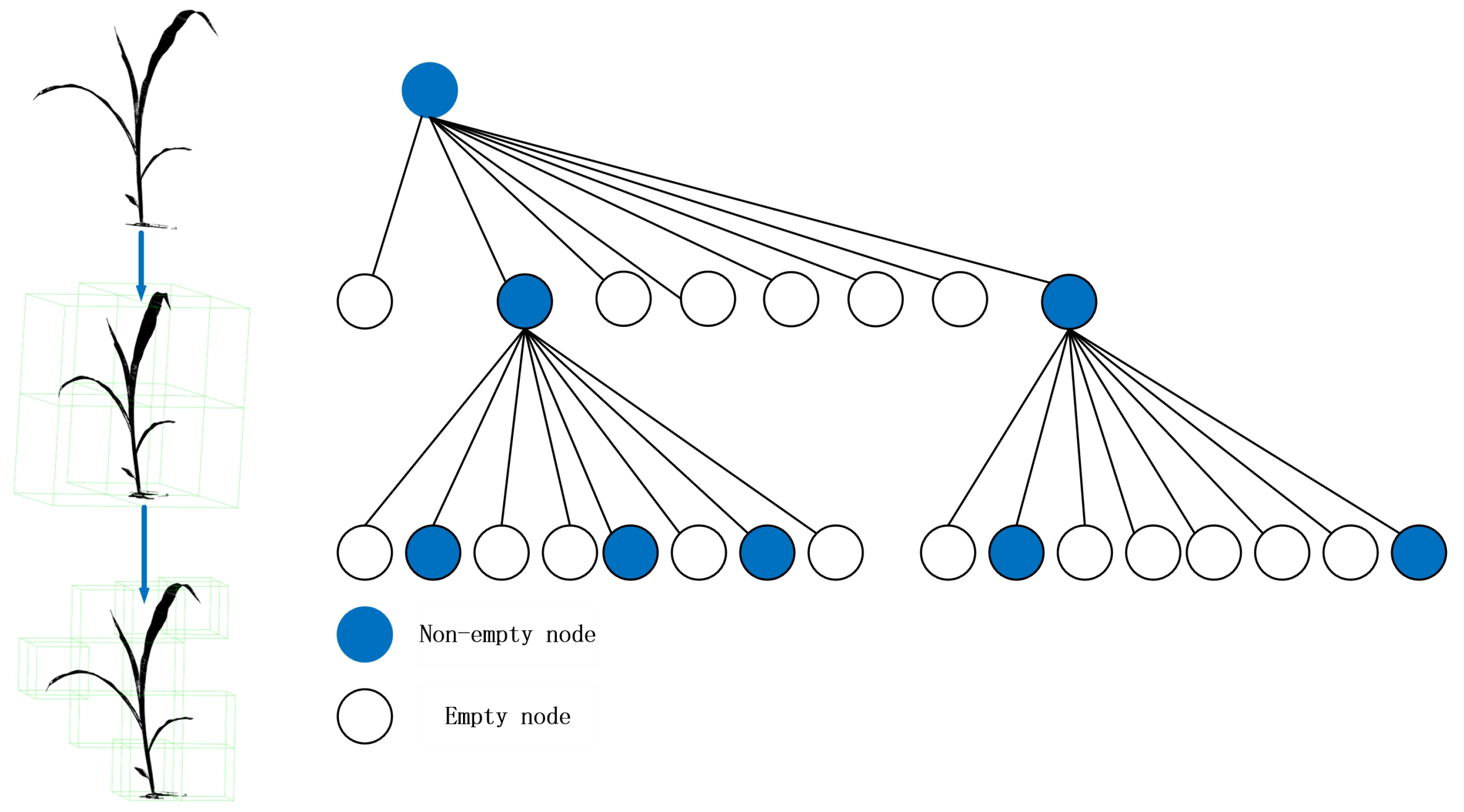
3.3.1. Octree-Based Voxelization
3.3.2. Coarse Segmentation
3.3.3. Refined Segmentation
3.3.4. Post-Processing Stage
3.4. Evaluation Metrics
4. Results
4.1. Parameter Sensitivity Analysis
4.2. Segmentation Results
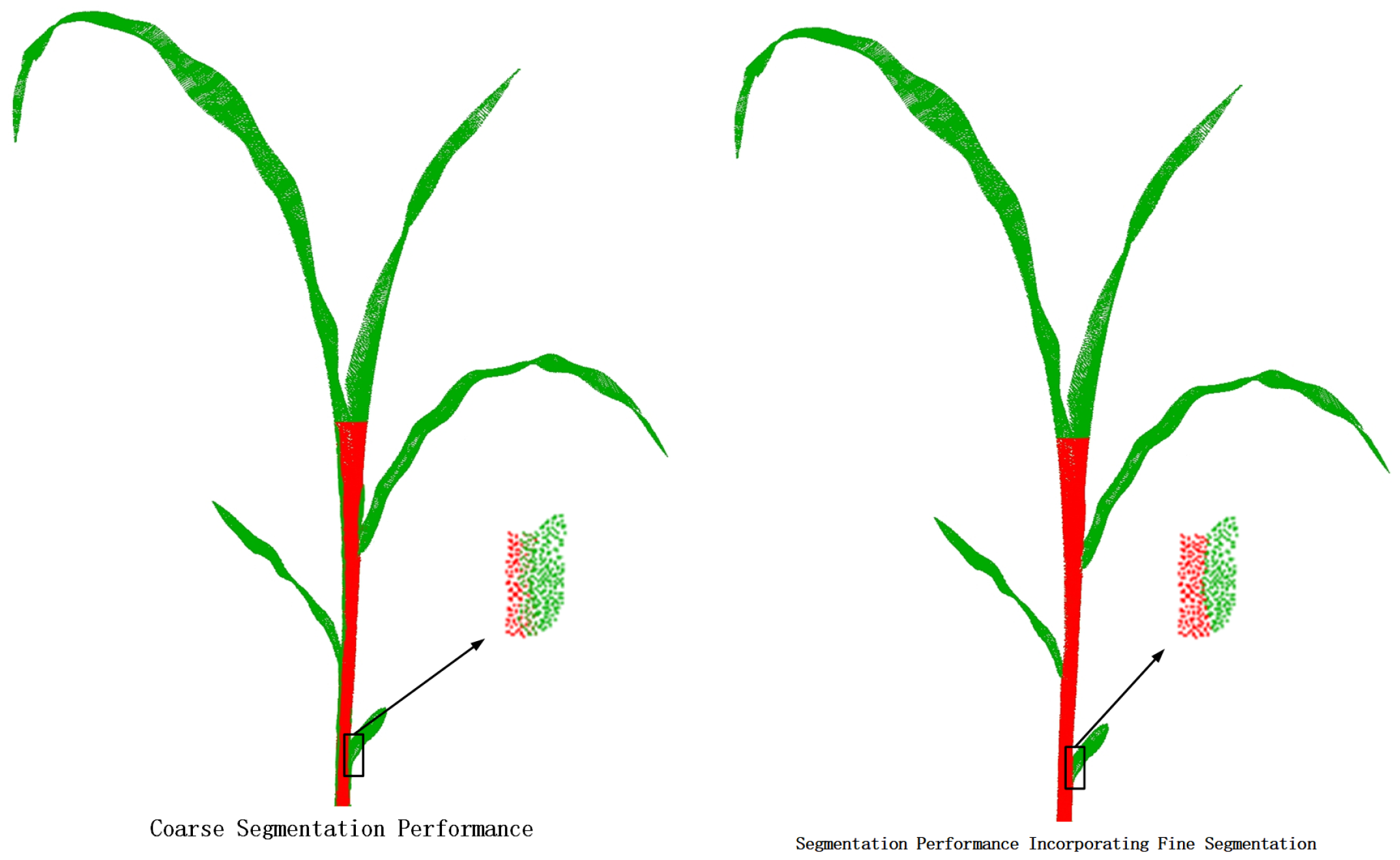
4.3. Ablation Analysis
4.4. Segmentation Efficiency
4.5. Comparison with Other Methods
5. Discussion
5.1. Performance on Other Plants
5.2. Adaptability and Robustness of Algorithm Parameters
5.3. Segmentation Accuracy in Complex Structures
5.4. Efficiency in Processing High-Density Point Clouds
5.5. Impact of Algorithm on Phenotypic Parameter Extraction
5.6. Advantages of Deep Learning Models and Future Application Prospects
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sun, B.; Zain, M.; Zhang, L.; Han, D.; Sun, C. Stem-Leaf Segmentation and Morphological Traits Extraction in Rapeseed Seedlings Using a Three-Dimensional Point Cloud. Agronomy 2025, 15, 276. [Google Scholar] [CrossRef]
- Husin, N.A.; Khairunniza-Bejo, S.; Abdullah, A.F.; Kassim, M.S.M.; Ahmad, D.; Aziz, M.H.A. Classification of basal stem rot disease in oil palm plantations using terrestrial laser scanning data and machine learning. Agronomy 2020, 10, 1624. [Google Scholar] [CrossRef]
- Young, T.J.; Chiranjeevi, S.; Elango, D.; Sarkar, S.; Singh, A.K.; Singh, A.; Ganapathysubramanian, B.; Jubery, T.Z. Soybean Canopy Stress Classification Using 3D Point Cloud Data. Agronomy 2024, 14, 1181. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, N.; Xia, J.; Chen, L.; Chen, S. Plant Height Estimation in Corn Fields Based on Column Space Segmentation Algorithm. Agriculture 2025, 15, 236. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Z. Segment Any Leaf 3D: A Zero-Shot 3D Leaf Instance Segmentation Method Based on Multi-View Images. Sensors 2025, 25, 526. [Google Scholar] [CrossRef]
- Cui, D.; Liu, P.; Liu, Y.; Zhao, Z.; Feng, J. Automated Phenotypic Analysis of Mature Soybean Using Multi-View Stereo 3D Reconstruction and Point Cloud Segmentation. Agriculture 2025, 15, 175. [Google Scholar] [CrossRef]
- Miao, Y.; Wang, L.; Peng, C.; Li, H.; Zhang, M. Single plant segmentation and growth parameters measurement of maize seedling stage based on point cloud intensity. Smart Agric. Technol. 2024, 9, 100665. [Google Scholar] [CrossRef]
- Zhu, Q.; Bai, M.; Yu, M. Maize Phenotypic Parameters Based on the Constrained Region Point Cloud Phenotyping Algorithm as a Developed Method. Agronomy 2024, 14, 2446. [Google Scholar] [CrossRef]
- Yang, X.; Miao, T.; Tian, X.; Wang, D.; Zhao, J.; Lin, L.; Zhu, C.; Yang, T.; Xu, T. Maize stem–leaf segmentation framework based on deformable point clouds. ISPRS J. Photogramm. Remote Sens. 2024, 211, 49–66. [Google Scholar] [CrossRef]
- Shamshiri, R.R.; Navas, E.; Käthner, J.; Höfner, N.; Koch, K.; Dworak, V.; Hameed, I.; Paraforos, D.S.; Fernández, R.; Weltzien, C. Agricultural robotics to revolutionize farming: Requirements and challenges. In Mobile Robots for Digital Farming; CRC Press: Boca Raton, FL, USA, 2025; pp. 107–155. [Google Scholar]
- Islam, M.M.; Himel, G.M.S.; Moazzam, M.G.; Uddin, M.S. Artificial Intelligence-based Rice Variety Classification: A State-of-the-Art Review and Future Directions. Smart Agric. Technol. 2025, 10, 100788. [Google Scholar] [CrossRef]
- Song, H.; Wen, W.; Wu, S.; Guo, X. Comprehensive review on 3D point cloud segmentation in plants. Artif. Intell. Agric. 2025, in press. [Google Scholar] [CrossRef]
- Yao, J.; Gong, Y.; Xia, Z.; Nie, P.; Xu, H.; Zhang, H.; Chen, Y.; Li, X.; Li, Z.; Li, Y. Facility of tomato plant organ segmentation and phenotypic trait extraction via deep learning. Comput. Electron. Agric. 2025, 231, 109957. [Google Scholar] [CrossRef]
- Zhang, L.; Shi, S.; Zain, M.; Sun, B.; Han, D.; Sun, C. Evaluation of Rapeseed Leave Segmentation Accuracy Using Binocular Stereo Vision 3D Point Clouds. Agronomy 2025, 15, 245. [Google Scholar] [CrossRef]
- Liang, X.; Yu, W.; Qin, L.; Wang, J.; Jia, P.; Liu, Q.; Lei, X.; Yang, M. Stem and Leaf Segmentation and Phenotypic Parameter Extraction of Tomato Seedlings Based on 3D Point. Agronomy 2025, 15, 120. [Google Scholar] [CrossRef]
- Navone, A.; Martini, M.; Ambrosio, M.; Ostuni, A.; Angarano, S.; Chiaberge, M. GPS-free autonomous navigation in cluttered tree rows with deep semantic segmentation. Robot. Auton. Syst. 2025, 183, 104854. [Google Scholar] [CrossRef]
- Zhang, W.; Dang, L.M.; Nguyen, L.Q.; Alam, N.; Bui, N.D.; Park, H.Y.; Moon, H. Adapting the Segment Anything Model for Plant Recognition and Automated Phenotypic Parameter Measurement. Horticulturae 2024, 10, 398. [Google Scholar] [CrossRef]
- Bhatti, M.A.; Zeeshan, Z.; Syam, M.S.; Bhatti, U.A.; Khan, A.; Ghadi, Y.Y.; Alsenan, S.; Li, Y.; Asif, M.; Afzal, T. Advanced plant disease segmentation in precision agriculture using optimal dimensionality reduction with fuzzy c-means clustering and deep learning. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2024, 17, 18264–18277. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, W.; Yang, G.; Lei, L.; Han, S.; Xu, W.; Chen, R.; Zhang, C.; Yang, H. Maize ear height and ear–plant height ratio estimation with LiDAR data and vertical leaf area profile. Remote Sens. 2023, 15, 964. [Google Scholar] [CrossRef]
- Miao, Y.; Li, S.; Wang, L.; Li, H.; Qiu, R.; Zhang, M. A single plant segmentation method of maize point cloud based on Euclidean clustering and K-means clustering. Comput. Electron. Agric. 2023, 210, 107951. [Google Scholar] [CrossRef]
- Gao, R.; Cui, S.; Xu, H.; Kong, Q.; Su, Z.; Li, J. A method for obtaining maize phenotypic parameters based on improved QuickShift algorithm. Comput. Electron. Agric. 2023, 214, 108341. [Google Scholar] [CrossRef]
- Wen, W.; Guo, X.; Tao, Y.; Zhao, D.; Teng, M.; Zhu, H.; Dong, C. Point cloud segmentation method of maize ear. J. Syst. Simul. 2020, 29, 3030–3035. [Google Scholar]
- Chao, Z.; Wu, W.; Liu, C.; Zhao, J.; Lin, L.; Tian, X.; Miao, T. Tassel segmentation of maize point cloud based on super voxels clustering and local features. Smart Agric. 2021, 3, 75. [Google Scholar]
- Zhu, C.; Miao, T.; Xu, T.; Yang, T.; Li, N. Stem-leaf segmentation and phenotypic trait extraction of maize shoots from three-dimensional point cloud. arXiv 2020, arXiv:2009.03108. [Google Scholar]
- Miao, T.; Zhu, C.; Xu, T.; Yang, T.; Li, N.; Zhou, Y.; Deng, H. Automatic stem-leaf segmentation of maize shoots using three-dimensional point cloud. Comput. Electron. Agric. 2021, 187, 106310. [Google Scholar] [CrossRef]
- Li, Y.; Liu, J.; Zhang, B.; Wang, Y.; Yao, J.; Zhang, X.; Fan, B.; Li, X.; Hai, Y.; Fan, X. Three-dimensional reconstruction and phenotype measurement of maize seedlings based on multi-view image sequences. Front. Plant Sci. 2022, 13, 974339. [Google Scholar] [CrossRef]
- Wang, D.; Song, Z.; Miao, T.; Zhu, C.; Yang, X.; Yang, T.; Zhou, Y.; Den, H.; Xu, T. DFSP: A fast and automatic distance field-based stem-leaf segmentation pipeline for point cloud of maize shoot. Front. Plant Sci. 2023, 14, 1109314. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, S.; Wen, W.; Lu, X.; Wang, C.; Gou, W.; Li, Y.; Guo, X.; Zhao, C. Three-dimensional branch segmentation and phenotype extraction of maize tassel based on deep learning. Plant Methods 2023, 19, 76. [Google Scholar] [CrossRef]
- Ao, Z.; Wu, F.; Hu, S.; Sun, Y.; Su, Y.; Guo, Q.; Xin, Q. Automatic segmentation of stem and leaf components and individual maize plants in field terrestrial LiDAR data using convolutional neural networks. Crop J. 2022, 10, 1239–1250. [Google Scholar] [CrossRef]
- Herrero-Huerta, M.; Gonzalez-Aguilera, D.; Yang, Y. Structural component phenotypic traits from individual maize skeletonization by UAS-based structure-from-motion photogrammetry. Drones 2023, 7, 108. [Google Scholar] [CrossRef]
- Guo, X.; Sun, Y.; Yang, H. FF-Net: Feature-Fusion-Based Network for Semantic Segmentation of 3D Plant Point Cloud. Plants 2023, 12, 1867. [Google Scholar] [CrossRef]
- Fan, Z.; Sun, N.; Qiu, Q.; Li, T.; Feng, Q.; Zhao, C. In situ measuring stem diameters of maize crops with a high-throughput phenotyping robot. Remote Sens. 2022, 14, 1030. [Google Scholar] [CrossRef]
- Sun, Y.; Guo, X.; Yang, H. Win-Former: Window-Based Transformer for Maize Plant Point Cloud Semantic Segmentation. Agronomy 2023, 13, 2723. [Google Scholar] [CrossRef]
- Luo, L.; Jiang, X.; Yang, Y.; Samy, E.R.A.; Lefsrud, M.; Hoyos-Villegas, V.; Sun, S. Eff-3dpseg: 3d organ-level plant shoot segmentation using annotation-efficient deep learning. Plant Phenomics 2023, 5, 0080. [Google Scholar] [CrossRef]
- Yan, J.; Wang, X. Unsupervised and semi-supervised learning: The next frontier in machine learning for plant systems biology. Plant J. 2022, 111, 1527–1538. [Google Scholar] [CrossRef]
- Schunck, D.; Magistri, F.; Rosu, R.A.; Cornelißen, A.; Chebrolu, N.; Paulus, S.; Léon, J.; Behnke, S.; Stachniss, C.; Kuhlmann, H.; et al. Pheno4D: A spatio-temporal dataset of maize and tomato plant point clouds for phenotyping and advanced plant analysis. PLoS ONE 2021, 16, e0256340. [Google Scholar] [CrossRef]
- Qi, C.R.; Su, H.; Mo, K.; Guibas, L.J. Pointnet: Deep learning on point sets for 3d classification and segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 652–660. [Google Scholar]
- Qi, C.R.; Yi, L.; Su, H.; Guibas, L.J. Pointnet++: Deep hierarchical feature learning on point sets in a metric space. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Phan, A.V.; Le Nguyen, M.; Nguyen, Y.L.H.; Bui, L.T. Dgcnn: A convolutional neural network over large-scale labeled graphs. Neural Netw. 2018, 108, 533–543. [Google Scholar] [CrossRef]
- Qin, Y.; Chen, J.; Jin, L.; Yao, R.; Gong, Z. Task offloading optimization in mobile edge computing based on a deep reinforcement learning algorithm using density clustering and ensemble learning. Sci. Rep. 2025, 15, 211. [Google Scholar] [CrossRef]
- Cui, J.; Tan, F.; Bai, N.; Fu, Y. Improving U-net network for semantic segmentation of corns and weeds during corn seedling stage in field. Front. Plant Sci. 2024, 15, 1344958. [Google Scholar] [CrossRef]
- Luo, Y.; Han, T.; Liu, Y.; Su, J.; Chen, Y.; Li, J.; Wu, Y.; Cai, G. CSFNet: Cross-modal Semantic Focus Network for Sematic Segmentation of Large-Scale Point Clouds. IEEE Trans. Geosci. Remote Sens. 2025, 63, 1–15. [Google Scholar] [CrossRef]
- Yang, H.-C.; Zhou, J.-P.; Zheng, C.; Wu, Z.; Li, Y.; Li, L.-G. PhenologyNet: A fine-grained approach for crop-phenology classification fusing convolutional neural network and phenotypic similarity. Comput. Electron. Agric. 2025, 229, 109728. [Google Scholar] [CrossRef]
- Thapa, S.; Zhu, F.; Walia, H.; Yu, H.; Ge, Y. A novel LiDAR-based instrument for high-throughput, 3D measurement of morphological traits in maize and sorghum. Sensors 2018, 18, 1187. [Google Scholar] [CrossRef] [PubMed]
- Zermas, D.; Morellas, V.; Mulla, D.; Papanikolopoulos, N. Extracting phenotypic characteristics of corn crops through the use of reconstructed 3D models. In Proceedings of the 2018 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), Madrid, Spain, 1–5 October 2008; IEEE: Piscataway, NJ, USA, 2018; pp. 8247–8254. [Google Scholar]
- Hämmerle, M.; Höfle, B. Mobile low-cost 3D camera maize crop height measurements under field conditions. Precis. Agric. 2018, 19, 630–647. [Google Scholar] [CrossRef]
- Vázquez-Arellano, M.; Reiser, D.; Paraforos, D.S.; Garrido-Izard, M.; Burce, M.E.C.; Griepentrog, H.W. 3-D reconstruction of maize plants using a time-of-flight camera. Comput. Electron. Agric. 2018, 145, 235–247. [Google Scholar] [CrossRef]
- Meiyan, S.; Mengyuan, S.; Qizhou, D.; Xiaohong, Y.; Baoguo, L.; Yuntao, M. Estimating the maize above-ground biomass by constructing the tridimensional concept model based on UAV-based digital and multi-spectral images. Field Crop. Res. 2022, 282, 108491. [Google Scholar] [CrossRef]
- Wang, D.; Zhao, M.; Li, Z.; Xu, S.; Wu, X.; Ma, X.; Liu, X. A survey of unmanned aerial vehicles and deep learning in precision agriculture. Eur. J. Agron. 2025, 164, 127477. [Google Scholar] [CrossRef]
- Filippi, P.; Han, S.Y.; Bishop, T.F.A. On crop yield modelling, predicting, and forecasting and addressing the common issues in published studies. Precis. Agric. 2025, 26, 1–19. [Google Scholar] [CrossRef]
- Li, Y.; Gao, G.; Wen, J.; Zhao, N.; Du, G.; Stanny, M. The measurement of agricultural disaster vulnerability in China and implications for land-supported agricultural resilience building. Land Use Policy 2025, 148, 107400. [Google Scholar] [CrossRef]



| Evaluation Metric | Segmentation Result (%) |
|---|---|
| Overall Accuracy (OA) | 98.15 |
| Precision (P) | 90.00 |
| Recall (R) | 91.70 |
| F1 Score | 90.80 |
| IoU | 83.17 |
| Method | OA (%) | P (%) | R (%) | F1 (%) | IoU (%) | Time (s) |
|---|---|---|---|---|---|---|
| Coarse Segmentation Only | 81.35 | 75.41 | 72.71 | 73.50 | 62.89 | 3.5 |
| Coarse Segmentation + Refined Segmentation | 94.45 | 91.91 | 93.34 | 92.15 | 80.30 | 5.1 |
| Growth Stage (Leaf Count) | Number of Input Points | Number of Downsampled Points | Segmentation Time (s) |
|---|---|---|---|
| V3 (3 leaves) | 500,000 | 100,000 | 1.8 |
| V6 (6 leaves) | 1,200,000 | 240,000 | 4.3 |
| V9 (9 leaves) | 2,500,000 | 500,000 | 8.9 |
| VT (Tasseling) | 3,800,000 | 760,000 | 13.6 |
| R1 (Maturity) | 5,000,000 | 1,200,000 | 17.8 |
| Method | OA (%) | P (%) | R (%) | F1 (%) | IoU (%) | Time (s) |
|---|---|---|---|---|---|---|
| PCL Region Growing | 89.67 | 82.31 | 83.50 | 82.90 | 75.80 | 6.2 |
| PointNet | 92.56 | 88.42 | 90.02 | 89.21 | 80.37 | 21.3 |
| PointNet++ | 93.84 | 89.11 | 91.43 | 90.25 | 82.40 | 11.9 |
| DGCNN | 95.67 | 92.79 | 93.85 | 93.32 | 84.81 | 10.7 |
| Our Method | 98.15 | 90.00 | 91.70 | 90.80 | 83.17 | 4.8 |
| Evaluation Metric | Segmentation Result (%) |
|---|---|
| Overall Accuracy (OA) | 95.31 |
| Precision (P) | 87.50 |
| Recall (R) | 89.20 |
| F1 Score | 88.35 |
| IoU | 81.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Q.; Yu, M. A Corn Point Cloud Stem-Leaf Segmentation Method Based on Octree Voxelization and Region Growing. Agronomy 2025, 15, 740. https://doi.org/10.3390/agronomy15030740
Zhu Q, Yu M. A Corn Point Cloud Stem-Leaf Segmentation Method Based on Octree Voxelization and Region Growing. Agronomy. 2025; 15(3):740. https://doi.org/10.3390/agronomy15030740
Chicago/Turabian StyleZhu, Qinzhe, and Ming Yu. 2025. "A Corn Point Cloud Stem-Leaf Segmentation Method Based on Octree Voxelization and Region Growing" Agronomy 15, no. 3: 740. https://doi.org/10.3390/agronomy15030740
APA StyleZhu, Q., & Yu, M. (2025). A Corn Point Cloud Stem-Leaf Segmentation Method Based on Octree Voxelization and Region Growing. Agronomy, 15(3), 740. https://doi.org/10.3390/agronomy15030740





