Engineering Synthetic Microbial Communities: Diversity and Applications in Soil for Plant Resilience
Abstract
1. Introduction
2. The Plant–Rhizosphere–Microbe Nexus: A Crucial Triangle
3. Synthetic Biology Tools and Approaches for Engineering SynComs
3.1. Conventional Approaches to Construct SynComs
3.2. Experimental Techniques to Construct SynComs
3.3. Computational Models and Genomic Databases to Construct SynComs
4. Leveraging Multifunctional Microbes in SynComs
| SynCom Candidates | Functional Traits | Reference |
|---|---|---|
| Arthrobacter sp. | Synthesis of IAA, which directly regulates plant growth and development | [68] |
| Enterobacter sp. | Release of IAA and ammonia, solubilizing phosphate as simple orthophosphate that plants can take up | [68] |
| Brevibacterium sp. | Release of ammonia, ultimately aiding healthy plant growth | [68] |
| Plantibacter sp. | Solubilizing phosphate as simple orthophosphate that plants can take up | [68] |
| Clostridium phytofermentans, Escherichia coli | Nutrient procurement through amino acid, organic acid, sugar and plant polymer catabolic pathways | [69,70] |
| Pseudomonas simiae WCS417r, Ralstonia sp. strain UNC404CL21Col and P. putida KT2440 | Production of phytase to catalyze mineralization | [71] |
| Bacillus spp., Acinetobacter spp., Enterobacter sp., Xanthomonas sp. and Burkholderia sp. | Release of IAA, which directly regulates plant growth and development | [20] |
| PGPR strains | Release of ACC lowers ethylene levels | [72] |
| Azotobacter, Microbacterium, Bacillus, Burkholderia, Enterobacter, Flavbacterium, Erwinia, Rhizobium and Serratia | Solubilization of phosphate enhances plant growth and yield | [73] |
| Azotobacter chroococcum, Enterobacter agglomerans, P. putida, Bradyrhizobium japonicum, Cladosporium herbarum and Rhizobium leguminosarum | Microbial species in potato, tomato, wheat and radish solubilize phosphorus | [74] |
| Bacillus subtilis, Trichoderma harzianum, Trichoderma asperellum and Aspergillus sp. | Foster plant growth and development by producing a variety of enzymes and signaling molecules, including organic acids, proteases, plant hormones, volatile organic compounds (VOCs) and amino acids | [75] |
| Pseudomonas chlororaphis subsp. piscium PS5, Bacillus velezensis BN8.2, and Trichoderma virens T2C1.4. | Delayed Fusarium wilt in Banana disease progress over time, with significant reductions in incidence and severity | [76] |
| 23 bacterial species, including Bacillus spp., Enterobacter spp., Pseudomonas spp., Serratia spp., and others, and 26 fungal species, including Acremonium spp., Aspergillus spp., Botryosporium sp., Cladosporium spp., Gibellulopsis spp., Penicillium spp., Trichoderma spp., Mortierella spp., and Wardomyces spp. | SynComs confer pronounced Fusarium wilt disease resistance to tomato plants compared to the controls during the entire growth period | [63] |
4.1. SynCom in Abiotic Stress Resilience
4.2. SynCom in Soil Health
4.3. SynComs in Biocontrol and Disease Suppression
| Plant | SynCom | Source | Pathogen | Reference |
|---|---|---|---|---|
| Astragalus mongholicus | 2 SynComs 13 bacterial strains 4 bacterial strains | diseased plant roots | Fusarium oxysporum | [113] |
| Zea mays | 6 different SynComs composed of Bacillus strains | roots and leaves | Rhizoctonia solani | [120] |
| Nicotiana attenuata | 6 bacterial strains | rhizospheric soil | Fusarium–Alternaria disease | [121] |
| Lycopersicum esculentum | Bacteria and fungi 4:1 | rhizospheric soil | Fusarium | [63] |
| Solanum lycopersicum | Many SynComs were tested, composed of 205 strains | rhizospheric soil | Fusarium oxysporum f. sp. lycopersici (FOL) | [63] |
| Gossypium spp. | 4 Bacillus strains | rhizospheric soil | Verticillium | [122] |
| Solanum tuberosum | 18 SynComs were tested | healthy leaf tissue | Fusarium solani | [123] |
| Lycopersicum esculentum | Flavobacteriaceae sp. TRM1 | rhizospheric soil | Ralstonia solanacearum | [117] |
| Musa paradisiaca | SynCom 1.0, 1.1 and 1.2 composed of 44, 11 and 03 isolates, respectively | rhizospheric soil | Fusarium oxysporum | [76] |
| Triticum aestivum | 7 SynComs composed of 14 strains in different ratios | rhizospheric soil | Rhizoctonia solani AG8 | [124] |
| Arachis hypogaea | Seed-borne bacterial strains | seed | Aspergillus flavus and Fusarium oxysporum | [125] |
| Arabidopsis thaliana | SynCom of 5-bacterial-strain | rhizospheric soil | Pseudomonas syringae | [52] |
| Beta vulgaris | 2-strain SynCom, i.e., Chitinophaga and Flavobacterium | plant roots | Rhizoctonia solani | [126] |
| Cucumis sativus | 10 strains (Pseudomonas, Bacillus, Stenotrophomonas and Bacillus spp.) | rhizospheric soil | Phytophthora capsici | [127] |
| Soybean | Pseudomonas protegens and Lysobacter enzymogenes via T4ASS | rhizospheric soil | Rhizoctonia solani | [128] |
5. Challenges and Information Gaps—SynComs in Sustainable Agriculture
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2,4-DAPG | 2,4-diacetylphloroglucinol |
| ACCD-1 | Aminocyclopropane-1-Carboxylate deoxygenase |
| AI | Artificial intelligence |
| AMF | Arbuscular mycorrhizal fungi |
| CAT | Catalase |
| Cd | Cadmium |
| Cr | Chromium |
| Cu | Copper |
| DOL | Division of labor |
| EPS | Extracellular polymeric compounds |
| HM | Heavy metal |
| IAA | Indole acetic acid |
| ISR | Induced systemic resistance |
| ML | Machine learning |
| Ni | Nickel |
| Pb | Lead |
| PS | Phosphate solubilization |
| SOD | Superoxide dismutase |
| Zn | Zinc |
References
- AbdelRahman, M.A.E. An Overview of Land Degradation, Desertification and Sustainable Land Management Using GIS and Remote Sensing Applications. Rend. Lincei. Sci. Fis. Nat. 2023, 34, 767–808. [Google Scholar] [CrossRef]
- Kumar, J.; Ramlal, A.; Mallick, D.; Mishra, V. An Overview of Some Biopesticides and Their Importance in Plant Protection for Commercial Acceptance. Plants 2021, 10, 1185. [Google Scholar] [CrossRef]
- Hamonts, K.; Trivedi, P.; Garg, A.; Janitz, C.; Grinyer, J.; Holford, P.; Botha, F.C.; Anderson, I.C.; Singh, B.K. Field Study Reveals Core Plant Microbiota and Relative Importance of Their Drivers. Environ. Microbiol. 2018, 20, 124–140. [Google Scholar] [CrossRef]
- Timm, C.M.; Pelletier, D.A.; Jawdy, S.S.; Gunter, L.E.; Henning, J.A.; Engle, N.; Aufrecht, J.; Gee, E.; Nookaew, I.; Yang, Z. Two Poplar-Associated Bacterial Isolates Induce Additive Favorable Responses in a Constructed Plant-Microbiome System. Front. Plant Sci. 2016, 7, 497. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.; Paulson, J.N.; Zheng, X.; Kolter, R. Simplified and Representative Bacterial Community of Maize Roots. Proc. Natl. Acad. Sci. USA 2017, 114, E2450–E2459. [Google Scholar] [CrossRef] [PubMed]
- Kareem, H.A.; Hao, X.; Shen, X. Collaborative Impact of Bacterial Exometabolites Governing Root Microbiota Formation. Stress Biol. 2023, 3, 38. [Google Scholar] [CrossRef] [PubMed]
- Wallenstein, M.D. Managing and Manipulating the Rhizosphere Microbiome for Plant Health: A Systems Approach. Rhizosphere 2017, 3, 230–232. [Google Scholar]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere Bacteriome Structure and Functions. Nat. Commun. 2022, 13, 836. [Google Scholar] [PubMed]
- Mitter, B.; Pfaffenbichler, N.; Sessitsch, A. Plant–Microbe Partnerships in 2020. Microb. Biotechnol. 2016, 9, 635–640. [Google Scholar] [CrossRef]
- Jain, A.; Chakraborty, J.; Das, S. Underlying Mechanism of Plant–Microbe Crosstalk in Shaping Microbial Ecology of the Rhizosphere. Acta Physiol. Plant. 2020, 42, 8. [Google Scholar] [CrossRef]
- Mukherjee, A.; Singh, B.N.; Kaur, S.; Sharma, M.; de Araújo, A.S.F.; de Araujo Pereira, A.P.; Morya, R.; Puopolo, G.; Melo, V.M.M.; Verma, J.P. Unearthing the Power of Microbes as Plant Microbiome for Sustainable Agriculture. Microbiol. Res. 2024, 286, 127780. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Chen, C.; Xiao, Y.-M.; Chen, K.-Y.; Wang, J.; Zhao, S.; Liu, N.; Li, J.-N.; Zhou, G.-Y. Trophic Relationships between Protists and Bacteria and Fungi Drive the Biogeography of Rhizosphere Soil Microbial Community and Impact Plant Physiological and Ecological Functions. Microbiol. Res. 2024, 280, 127603. [Google Scholar] [CrossRef]
- Qian, X.; Chen, L.; Sui, Y.; Chen, C.; Zhang, W.; Zhou, J.; Dong, W.; Jiang, M.; Xin, F.; Ochsenreither, K. Biotechnological Potential and Applications of Microbial Consortia. Biotechnol. Adv. 2020, 40, 107500. [Google Scholar] [CrossRef] [PubMed]
- Nuccio, E.E.; Starr, E.; Karaoz, U.; Brodie, E.L.; Zhou, J.; Tringe, S.G.; Malmstrom, R.R.; Woyke, T.; Banfield, J.F.; Firestone, M.K. Niche Differentiation Is Spatially and Temporally Regulated in the Rhizosphere. ISME J. 2020, 14, 999–1014. [Google Scholar] [CrossRef]
- Farhat, F.; Tariq, A.; Waseem, M.; Masood, A.; Raja, S.; Ajmal, W.; Iftikhar, I.; Zulfiqar, U.; Maqsood, M.F. Plant Growth Promoting Rhizobacteria (PGPR) Induced Improvements in the Growth, Photosynthesis, Antioxidants, and Nutrient Uptake of Rapeseed (Brassica napus L.). Gesunde Pflanz. 2023, 75, 2075–2088. [Google Scholar] [CrossRef]
- Mataigne, V.; Vannier, N.; Vandenkoornhuyse, P.; Hacquard, S. Multi-Genome Metabolic Modeling Predicts Functional Inter-Dependencies in the Arabidopsis Root Microbiome. Microbiome 2022, 10, 217. [Google Scholar] [CrossRef]
- Vorholt, J.A.; Vogel, C.; Carlström, C.I.; Müller, D.B. Establishing Causality: Opportunities of Synthetic Communities for Plant Microbiome Research. Cell Host Microbe 2017, 22, 142–155. [Google Scholar] [CrossRef]
- van Leeuwen, P.T.; Brul, S.; Zhang, J.; Wortel, M.T. Synthetic Microbial Communities (SynComs) of the Human Gut: Design, Assembly, and Applications. FEMS Microbiol. Rev. 2023, 47, fuad012. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.T. Consortia-Based Microbial Inoculants for Sustaining Agricultural Activities. Appl. Soil Ecol. 2022, 176, 104503. [Google Scholar] [CrossRef]
- Liu, H.; Qiu, Z.; Ye, J.; Verma, J.P.; Li, J.; Singh, B.K. Effective Colonisation by a Bacterial Synthetic Community Promotes Plant Growth and Alters Soil Microbial Community. J. Sustain. Agric. Environ. 2022, 1, 30–42. [Google Scholar] [CrossRef]
- Fields, B.; Friman, V.-P. Microbial Eco-Evolutionary Dynamics in the Plant Rhizosphere. Curr. Opin. Microbiol. 2022, 68, 102153. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Zhu, G.; Guo, H.; Zhang, Y.; Pan, H.; Yong, L.; Ma, H. Influence of Vegetation Coverage and Climate Environment on Soil Organic Carbon in the Qilian Mountains. Sci. Rep. 2019, 9, 17623. [Google Scholar] [CrossRef]
- Wu, J.; Ma, W.; Li, G.; Alhassan, A.-R.M.; Wang, H.; Chen, G. Vegetation Degradation along Water Gradient Leads to Soil Active Organic Carbon Loss in Gahai Wetland. Ecol. Eng. 2020, 145, 105666. [Google Scholar] [CrossRef]
- Yang, R.; Fang, J.; Cao, Q.; Zhao, D.; Dong, J.; Wang, R.; Liu, J. The Content, Composition, and Influencing Factors of Organic Carbon in the Sediments of Two Types of Constructed Wetlands. Environ. Sci. Pollut. Res. 2021, 28, 49206–49219. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Chen, Y.; Sun, L.; Yu, M.; Li, R.; Li, S.; Su, J.; Zhu, B. Linking Rhizosphere Soil Microbial Activity and Plant Resource Acquisition Strategy. J. Ecol. 2023, 111, 875–888. [Google Scholar] [CrossRef]
- Yang, Y.; Qiu, K.; Xie, Y.; Li, X.; Zhang, S.; Liu, W.; Huang, Y.; Cui, L.; Wang, S.; Bao, P. Geographical, Climatic, and Soil Factors Control the Altitudinal Pattern of Rhizosphere Microbial Diversity and Its Driving Effect on Root Zone Soil Multifunctionality in Mountain Ecosystems. Sci. Total Environ. 2023, 904, 166932. [Google Scholar] [CrossRef]
- Brenner, K.; You, L.; Arnold, F.H. Engineering Microbial Consortia: A New Frontier in Synthetic Biology. Trends Biotechnol. 2008, 26, 483–489. [Google Scholar] [CrossRef] [PubMed]
- McCarty, N.S.; Ledesma-Amaro, R. Synthetic Biology Tools to Engineer Microbial Communities for Biotechnology. Trends Biotechnol. 2019, 37, 181–197. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, F.; Li, X.; Li, C.; Zhao, Y.; Gao, Y.; Liu, J. Effects of Plants and Soil Microorganisms on Organic Carbon and the Relationship between Carbon and Nitrogen in Constructed Wetlands. Environ. Sci. Pollut. Res. 2023, 30, 62249–62261. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Liu, Y.; Yin, Y.; Chen, Y.; Jia, S.; Wu, T.; Liao, J.; Jiang, X.; Kareem, H.A.; Li, X. Unveiling the Multifaceted Potential of Pseudomonas Khavaziana Strain SR9: A Promising Biocontrol Agent for Wheat Crown Rot. Microbiol. Spectr. 2024, 12, e00712-24. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Ahmad, M.; Bushra; Hussain, A.; Mumtaz, M.Z.; Abbasi, G.H.; Nazli, F.; Pataczek, L.; Ali, H.M. Mineral-Solubilizing Bacteria-Mediated Enzymatic Regulation and Nutrient Acquisition Benefit Cotton’s (Gossypium hirsutum L.) Vegetative and Reproductive Growth. Microorganisms 2023, 11, 861. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, H.; Liu, H.; Wu, C.; Wan, Y.; Zhu, L.; Yang, J.; Cai, P.; Chen, J.; Ge, T. Combating Wheat Yellow Mosaic Virus through Microbial Interactions and Hormone Pathway Modulations. Microbiome 2024, 12, 200. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; He, W.; Mao, X.; Liao, S.; Wang, Q.; Wang, Z.; Tang, M.; Xu, T.; Chen, H. Arbuscular Mycorrhizal Fungi and Exogenous Ca2+ Application Synergistically Enhance Salt and Alkali Resistance in Perennial Ryegrass through Diverse Adaptive Strategies. Microbiol. Res. 2024, 289, 127906. [Google Scholar] [CrossRef]
- Yue, H.; Sun, X.; Wang, T.; Zhang, A.; Han, D.; Wei, G.; Song, W.; Shu, D. Host Genotype-Specific Rhizosphere Fungus Enhances Drought Resistance in Wheat. Microbiome 2024, 12, 44. [Google Scholar]
- Ding, C.; Zhao, Y.; Zhang, Q.; Lin, Y.; Xue, R.; Chen, C.; Zeng, R.; Chen, D.; Song, Y. Cadmium Transfer between Maize and Soybean Plants via Common Mycorrhizal Networks. Ecotoxicol. Environ. Saf. 2022, 232, 113273. [Google Scholar] [CrossRef] [PubMed]
- Toju, H.; Abe, M.S.; Ishii, C.; Hori, Y.; Fujita, H.; Fukuda, S. Scoring Species for Synthetic Community Design: Network Analyses of Functional Core Microbiomes. Front. Microbiol. 2020, 11, 1361. [Google Scholar] [CrossRef]
- Mahmud, K.; Makaju, S.; Ibrahim, R.; Missaoui, A. Current Progress in Nitrogen Fixing Plants and Microbiome Research. Plants 2020, 9, 97. [Google Scholar] [CrossRef] [PubMed]
- Liu-Xu, L.; González-Hernández, A.I.; Camañes, G.; Vicedo, B.; Scalschi, L.; Llorens, E. Harnessing Green Helpers: Nitrogen-Fixing Bacteria and Other Beneficial Microorganisms in Plant–Microbe Interactions for Sustainable Agriculture. Horticulturae 2024, 10, 621. [Google Scholar] [CrossRef]
- Bender, F.R.; Alves, L.C.; da Silva, J.F.M.; Ribeiro, R.A.; Pauli, G.; Nogueira, M.A.; Hungria, M. Microbiome of Nodules and Roots of Soybean and Common Bean: Searching for Differences Associated with Contrasting Performances in Symbiotic Nitrogen Fixation. Int. J. Mol. Sci. 2022, 23, 12035. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.K.; Habtewold, J.Z. Evaluation of Legume–Rhizobial Symbiotic Interactions beyond Nitrogen Fixation That Help the Host Survival and Diversification in Hostile Environments. Microorganisms 2023, 11, 1454. [Google Scholar] [CrossRef] [PubMed]
- Pathania, P.; Rajta, A.; Singh, P.C.; Bhatia, R. Role of Plant Growth-Promoting Bacteria in Sustainable Agriculture. Biocatal. Agric. Biotechnol. 2020, 30, 101842. [Google Scholar] [CrossRef]
- Johns, N.I.; Blazejewski, T.; Gomes, A.L.C.; Wang, H.H. Principles for Designing Synthetic Microbial Communities. Curr. Opin. Microbiol. 2016, 31, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Herrera Paredes, S.; Gao, T.; Law, T.F.; Finkel, O.M.; Mucyn, T.; Teixeira, P.J.P.L.; Salas González, I.; Feltcher, M.E.; Powers, M.J.; Shank, E.A. Design of Synthetic Bacterial Communities for Predictable Plant Phenotypes. PLoS Biol. 2018, 16, e2003962. [Google Scholar] [CrossRef] [PubMed]
- Kehe, J.; Kulesa, A.; Ortiz, A.; Ackerman, C.M.; Thakku, S.G.; Sellers, D.; Kuehn, S.; Gore, J.; Friedman, J.; Blainey, P.C. Massively Parallel Screening of Synthetic Microbial Communities. Proc. Natl. Acad. Sci. USA 2019, 116, 12804–12809. [Google Scholar] [CrossRef]
- Laurent, P.; Griffiths, B.S.; Silke, L. Microbial Community Resilience across Ecosystems and Multiple Disturbances. Microbiol. Mol. Biol. Rev. 2021, 85, e00026-20. [Google Scholar]
- Ruan, Z.; Chen, K.; Cao, W.; Meng, L.; Yang, B.; Xu, M.; Xing, Y.; Li, P.; Freilich, S.; Chen, C. Engineering Natural Microbiomes toward Enhanced Bioremediation by Microbiome Modeling. Nat. Commun. 2024, 15, 4694. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, X.; Tang, C.; Li, P.; Zhu, R.; Sun, J.; Zhang, Y.; Cui, H.; Ma, J.; Song, X. Engineering Consortia by Polymeric Microbial Swarmbots. Nat. Commun. 2022, 13, 3879. [Google Scholar] [CrossRef] [PubMed]
- De Souza, R.S.C.; Armanhi, J.S.L.; Damasceno, N.D.B.; Imperial, J.; Arruda, P. Genome Sequences of a Plant Beneficial Synthetic Bacterial Community Reveal Genetic Features for Successful Plant Colonization. Front. Microbiol. 2019, 10, 436354. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.; Salas Gonzalez, I.; Mittelviefhaus, M.; Clingenpeel, S.; Herrera Paredes, S.; Miao, J.; Wang, K.; Devescovi, G.; Stillman, K.; Monteiro, F. Genomic Features of Bacterial Adaptation to Plants. Nat. Genet. 2018, 50, 138–150. [Google Scholar] [CrossRef]
- Toju, H.; Peay, K.G.; Yamamichi, M.; Narisawa, K.; Hiruma, K.; Naito, K.; Fukuda, S.; Ushio, M.; Nakaoka, S.; Onoda, Y. Core Microbiomes for Sustainable Agroecosystems. Nat. Plants 2018, 4, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Amyntas, A.; Berti, E.; Gauzens, B.; Albert, G.; Yu, W.; Werner, A.; Eisenhauer, N.; Brose, U. The Role of Niche Complementarity in the Strengthening of the Diversity-Ecosystem Functioning Relationship over Time. Authorea 2023. [Google Scholar]
- Gonçalves, O.S.; Creevey, C.J.; Santana, M.F. Designing a Synthetic Microbial Community through Genome Metabolic Modeling to Enhance Plant–Microbe Interaction. Environ. Microbiome 2023, 18, 81. [Google Scholar] [CrossRef] [PubMed]
- Karkaria, B.D.; Fedorec, A.J.H.; Barnes, C.P. Automated Design of Synthetic Microbial Communities. Nat. Commun. 2021, 12, 672. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.; Tyagi, R.; Sharma, S. Combating Biotic Stresses in Plants by Synthetic Microbial Communities: Principles, Applications and Challenges. J. Appl. Microbiol. 2022, 133, 2742–2759. [Google Scholar] [CrossRef]
- Basso, B.; Antle, J. Digital Agriculture to Design Sustainable Agricultural Systems. Nat. Sustain. 2020, 3, 254–256. [Google Scholar] [CrossRef]
- Espinel, R.; Herrera-Franco, G.; Rivadeneira García, J.L.; Escandón-Panchana, P. Artificial Intelligence in Agricultural Mapping: A Review. Agriculture 2024, 14, 1071. [Google Scholar] [CrossRef]
- MacPherson, J.; Voglhuber-Slavinsky, A.; Olbrisch, M.; Schöbel, P.; Dönitz, E.; Mouratiadou, I.; Helming, K. Future Agricultural Systems and the Role of Digitalization for Achieving Sustainability Goals. A Review. Agron. Sustain. Dev. 2022, 42, 70. [Google Scholar] [CrossRef] [PubMed]
- Kavusi, E.; Shahi Khalaf Ansar, B.; Dehghanian, Z.; Asgari Lajayer, B.; Nobaharan, K.; Ma, Y.; Glick, B.R. Delivery of Beneficial Microbes via Seed Coating for Medicinal and Aromatic Plant Production: A Critical Review. J. Plant Growth Regul. 2023, 42, 575–597. [Google Scholar] [CrossRef]
- Tang, A.; Haruna, A.O.; Majid, N.M.A.; Jalloh, M.B. Potential PGPR Properties of Cellulolytic, Nitrogen-Fixing, Phosphate-Solubilizing Bacteria in Rehabilitated Tropical Forest Soil. Microorganisms 2020, 8, 442. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, L.; Rajendran, S.; Subramanian, N. Metal Stress Impacting Plant Growth in Contaminated Soil Is Alleviated by Microbial Siderophores. Role Microb. Communities Sustain. 2021, 41, 317–332. [Google Scholar]
- Kunal; Pranaw, K.; Kumawat, K.C.; Meena, V.S. Plant Growth-Promoting Rhizobacteria (PGPR) and Plant Hormones: An Approach for Plant Abiotic Stress Management and Sustainable Agriculture. Front. Microbiol. 2023, 14, 1285756. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.J.; Pasche, J.; Silva, H.A.O.; Selten, G.; Savastano, N.; Abreu, L.M.; Bais, H.P.; Garrett, K.A.; Kraisitudomsook, N.; Pieterse, C.M.J. The Use of Synthetic Microbial Communities to Improve Plant Health. Phytopathology® 2023, 113, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, J.; Liu, F.; Liang, J.; Zhao, P.; Tsui, C.K.M.; Cai, L. Cross-Kingdom Synthetic Microbiota Supports Tomato Suppression of Fusarium Wilt Disease. Nat. Commun. 2022, 13, 7890. [Google Scholar] [CrossRef]
- Gou, X.; Kong, W.; Sadowsky, M.J.; Chang, X.; Qiu, L.; Liu, W.; Shao, M.; Wei, X. Global Responses of Soil Bacteria and Fungi to Inoculation with Arbuscular Mycorrhizal Fungi. Catena 2024, 237, 107817. [Google Scholar] [CrossRef]
- Xu, Y.; Yan, Y.; Zhou, T.; Lu, Y.; Yang, X.; Tang, K.; Liu, F. Synergy between Arbuscular Mycorrhizal Fungi and Rhizosphere Bacterial Communities Increases the Utilization of Insoluble Phosphorus and Potassium in the Soil by Maize. J. Agric. Food Chem. 2024, 72, 23631–23642. [Google Scholar] [CrossRef] [PubMed]
- Finkel, O.M.; Castrillo, G.; Paredes, S.H.; González, I.S.; Dangl, J.L. Understanding and Exploiting Plant Beneficial Microbes. Curr. Opin. Plant Biol. 2017, 38, 155–163. [Google Scholar] [CrossRef] [PubMed]
- de Souza, R.S.C.; Okura, V.K.; Armanhi, J.S.L.; Jorrín, B.; Lozano, N.; Da Silva, M.J.; González-Guerrero, M.; de Araújo, L.M.; Verza, N.C.; Bagheri, H.C. Unlocking the Bacterial and Fungal Communities Assemblages of Sugarcane Microbiome. Sci. Rep. 2016, 6, 28774. [Google Scholar] [CrossRef]
- Kaur, S.; Egidi, E.; Qiu, Z.; Macdonald, C.A.; Verma, J.P.; Trivedi, P.; Wang, J.; Liu, H.; Singh, B.K. Synthetic Community Improves Crop Performance and Alters Rhizosphere Microbial Communities. J. Sustain. Agric. Environ. 2022, 1, 118–131. [Google Scholar] [CrossRef]
- Zomorrodi, A.R.; Segrè, D. Synthetic Ecology of Microbes: Mathematical Models and Applications. J. Mol. Biol. 2016, 428, 837–861. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Patel, A.; Hunt, K.A.; Henson, M.A.; Carlson, R.P. Artificial Consortium Demonstrates Emergent Properties of Enhanced Cellulosic-Sugar Degradation and Biofuel Synthesis. NPJ Biofilms Microbiomes 2020, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Shulse, C.N.; Chovatia, M.; Agosto, C.; Wang, G.; Hamilton, M.; Deutsch, S.; Yoshikuni, Y.; Blow, M.J. Engineered Root Bacteria Release Plant-Available Phosphate from Phytate. Appl. Environ. Microbiol. 2019, 85, e01210-19. [Google Scholar] [CrossRef] [PubMed]
- Ratnaningsih, H.R.; Noviana, Z.; Dewi, T.K.; Loekito, S.; Wiyono, S.; Gafur, A.; Antonius, S. IAA and ACC Deaminase Producing-Bacteria Isolated from the Rhizosphere of Pineapple Plants Grown under Different Abiotic and Biotic Stresses. Heliyon 2023, 9, e16306. [Google Scholar] [CrossRef] [PubMed]
- Flores Clavo, R.; Valladolid-Suyón, E.; Reinoza-Farroñan, K.; Asmat Ortega, C.; Riboldi Monteiro, P.H.; Apaza-Castillo, G.A.; Zuñiga-Valdera, G.; Fantinatti Garboggini, F.; Iglesias-Osores, S.; Carreño-Farfán, C.R. Rhizobacterial Isolates from Prosopis limensis Promote the Growth of Raphanus Sativus L. under Salt Stress. Curr. Microbiol. 2023, 80, 269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Fan, C.; Wang, Y.; Xia, Y.; Xiao, W.; Cui, X. Salt-Tolerant and Plant-Growth-Promoting Bacteria Isolated from High-Yield Paddy Soil. Can. J. Microbiol. 2018, 64, 968–978. [Google Scholar] [CrossRef]
- You, T.; Liu, Q.; Chen, M.; Tang, S.; Ou, L.; Li, D. Synthetic Microbial Communities Enhance Pepper Growth and Root Morphology by Regulating Rhizosphere Microbial Communities. Microorganisms 2025, 13, 148. [Google Scholar] [CrossRef]
- Prigigallo, M.I.; Gómez-Lama Cabanás, C.; Mercado-Blanco, J.; Bubici, G. Designing a Synthetic Microbial Community Devoted to Biological Control: The Case Study of Fusarium Wilt of Banana. Front. Microbiol. 2022, 13, 967885. [Google Scholar] [CrossRef] [PubMed]
- Armanhi, J.S.L.; de Souza, R.S.C.; Biazotti, B.B.; Yassitepe, J.E.D.C.T.; Arruda, P. Modulating Drought Stress Response of Maize by a Synthetic Bacterial Community. Front. Microbiol. 2021, 12, 747541. [Google Scholar] [CrossRef] [PubMed]
- Mažylytė, R.; Kailiuvienė, J.; Mažonienė, E.; Orola, L.; Kaziūnienė, J.; Mažylytė, K.; Lastauskienė, E.; Gegeckas, A. The Co-Inoculation Effect on Triticum aestivum Growth with Synthetic Microbial Communities (SynComs) and Their Potential in Agrobiotechnology. Plants 2024, 13, 1716. [Google Scholar] [CrossRef] [PubMed]
- Styer, A.; Pettinga, D.; Caddell, D.F.; Coleman-Derr, D. Improving Rice Drought Tolerance through Host-Mediated Microbiome Selection. bioRxiv 2024, 2002–2024. [Google Scholar]
- Schmitz, L.; Yan, Z.; Schneijderberg, M.; de Roij, M.; Pijnenburg, R.; Zheng, Q.; Franken, C.; Dechesne, A.; Trindade, L.M.; van Velzen, R.; et al. Synthetic Bacterial Community Derived from a Desert Rhizosphere Confers Salt Stress Resilience to Tomato in the Presence of a Soil Microbiome. ISME J. 2022, 16, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Flores-Duarte, N.J.; Navarro-Torre, S.; Mateos-Naranjo, E.; Redondo-Gómez, S.; Pajuelo, E.; Rodríguez-Llorente, I.D. Nodule Synthetic Bacterial Community as Legume Biofertilizer under Abiotic Stress in Estuarine Soils. Plants 2023, 12, 2083. [Google Scholar] [CrossRef]
- Liu, L.; Cheng, L.; Liu, K.; Yu, T.; Liu, Q.; Gong, Z.; Cai, Z.; Liu, J.; Zhao, X.; Nian, H. Transgenic Soybean of GsMYB10 Shapes Rhizosphere Microbes to Promote Resistance to Aluminum (Al) Toxicity. J. Hazard. Mater. 2023, 455, 131621. [Google Scholar] [CrossRef]
- Kumar, A.; Verma, J.P. Does Plant—Microbe Interaction Confer Stress Tolerance in Plants: A Review? Microbiol. Res. 2018, 207, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Jha, Y.; Subramanian, R.B. PGPR Regulate Caspase-like Activity, Programmed Cell Death, and Antioxidant Enzyme Activity in Paddy under Salinity. Physiol. Mol. Biol. Plants 2014, 20, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Sahni, D. Halophilic Microbe Interactions with Plants to Mitigate Salt Stress. Salt Stress. Microbes Plant Interact. Causes Solut. 2019, 1, 249–272. [Google Scholar]
- Yavuz, A.; Erdogan, U.; Turan, M.; Argın, S.; Kocaman, A. Synergistic Strategies for Overcoming Salt Stress in Strawberry Farming: The Use of Organic Fertilizers and Plant Growth Promoting Rhizobacteria (PGPR). Appl. Fruit Sci. 2024, 66, 1787–1797. [Google Scholar] [CrossRef]
- Sheteiwy, M.S.; Ali, D.F.I.; Xiong, Y.-C.; Brestic, M.; Skalicky, M.; Hamoud, Y.A.; Ulhassan, Z.; Shaghaleh, H.; AbdElgawad, H.; Farooq, M.; et al. Physiological and Biochemical Responses of Soybean Plants Inoculated with Arbuscular Mycorrhizal Fungi and Bradyrhizobium under Drought Stress. BMC Plant Biol. 2021, 21, 195. [Google Scholar] [CrossRef]
- Barnawal, D.; Bharti, N.; Maji, D.; Chanotiya, C.S.; Kalra, A. 1-Aminocyclopropane-1-Carboxylic Acid (ACC) Deaminase-Containing Rhizobacteria Protect Ocimum sanctum Plants during Waterlogging Stress via Reduced Ethylene Generation. Plant Physiol. Biochem. 2012, 58, 227–235. [Google Scholar] [CrossRef]
- Sheikh, S.; Amin, A.R.; Asra, M.; Bhagyalakshmi, N. Microbes and Their Role in Alleviation of Abiotic and Biotic Stress Tolerance in Crop Plants. In Microbial Symbionts and Plant Health: Trends and Applications for Changing Climate; Springer: Singapore, 2023; pp. 109–126. [Google Scholar]
- Poveda, J. Trichoderma as Biocontrol Agent against Pests: New Uses for a Mycoparasite. Biol. Control 2021, 159, 104634. [Google Scholar] [CrossRef]
- Lehmann, J.; Bossio, D.A.; Kögel-Knabner, I.; Rillig, M.C. The Concept and Future Prospects of Soil Health. Nat. Rev. Earth Environ. 2020, 1, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Kumar, U.; Nayak, A.K.; Shahid, M.; Gupta, V.V.S.R.; Panneerselvam, P.; Mohanty, S.; Kaviraj, M.; Kumar, A.; Chatterjee, D.; Lal, B. Continuous Application of Inorganic and Organic Fertilizers over 47 Years in Paddy Soil Alters the Bacterial Community Structure and Its Influence on Rice Production. Agric. Ecosyst. Environ. 2018, 262, 65–75. [Google Scholar] [CrossRef]
- Zhaoyu, K.; Ye, J.; Pei, K.; He, Y.; Wang, B.; Huang, S.; Cai, Q.; Liu, Y.; Ge, G.; Wu, L. A Synthetic Bacterial Community Engineered from Miscanthus floridulus Roots Enhances Ammonia Nitrogen Removal in Ionic Rare Earth Mine Tailings. Chemosphere 2024, 367, 143650. [Google Scholar] [CrossRef] [PubMed]
- Ju, W.; Liu, L.; Fang, L.; Cui, Y.; Duan, C.; Wu, H. Impact of Co-Inoculation with Plant-Growth-Promoting Rhizobacteria and Rhizobium on the Biochemical Responses of Alfalfa-Soil System in Copper Contaminated Soil. Ecotoxicol. Environ. Saf. 2019, 167, 218–226. [Google Scholar] [CrossRef]
- Fahmy, M.A.; Salem, S.H.; El-Fattah, H.I.A.; Akl, B.A.; Fayez, M.; Maher, M.; Aioub, A.A.A.; Sitohy, M. Insights into the Role of Hexa-Bacterial Consortium for Bioremediation of Soil Contaminated with Chlorantraniliprole. Environ. Sci. Eur. 2024, 36, 197. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, M.; Zhao, X.; Zhang, X.; Zhou, F. Paracoccus and Achromobacter Bacteria Contribute to Rapid Biodegradation of Imidacloprid in Soils. Ecotoxicol. Environ. Saf. 2021, 225, 112785. [Google Scholar] [CrossRef]
- Sharma, U.C.; Datta, M.; Sharma, V. Soil Microbes and Biofertilizers. In Soils in the Hindu Kush Himalayas: Management for Agricultural Land Use; Springer: Cham, Switzerland; Berlin/Heidelberg, Germany, 2023; pp. 117–144. [Google Scholar]
- Antje, B.; Jeroen, H.T.Z.; Doreen, B.; Erik, L.H.C.; Albert, S.; Roberto, L.; Lourdes, L.; Joseph, N.; Søren, J.S.; Karsten, K.; et al. Importance of Microbial Communities at the Root-Soil Interface for Extracellular Polymeric Substances and Soil Aggregation in Semiarid Grasslands. Soil Biol. Biochem. 2021, 159, 108301. [Google Scholar]
- Hao, X.; Wang, X.; Chen, C.; Liu, R.; Yin, Y.; Yao, J.; Xiao, Z.; Liu, X.; Shen, X.; Liu, X. Synthetic Bacterial Communities Reshape Microbial Communities and Enhance Nutrient Supply in Desertified Land of Northwest China. Appl. Soil Ecol. 2023, 189, 104972. [Google Scholar] [CrossRef]
- Diallo, M.M.; Vural, C.; Cay, H.; Ozdemir, G. Enhanced Biodegradation of Crude Oil in Soil by a Developed Bacterial Consortium and Indigenous Plant Growth Promoting Bacteria. J. Appl. Microbiol. 2021, 130, 1192–1207. [Google Scholar] [CrossRef]
- Cherni, Y.; Botta, C.; Kasmi, M.; Franciosa, I.; Cocolin, L.; Chatti, A.; Trabelsi, I.; Elleuch, L. Mixed Culture of Lactococcus lactis and Kluyveromyces marxianus Isolated from Kefir Grains for Pollutants Load Removal from Jebel Chakir Leachate. Water Environ. Res. 2020, 92, 2041–2048. [Google Scholar] [CrossRef]
- Shah, T.; Khan, Z.; Asad, M.; D’amato, R.; Alsahli, A.A.; Ahmad, P. Synthetic Bacterial Community Derived from Astragalus mongholicus and Plant-Plant Interactions Inhibit Cadmium Uptake by Modulating Gene Expression, Antioxidant System and Carbohydrate Metabolism under Cadmium Contaminated Soil. J. Environ. Chem. Eng. 2024, 12, 111619. [Google Scholar] [CrossRef]
- Zhang, L.; Hang, P.; Zhou, X.; Dai, C.; He, Z.; Jiang, J. Mineralization of the Herbicide Swep by a Two-Strain Consortium and Characterization of a New Amidase for Hydrolyzing Swep. Microb. Cell Fact. 2020, 19, 4. [Google Scholar] [CrossRef]
- Laothamteep, N.; Kawano, H.; Vejarano, F.; Suzuki-Minakuchi, C.; Shintani, M.; Nojiri, H.; Pinyakong, O. Effects of Environmental Factors and Coexisting Substrates on PAH Degradation and Transcriptomic Responses of the Defined Bacterial Consortium OPK. Environ. Pollut. 2021, 277, 116769. [Google Scholar] [CrossRef]
- Mohanty, S.S.; Kumar, A. Enhanced Degradation of Anthraquinone Dyes by Microbial Monoculture and Developed Consortium through the Production of Specific Enzymes. Sci. Rep. 2021, 11, 7678. [Google Scholar] [CrossRef] [PubMed]
- Roszak, M.; Jabłońska, J.; Stachurska, X.; Dubrowska, K.; Kajdanowicz, J.; Gołębiewska, M.; Kiepas-Kokot, A.; Osińska, B.; Augustyniak, A.; Karakulska, J. Development of an Autochthonous Microbial Consortium for Enhanced Bioremediation of PAH-Contaminated Soil. Int. J. Mol. Sci. 2021, 22, 13469. [Google Scholar] [CrossRef]
- Chaudhary, P.; Beniwal, V.; Umar, A.; Kumar, R.; Sharma, P.; Kumar, A.; Al-Hadeethi, Y.; Chhokar, V. In Vitro Microcosm of Co-Cultured Bacteria for the Removal of Hexavalent Cr and Tannic Acid: A Mechanistic Approach to Study the Impact of Operational Parameters. Ecotoxicol. Environ. Saf. 2021, 208, 111484. [Google Scholar] [CrossRef]
- Li, L.; Guo, S.; Sun, Y.; Li, X.; Gao, Y.; Xu, H.; Li, Y. Detoxification effect of single inoculation and co-inoculation of Oudemansiella radicata and Serratia marcescens on Pb and fluoranthene co-contaminated soil. J. Soils Sediments 2019, 19, 3008–3017. [Google Scholar] [CrossRef]
- Gupta, S.; Pathak, B.; Ravi, R.K. Biodegradation of Naphthalene Using Biosurfactant Producing Fusarium proliferatum WC416 Isolated from Refinery Effluent. Appl. Biochem. Biotechnol. 2024, 196, 2549–2565. [Google Scholar] [CrossRef]
- John, E.M.; Sreekumar, J.; Jisha, M.S. Optimization of Chlorpyrifos Degradation by Assembled Bacterial Consortium Using Response Surface Methodology. Soil Sediment Contam. Int. J. 2016, 25, 668–682. [Google Scholar] [CrossRef]
- Jin, L.; Sun, X.; Zhang, X.; Guo, Y.; Shi, H. Co-Metabolic Biodegradation of DBP by Paenibacillus Sp. S-3 and H-2. Curr. Microbiol. 2014, 68, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Lama Cabanás, C.; Wentzien, N.M.; Zorrilla-Fontanesi, Y.; Valverde-Corredor, A.; Fernández-González, A.J.; Fernández-López, M.; Mercado-Blanco, J. Impacts of the Biocontrol Strain Pseudomonas simiae PICF7 on the Banana Holobiont: Alteration of Root Microbial Co-Occurrence Networks and Effect on Host Defense Responses. Front. Microbiol. 2022, 13, 809126. [Google Scholar] [CrossRef]
- Li, Z.; Bai, X.; Jiao, S.; Li, Y.; Li, P.; Yang, Y.; Zhang, H.; Wei, G. A Simplified Synthetic Community Rescues Astragalus mongholicus from Root Rot Disease by Activating Plant-Induced Systemic Resistance. Microbiome 2021, 9, 1–20. [Google Scholar] [CrossRef]
- Ke, J.; Wang, B.; Yoshikuni, Y. Microbiome Engineering: Synthetic Biology of Plant-Associated Microbiomes in Sustainable Agriculture. Trends Biotechnol. 2021, 39, 244–261. [Google Scholar] [CrossRef]
- Khan, R.S.; Iqbal, A.; Bibi, A.; Khalil, I.; Ul Islam, Z.; Jan, F.; Khalid, A.; Abdalla, A.N.; Wadood, A. Plant Chitinases: Types, Structural Classification, Antifungal Potential and Transgenic Expression in Plants for Enhanced Disease Resistance. Plant Cell Tissue Organ Cult. 2024, 156, 75. [Google Scholar] [CrossRef]
- Zhou, H.; Dong, K.; Du, Q.; Wei, Q.; Wu, J.; Deng, J.; Wang, F. Biofilm-Forming of Bacillus tequilensis DZY 6715 Enhanced Suppression the Camellia oleifera Anthracnose Caused by Colletotrichum fructicola and Its Mechanism. Sci. Hortic. 2024, 338, 113676. [Google Scholar] [CrossRef]
- Gomaa, E.Z. Microbial Chitinases: Properties, Enhancement and Potential Applications. Protoplasma 2021, 258, 695–710. [Google Scholar] [CrossRef]
- Wei, X.; Xie, B.; Wan, C.; Song, R.; Zhong, W.; Xin, S.; Song, K. Enhancing Soil Health and Plant Growth through Microbial Fertilizers: Mechanisms, Benefits, and Sustainable Agricultural Practices. Agronomy 2024, 14, 609. [Google Scholar] [CrossRef]
- Kwak, M.-J.; Kong, H.G.; Choi, K.; Kwon, S.-K.; Song, J.Y.; Lee, J.; Lee, P.A.; Choi, S.Y.; Seo, M.; Lee, H.J. Rhizosphere Microbiome Structure Alters to Enable Wilt Resistance in Tomato. Nat. Biotechnol. 2018, 36, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.A.M.; Sayyed, R.Z.; Mir, M.I.; Khan, M.Y.; Hameeda, B.; Alkhanani, M.F.; Haque, S.; Mohammad Al Tawaha, A.R.; Poczai, P. Induction of Systemic Resistance in Maize and Antibiofilm Activity of Surfactin from Bacillus velezensis MS20. Front. Microbiol. 2022, 13, 879739. [Google Scholar] [CrossRef]
- Santhanam, R.; Luu, V.T.; Weinhold, A.; Goldberg, J.; Oh, Y.; Baldwin, I.T. Native Root-Associated Bacteria Rescue a Plant from a Sudden-Wilt Disease That Emerged during Continuous Cropping. Proc. Natl. Acad. Sci. USA 2015, 112, E5013–E5020. [Google Scholar] [CrossRef]
- Wu, C.-D.; Fan, Y.-B.; Chen, X.; Cao, J.-W.; Ye, J.-Y.; Feng, M.-L.; Liu, X.-X.; Sun, W.-J.; Liu, R.-N.; Wang, A.-Y. Analysis of Endophytic Bacterial Diversity in Seeds of Different Genotypes of Cotton and the Suppression of Verticillium Wilt Pathogen Infection by a Synthetic Microbial Community. BMC Plant Biol. 2024, 24, 263. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Li, W.; Chen, H.; Meng, Y.; Wu, H.; Wang, J.; Shen, S. Synthetic Microbial Community Members Interact to Metabolize Caproic Acid to Inhibit Potato Dry Rot Disease. Int. J. Mol. Sci. 2024, 25, 4437. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Hagerty, C.H.; Paulitz, T.C. Synthetic Microbial Consortia Derived from Rhizosphere Soil Protect Wheat against a Soilborne Fungal Pathogen. Front. Microbiol. 2022, 13, 908981. [Google Scholar] [CrossRef]
- Syed, S.; Tollamadugu, N.P.; Lian, B. Aspergillus and Fusarium Control in the Early Stages of Arachis hypogaea (Groundnut Crop) by Plant Growth-Promoting Rhizobacteria (PGPR) Consortium. Microbiol. Res. 2020, 240, 126562. [Google Scholar] [CrossRef] [PubMed]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; De Hollander, M.; Ruiz-Buck, D.; Mendes, L.W.; van Ijcken, W.F.J.; Gomez-Exposito, R.; Elsayed, S.S. Pathogen-Induced Activation of Disease-Suppressive Functions in the Endophytic Root Microbiome. Science 2019, 366, 606–612. [Google Scholar] [CrossRef]
- Islam, S.; Akanda, A.M.; Prova, A.; Islam, M.T.; Hossain, M.M. Isolation and Identification of Plant Growth Promoting Rhizobacteria from Cucumber Rhizosphere and Their Effect on Plant Growth Promotion and Disease Suppression. Front. Microbiol. 2016, 6, 165532. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, Z.; Xu, F.; Yang, Z.; Li, Z.; Shen, D.; Wang, L.; Wu, H.; Li, T.; Yan, Q. Soil Bacterium Manipulates Antifungal Weapons by Sensing Intracellular Type IVA Secretion System Effectors of a Competitor. ISME J. 2023, 17, 2232–2246. [Google Scholar] [CrossRef]
- Jing, J.; Garbeva, P.; Raaijmakers, J.M.; Medema, M.H. Strategies for Tailoring Functional Microbial Synthetic Communities. ISME J. 2024, 18, wrae049. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, C.; Yuan, Z.; Wang, R.; Angelidaki, I.; Zhu, G. Syntrophy Mechanism, Microbial Population, and Process Optimization for Volatile Fatty Acids Metabolism in Anaerobic Digestion. Chem. Eng. J. 2023, 452, 139137. [Google Scholar] [CrossRef]
- Emmenegger, B.; Massoni, J.; Pestalozzi, C.M.; Bortfeld-Miller, M.; Maier, B.A.; Vorholt, J.A. Identifying Microbiota Community Patterns Important for Plant Protection Using Synthetic Communities and Machine Learning. Nat. Commun. 2023, 14, 7983. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.-X.; Guo, X.; Qin, Y.; Garrido-Oter, R.; Schulze-Lefert, P.; Bai, Y. High-Throughput Cultivation and Identification of Bacteria from the Plant Root Microbiota. Nat. Protoc. 2021, 16, 988–1012. [Google Scholar] [CrossRef]
- Han, L. Optimizing Synthetic Microbial Communities for Sustainable Agriculture: Design, Functionality, and Field Performance. Mol. Microbiol. Res. 2024, 14, 31–38. [Google Scholar] [CrossRef]
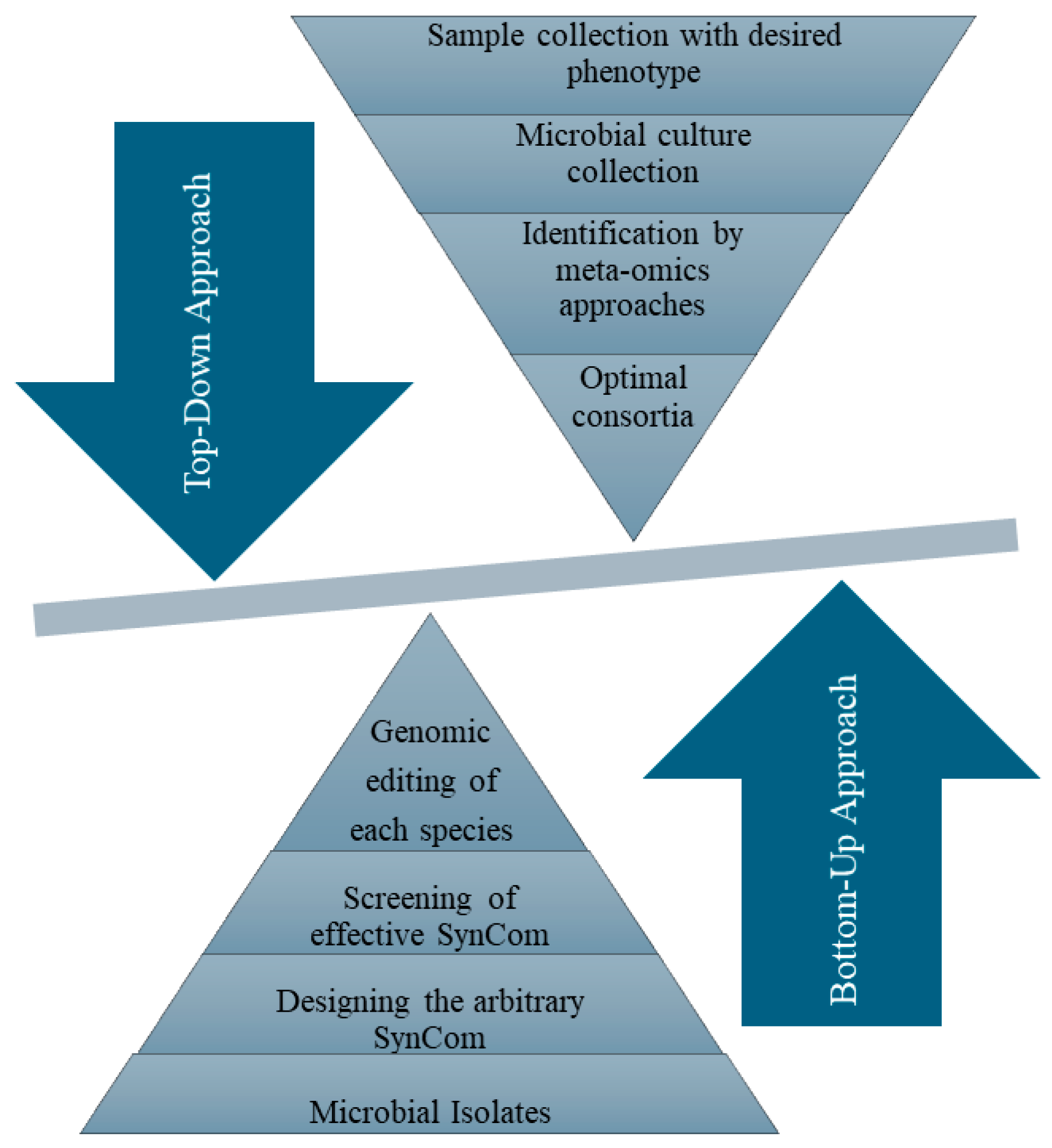
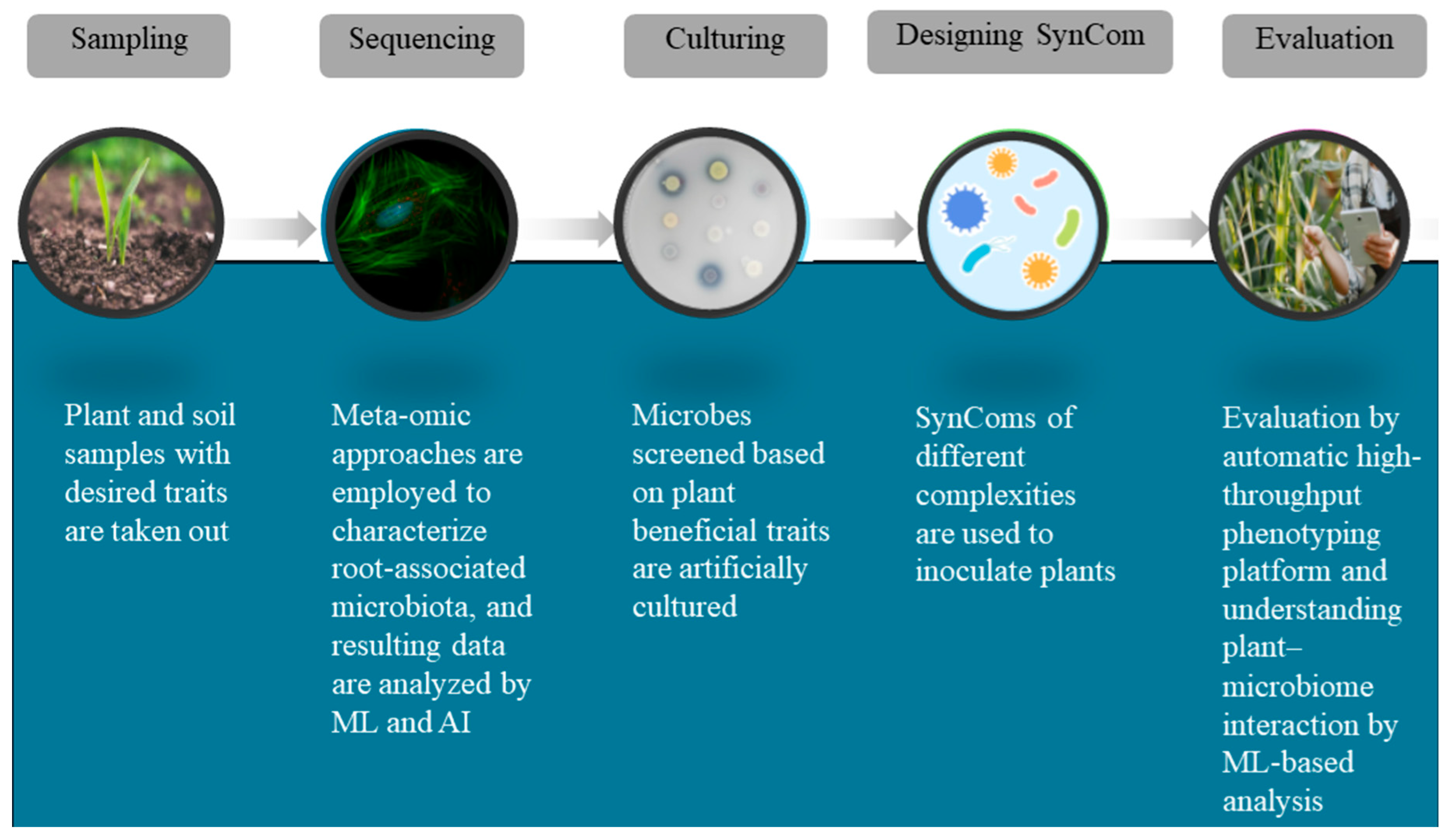
| Microorganism | Plant Species | Region/Source | Possible Interaction | Reference |
|---|---|---|---|---|
| Pseudomonas khavaziana | Arabidopsis thaliana; Triticum aestivum | Wheat | Promote root growth and synthesize plant hormones | [30] |
| Bacillus subtilis; Paenibacillus polymyxa; Bacillus aryabhattai | Gossypium hirsutum | Cotton | Improves plant nutrient absorption | [31] |
| Sphingomonas azotifigens; Rhizobium deserti | Triticum aestivum | Wheat | Antagonism of pathogenic microorganisms | [32] |
| Rhizophagus irregularis | Lolium perenne | Maize | Improves water and nutrient absorption by plants | [33] |
| Mortierella alpine | Triticum aestivum | Wheat | Promotes water absorption and regulates water balance | [34] |
| Arbuscular mycorrhizal fungi (AMF) | Zea mays; Glycine max | Maize | Heavy metal detoxification | [35] |
| Microbial Strains | Mechanism of Action | Plant Resistance | Reference |
|---|---|---|---|
| Deltaproteobacteria, Acidobacteria and Actinobacteria | Upregulate the synthesis of phytohormones involved in plant’s cell division and growth | Boost rice endurance in water-scarce conditions | [83] |
| P. pseudoalcaligenes and B. pumilus | Diminish caspase activity, malondialdehyde content and programmed cell death and increase antioxidant capacity | Salinity endurance of rice | [84] |
| Bacillales, Actinomycetales, Rhizobiales and Oceanospirillales | 1-Aminocyclopropane-1-carboxycarboxylate (ACC) deaminase production under salt stress | Enhanced seed germination and root growth against salt stress in Oryza sativa | [74] |
| PGP bacteria | Associated with plant roots and producing some osmolytes (e.g., carbohydrates) | Alleviate osmotic stress | [85] |
| Kocuria erythromyxa EY43 and Staphylococcus kloosii EY37 | Reduce the absorption of excess ions (sodium and chloride) from saline soils | Improve the growth of strawberry plants | [86] |
| Streptomyces species AMF and Bradyrhizobium | In dried soil, the diffusion pathways become reduced, leading to nutrient deficiency. Microbes must accumulate osmolytes inside their cells to lower the internal solute potential to avoid water loss to their environment | Resistant to drought stress | [87] |
| Pseudomonas putida UW4 | ACC-deaminase-producing bacteria decrease ethylene levels. This enzyme regulates the protein profile, which plays a significant role in nutrient metabolism, defense stress and antioxidant activity | Enhanced growth of basil (Ocimum sanctum) under anoxic conditions | [88] |
| Pseudomonas cedrina, Brevundimonas terrae and Arthrobacter nicotianae | Release of enzymes and osmolyte accumulation | Have the potential to maintain plant health under low temperatures | [89] |
| M. alpina, E. nigrum | Together, these have a negative effect on the lateral roots and root hairs of wheat. | Leads to more sensitivity of wheat to drought stress | [34] |
| Trichoderma sp. | Induce plant systemic resistance against pathogens and pests. Produce multiple volatile compounds that mediate numerous activities | Improve plant growth and tolerance to abiotic stresses | [90] |
| SynCom Composition | Pollutant | Nature | Mechanism/Enzymes | Reference |
|---|---|---|---|---|
| Acinetobacter and Pseudomonas spp. | Hydrocarbons | Organic | The release of hydroxylase and dioxygenase enzymes leads to the degradation of aromatic hydrocarbons | [100] |
| Lactococcus lactis and Kluyveromyces marxianus | Ni, Cu, Cd and Pb | Inorganic | Biosorption and reduction leads to removal of pollutants | [101] |
| Pseudomonas sp., Achromobacter sp., Delftia sp., Enterobacter sp., Advenella sp., Flavobacterium sp., Duganella sp., Stenotrophomonas sp., Ochrobactrum sp., Phyllobacterium sp., Comamonas sp., Oerskovia sp. and Rhizobium sp. | Cd | Inorganic | Increased cytoplasmic invertase, vacuolar invertase, hexokinase, phosphoglucoisomerase, glucose 6-phosphate dehydrogenase, phosphofructokinase; maintained ROS balance; downregulation of HM-related genes | [102] |
| Comamonas sp. and Alicycliphilus sp. | Herbicide swep | Organic | Degradation by amidase | [103] |
| Mycobacterium sp., Novosphingobium pentaromativorans and Bacillus sp. | Pyrene | Organic | Degradation by pyrene-degrading enzymes | [104] |
| B. flexus, Proteus mirabilis and Pseudomonas aeruginosa | 2-Naphthol indanthrene blue RS dye | Organic | Degradation by lignin peroxidase, laccase, tyrosinase and NADH–DCIP reductase | [105] |
| Microbacterium, Pseudomonas, Streptomyces, Arthrobacter and Rhodococcus | Polycyclic aromatic hydrocarbons (PAHs) | Organic | Degradation by dioxygenases and monooxygenases | [106] |
| B. subtilis and B. safensis | Cr, Zn, Pb, Cd and Ni | Inorganic | Adsorption and reduction resulting in HM bioremediation | [107] |
| Oudemansiella radicata and Serratia marcescens | Fluoranthene and Pb | Organic/inorganic | Bioaccumulation/microbial ligninolytic enzymes (laccase and MnP), and soil enzymes (dehydrogenase and acid phosphatase), leading to degradation of pollutants | [108] |
| Funneliformis mosseae and Enterobacter sp. EG16 | Cd | Inorganic | Biosorption, chelation and bioaccumulation | [108] |
| Mycobacterium spp. Novosphingobium pentaromativorans and Bacillus sp. | Benzopyrene | Organic | Fluoranthene dioxygenase and putative 9-fluorenone-1-carboxylic acid dioxygenase | [104] |
| Fusarium proliferatum | Naphthalene | Organic | Dioxygenase | [109] |
| Comamonas sp. and Alicycliphilus sp. | Carbamate | Organic | Amidase | [93] |
| Staphylococcus warneri, P. putida and Stenotrophomonas maltophilia | Chlorpyrifos | Organic | Organophosphorus hydrolase | [110] |
| Paenibacillus spp. | Di-n-butyl phthalate | Organic | 3,4-Phthalate dioxygenase and carboxyesterase | [111] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tariq, A.; Guo, S.; Farhat, F.; Shen, X. Engineering Synthetic Microbial Communities: Diversity and Applications in Soil for Plant Resilience. Agronomy 2025, 15, 513. https://doi.org/10.3390/agronomy15030513
Tariq A, Guo S, Farhat F, Shen X. Engineering Synthetic Microbial Communities: Diversity and Applications in Soil for Plant Resilience. Agronomy. 2025; 15(3):513. https://doi.org/10.3390/agronomy15030513
Chicago/Turabian StyleTariq, Arneeb, Shengzhi Guo, Fozia Farhat, and Xihui Shen. 2025. "Engineering Synthetic Microbial Communities: Diversity and Applications in Soil for Plant Resilience" Agronomy 15, no. 3: 513. https://doi.org/10.3390/agronomy15030513
APA StyleTariq, A., Guo, S., Farhat, F., & Shen, X. (2025). Engineering Synthetic Microbial Communities: Diversity and Applications in Soil for Plant Resilience. Agronomy, 15(3), 513. https://doi.org/10.3390/agronomy15030513









