Identification and Analysis of Melon (Cucumis melo L.) SHMT Gene Family Members and Their Functional Studies on Tolerance to Low-Temperature Stress
Abstract
1. Introduction
2. Preparation of Materials and Analytical Methods
2.1. Plant Materials and Treatments
2.2. Identifying and Analyzing the Physicochemical Properties of CmSHMT Members in Melon
2.3. Chromosome Distribution, Gene Structure, Conserved Motifs, Gene Repetition Events, and Interspecific Homology Analysis of CmSHMT Genes
2.4. The CmSHMT Protein–Protein Interaction Network
2.5. The CmSHMT Gene Phylogenetic
2.6. Prediction of Promoter Cis-Acting Elements of CmSHMTs
2.7. The Analysis of Tissue-Specific Expression of CmSHMT Genes
2.8. The Melon RNA Isolation and Analysis via qRT-PCR
3. Results
3.1. Identification and Physicochemical Property Analysis of SHMT Gene in Melon
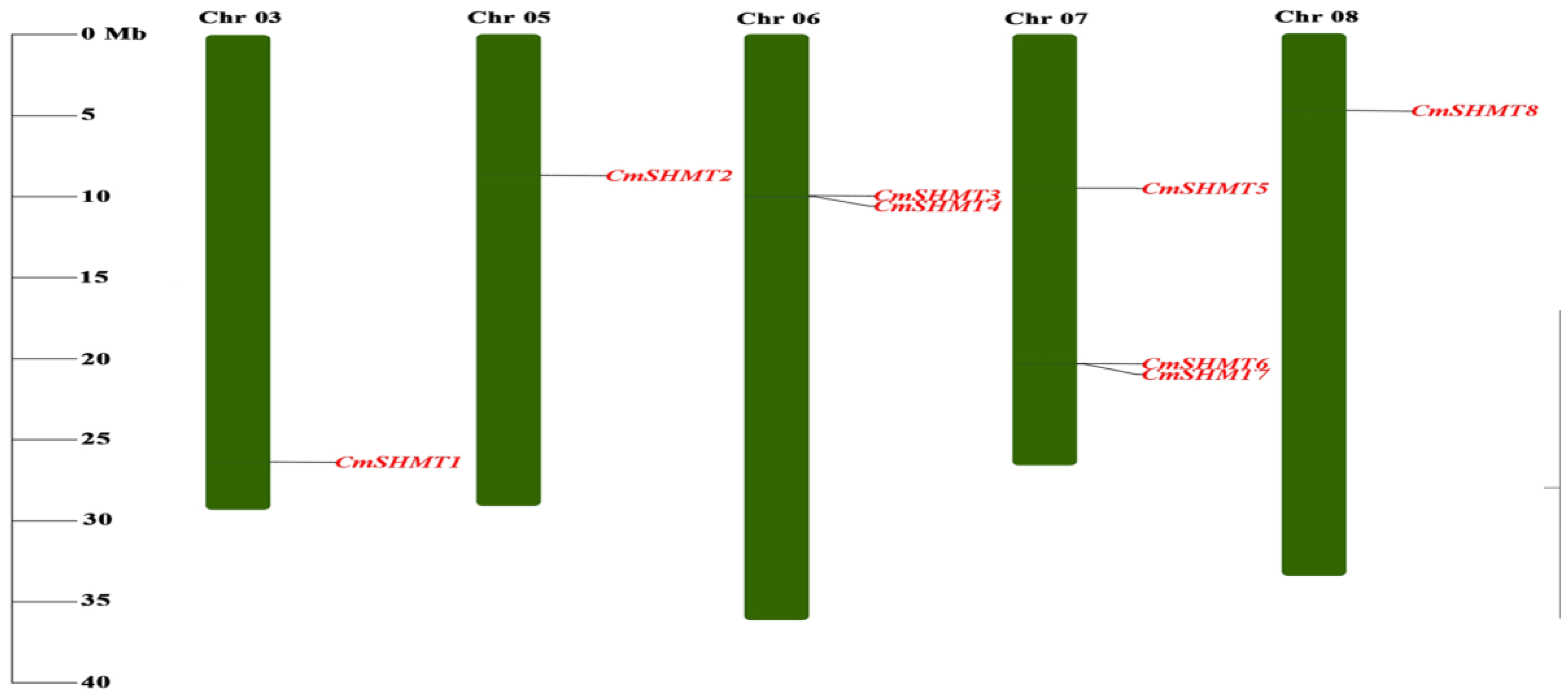
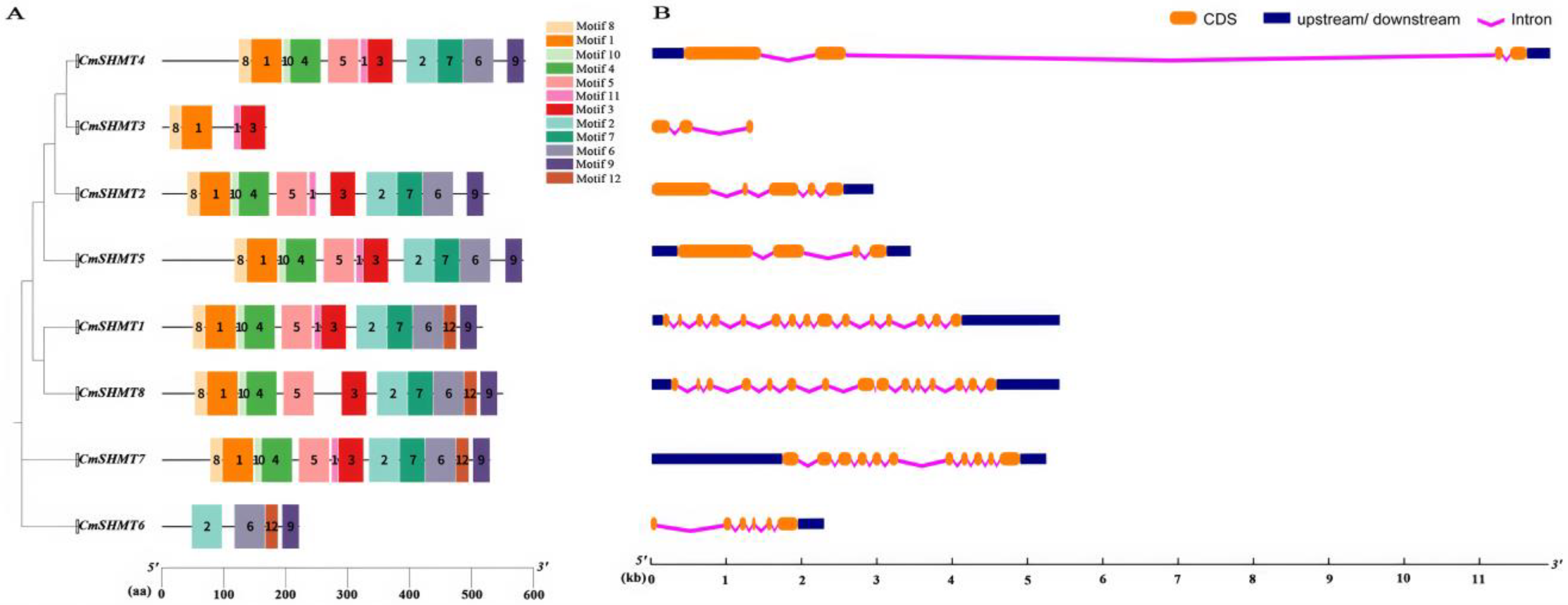
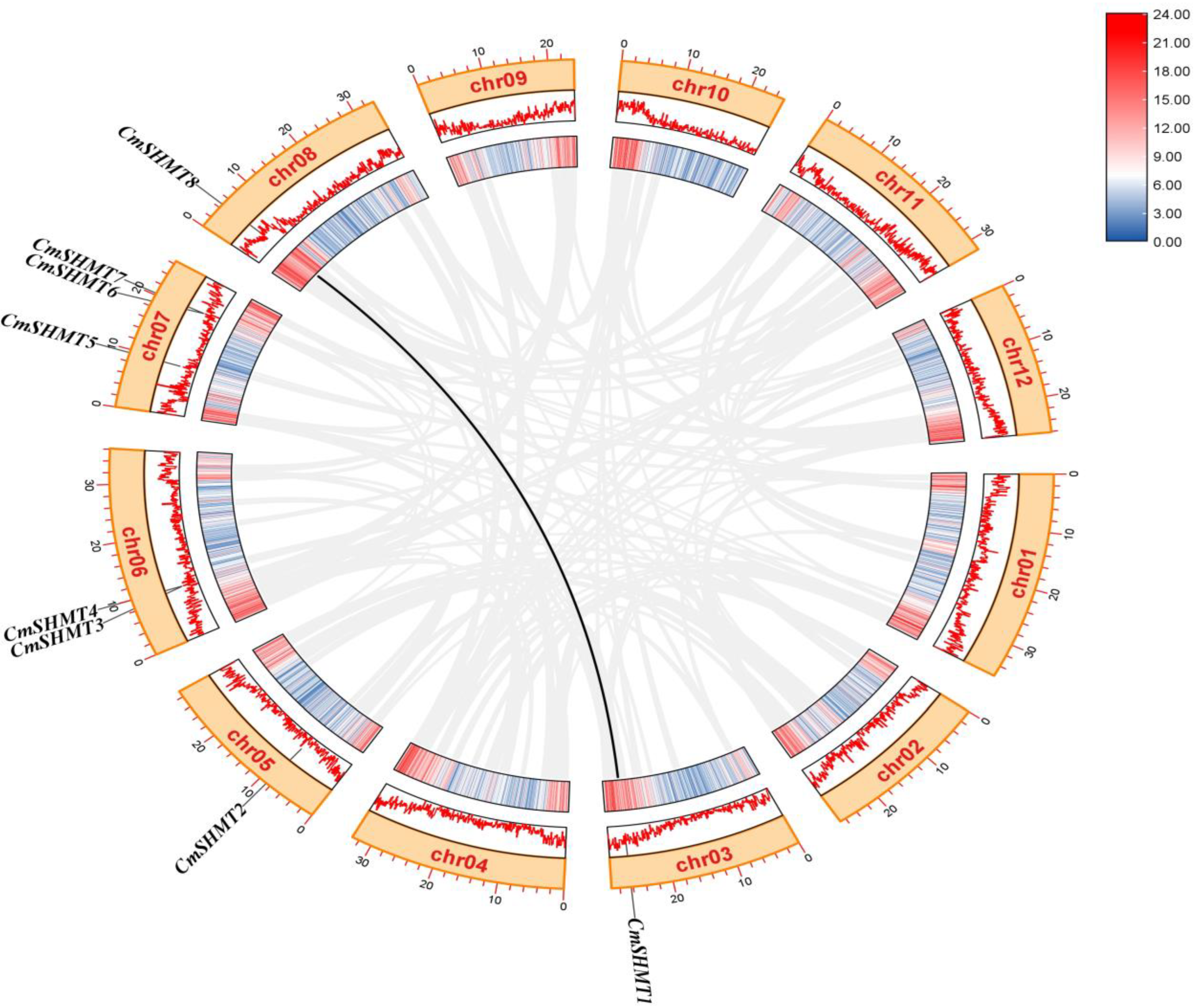
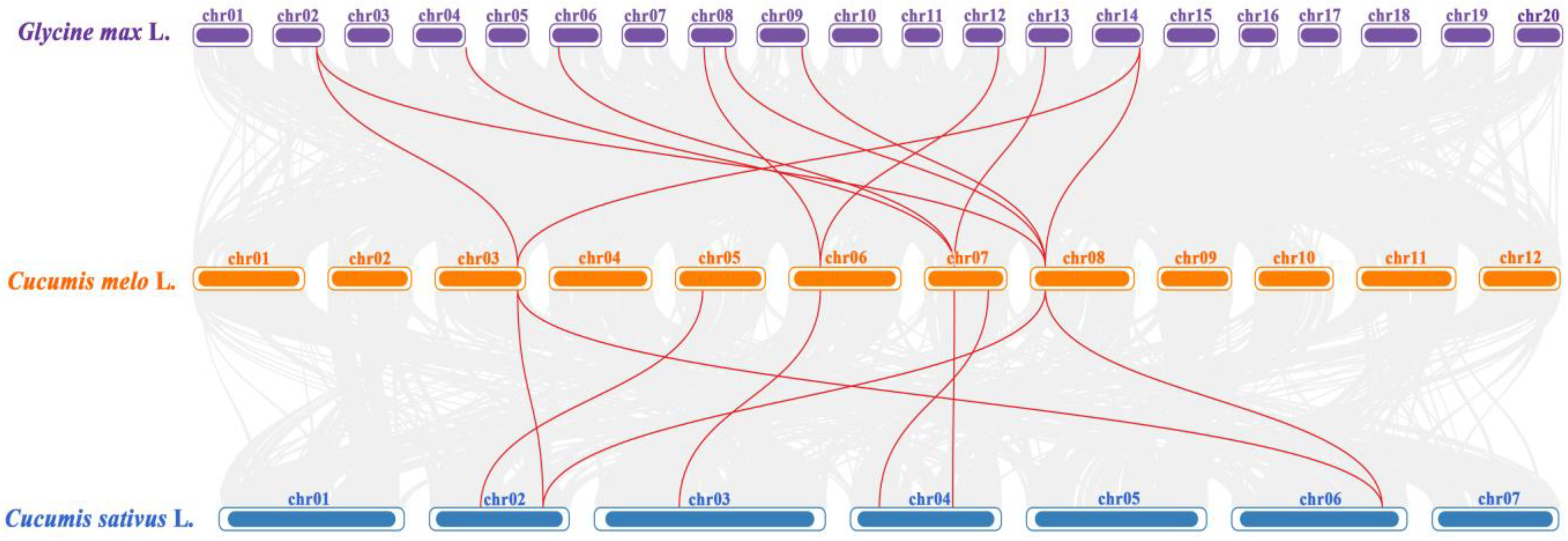
3.2. Chromosome Distribution, Gene Structure, Conserved Motifs, Gene Repetition Events, and Interspecific Homology Analysis of CmSHMTs Gene
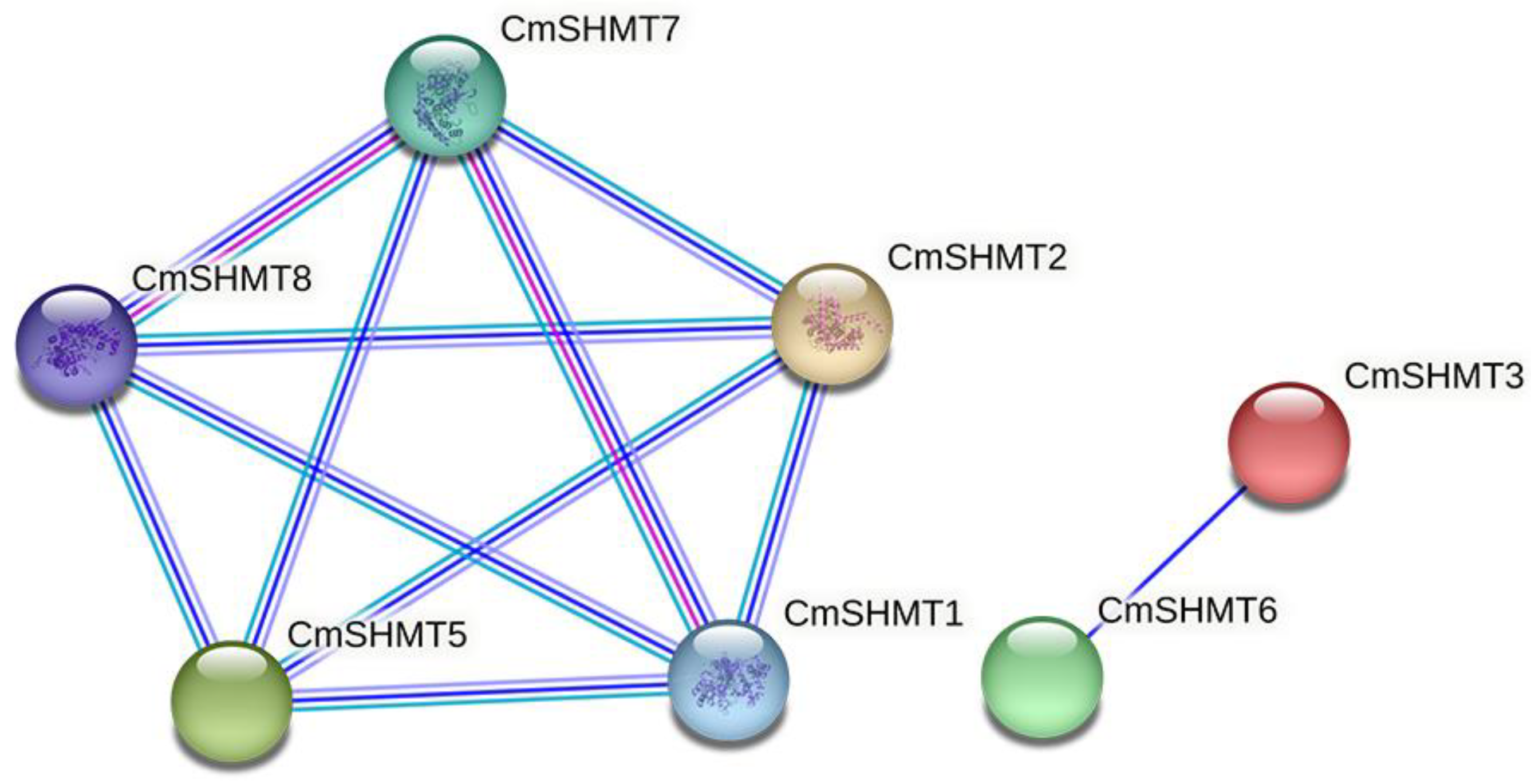
3.3. Network Analysis of Protein–Protein Interactions Involving CmSHMT
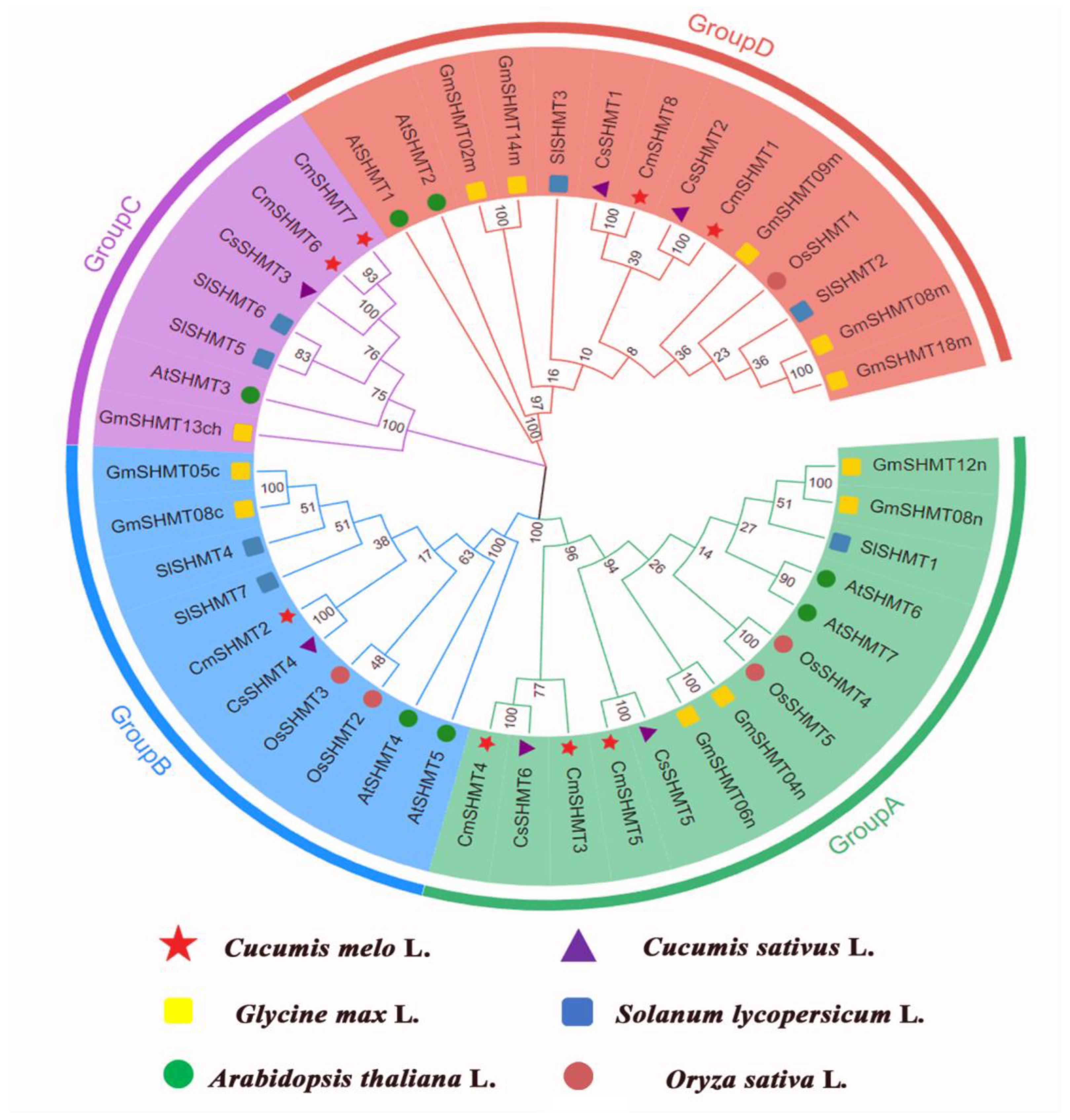
3.4. The Phylogenetic Analysis of CmSHMT Genes
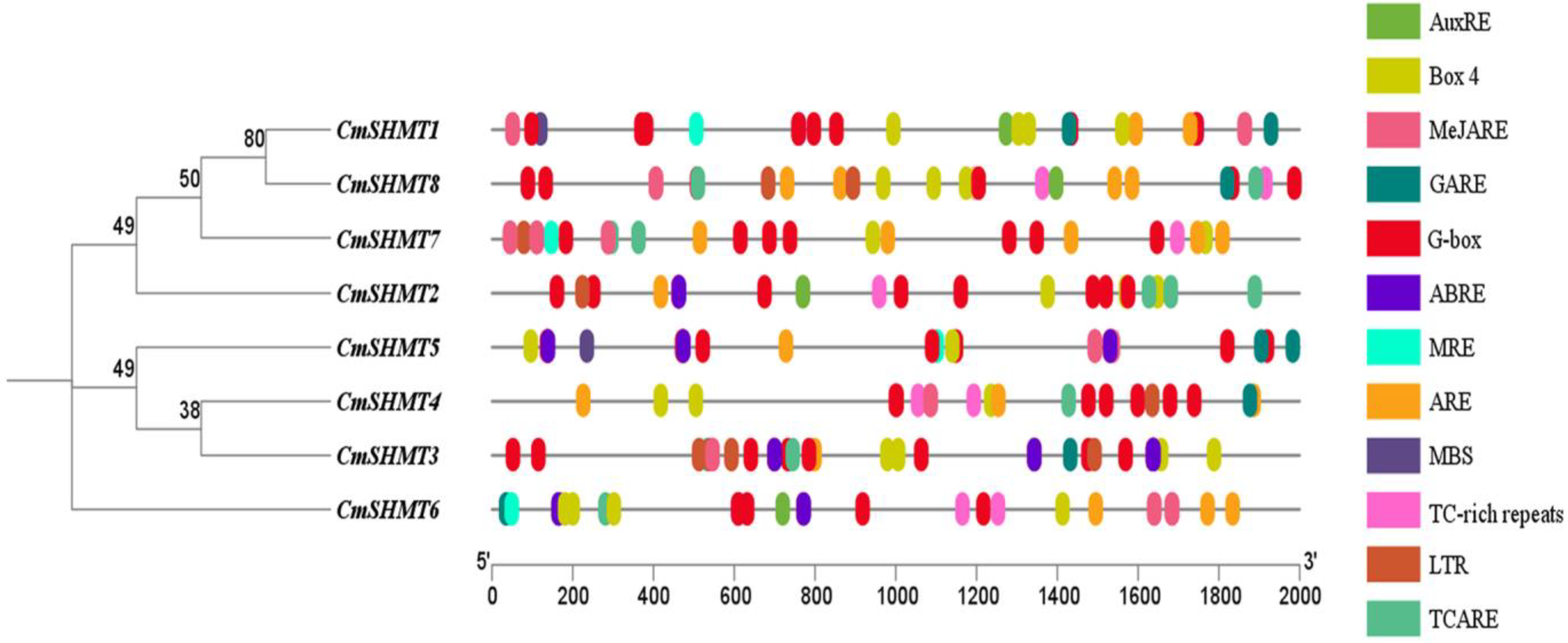
3.5. Prediction and Analysis of Cis-Acting Elements in the Promoter Region of CmSHMTs
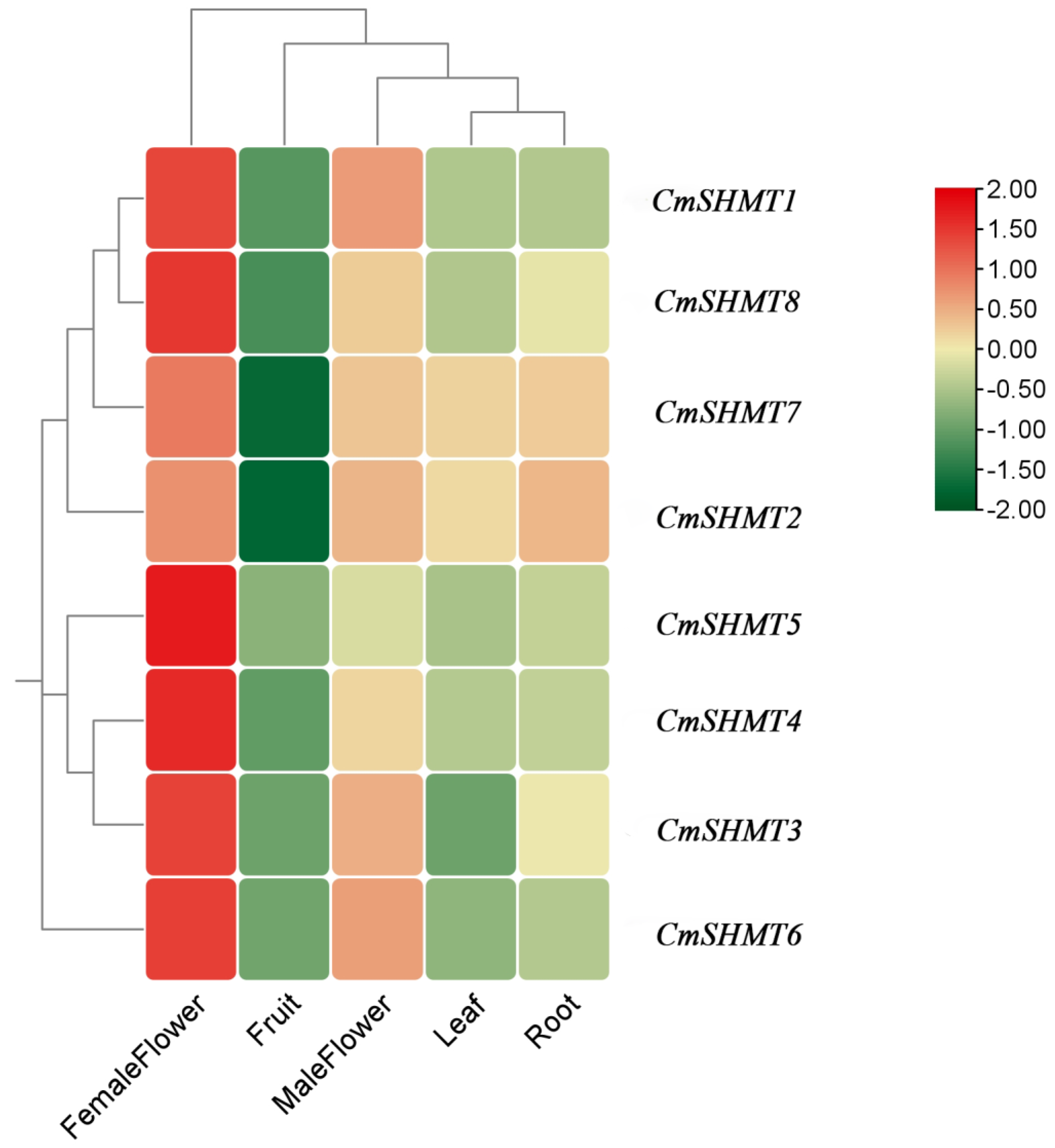
3.6. Analysis of Tissue-Specific Expression of CmSHMT Genes
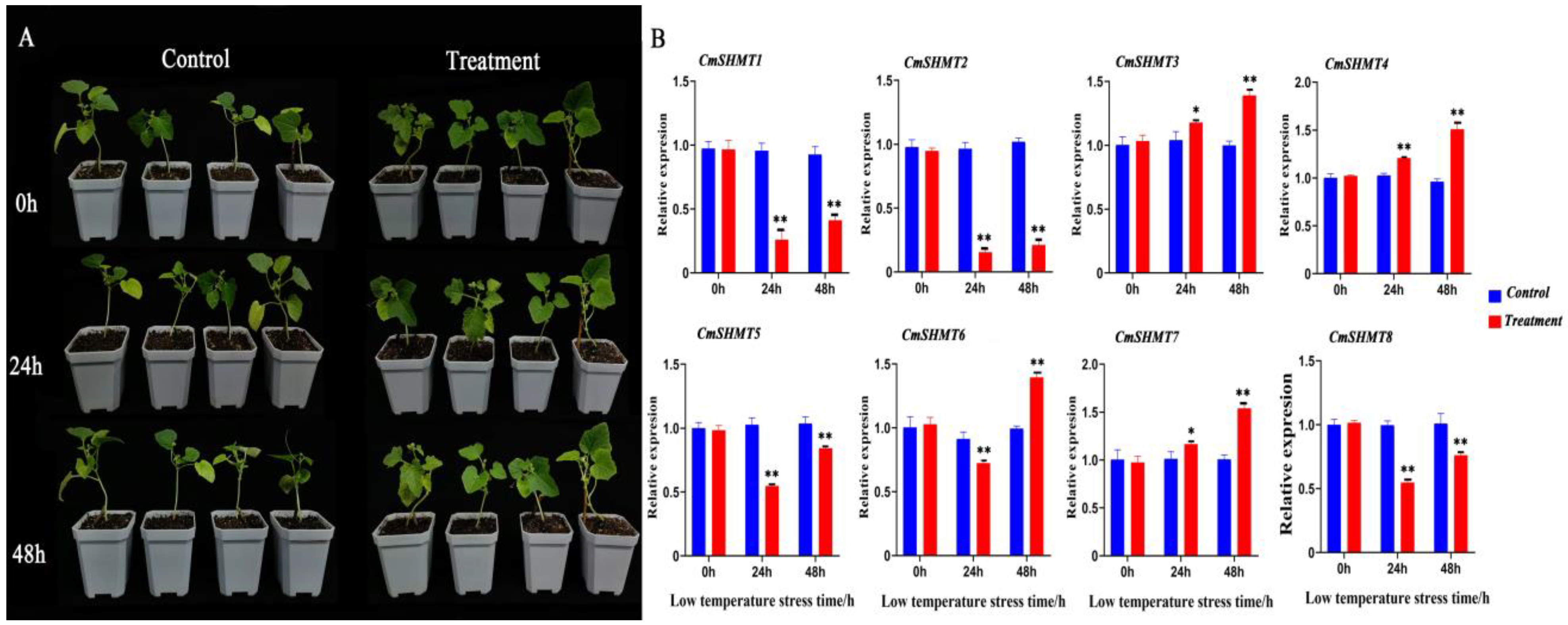
3.7. RNA Extraction and qRT-PCR Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, J.; Li, M.; Bryan, A.C.; Yoo, C.G.; Rottmann, W.; Winkeler, K.A.; Chen, J.G. Overexpression of a serine hydroxymethyltransferase increases biomass production and reduces recalcitrance in the bioenergy crop Populus. Sustain. Energ. Fuels 2019, 3, 195–207. [Google Scholar] [CrossRef]
- Scaletti, E.; Jemth, A.S.; Helleday, T.; Stenmark, P. Structural basis of inhibition of the human serine hydroxymethyltransferase SHMT 2 by antifolate drugs. FEBS Lett. 2019, 593, 1863–1873. [Google Scholar] [CrossRef] [PubMed]
- Pieroth, R.; Paver, S.; Day, S.; Lammersfeld, C. Folate and its impact on cancer risk. Curr. Nutr. Rep. 2018, 7, 70–84. [Google Scholar] [CrossRef] [PubMed]
- García-Cañaveras, J.C.; Lancho, O.; Ducker, G.S.; Ghergurovich, J.M.; Xu, X.; da Silva-Diz, V.; Rabinowitz, J.D. SHMT inhibition is effective and synergizes with methotrexate in T-cell acute lymphoblastic leukemia. Leukemia 2021, 35, 377–388. [Google Scholar] [CrossRef]
- Troesch, B.; Weber, P.; Mohajeri, M.H. Potential Links between Impaired One-Carbon Metabolism Due to Polymorphisms, Inadequate B-Vitamin Status, and the Development of Alzheimer’s Disease. Nutrients 2016, 8, 803. [Google Scholar] [CrossRef]
- Florio, R.; di Salvo, M.L.; Vivoli, M.; Contestabile, R. Serine hydroxymethyltransferase: A model enzyme for mechanistic, structural, and evolutionary studies. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2011, 1814, 1489–1496. [Google Scholar] [CrossRef]
- Waditee-Sirisattha, R.; Sittipol, D.; Tanaka, Y.; Takabe, T. Overexpression of serine hydroxymethyltransferase from halotolerant cyanobacterium in Escherichia coliresults in increased accumulation of choline precursors and enhanced salinity tolerance. FEMS Microbiol. Lett. 2012, 333, 46–53. [Google Scholar] [CrossRef]
- Shen, T.; Rui, B.; Zhou, H.; Zhang, X.; Yi, Y.; Wen, H.; Shi, Y. Metabolic flux ratio analysis and multi-objective optimization revealed a globally conserved and coordinated metabolic response of E. coli to paraquat-induced oxidative stress. Mol. BioSyst. 2013, 9, 121–132. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, K.; Sandoval, F.J.; Santiago, K.; Roje, S. One-carbon metabolism in plants: Characterization of a plastid serine hydroxymethyltransferase. Biochem. J. 2010, 430, 97–105. [Google Scholar] [CrossRef]
- Liu, Z.; Pan, X.; Wang, C.; Yun, F.; Huang, D.; Yao, Y.; Gao, R.; Ye, F.; Liu, X.; Liao, W. Genome-wide identification and expression analysis of serine hydroxymethyltransferase (SHMT) gene family in tomato (Solanum lycopersicum). PeerJ 2022, 10, e12943. [Google Scholar] [CrossRef]
- Ye, J.; Chen, W.; Feng, L.; Liu, G.; Wang, Y.; Li, H.; Zhang, Y. The chaperonin 60 protein SlCpn60α1 modulates photosynthesis and photorespiration in tomato. J. Exp. Bot. 2020, 71, 7224–7240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dai, L.; Liu, Y.; Zhang, Y.; Wang, S. Identifying novel fruit-related genes in Arabidopsis thaliana based on the random walk with restart algorithm. PLoS ONE 2017, 12, e0177017. [Google Scholar] [CrossRef] [PubMed]
- Lakhssassi, N.; Knizia, D.; El Baze, A.; Lakhssassi, A.; Meksem, J.; Meksem, K. Proteomic, Transcriptomic, Mutational, and Functional Assays Reveal the Involvement of Both THF and PLP Sites at the GmSHMT08 in Resistance to Soybean Cyst Nematode. Int. J. Mol. Sci. 2022, 23, 11278. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Shu, X.; Jing, X.; Jiao, C.; Chen, L.; Zhang, J.; Zheng, A. Identification of rice (Oryza sativa L.) genes involved in sheath blight resistance via a genome-wide association study. Plant Biotechnol. J. 2021, 19, 1553–1566. [Google Scholar] [CrossRef]
- Hu, P.; Song, P.; Xu, J.; Wei, Q.; Tao, Y.; Ren, Y.; Li, C. Genome-wide analysis of serine hydroxymethyltransferase genes in triticeae species reveals that TaSHMT3A-1 regulates fusarium head blight resistance in wheat. Front. Plant Sci. 2022, 13, 847087. [Google Scholar] [CrossRef]
- Gao, R.; Chen, L.; Chen, F.; Ma, H. Genome-wide identification of SHMT family genes in alfalfa (Medicago sativa) and its functional analyses under various abiotic stresses. BMC Genom. 2024, 25, 781. [Google Scholar] [CrossRef]
- Gao, R.; Luo, Y.; Pan, X.; Wang, C.; Liao, W. Genome-wide identification of SHMT family genes in cucumber (Cucumis sativus L.) and functional analyses of CsSHMTs in response to hormones and abiotic stresses. 3 Biotech 2022, 12, 305. [Google Scholar] [CrossRef]
- Moreno, J.I.; Martín, R.; Castresana, C. Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J. 2005, 41, 451–463. [Google Scholar] [CrossRef]
- Mishra, P.; Jain, A.; Takabe, T.; Tanaka, Y.; Negi, M.; Singh, N.; Rai, V. Heterologous Expression of Serine Hydroxymethyltransferase-3 from Rice Confers Tolerance to Salinity Stress in E. coli and Arabidopsis. Front. Plant Sci. 2019, 10, 217. [Google Scholar] [CrossRef]
- Zhou, H.; Zhao, J.; Yang, Y.; Chen, C.; Liu, Y.; Jin, X.; Guo, Y. Ubiquitin-specific protease16 modulates salt tolerance in Arabidopsis by regulating Na(+)/H(+) antiport activity and serine hydroxymethyltransferase stability. Plant Cell 2012, 24, 5106–5122. [Google Scholar] [CrossRef]
- Fang, C.; Zhang, P.; Li, L.; Yang, L.; Mu, D.; Yan, X.; Li, Z.; Lin, W. Serine Hydroxymethyltransferase Localised in the Endoplasmic Reticulum Plays a Role in Scavenging H2O2 to Enhance Rice Chilling Tolerance. BMC Plant Biol. 2020, 20, 236. [Google Scholar] [CrossRef] [PubMed]
- Lija, M.; Beevy, S.S. A Review on the diversity of Melon. Plant Sci. Today 2021, 8, 995–1003. [Google Scholar] [CrossRef]
- Duan, X.; Yuan, Y.; Real, N.; Tang, M.; Ren, J.; Wei, J.; Zhang, X. Fine mapping and identification of candidate genes associated with powdery mildew resistance in melon (Cucumis melo L.). Hortic. Res. 2024, 11, uhae222. [Google Scholar] [CrossRef] [PubMed]
- Chikh-Rouhou, H.; Tlili, I.; Ilahy, R.; R’him, T.; Sta-Baba, R. Fruit quality assessment and characterization of melon genotypes. Int. J. Veg. Sci. 2021, 27, 3–19. [Google Scholar] [CrossRef]
- Li, L.; Li, Q.; Chen, B.; Wang, J.; Ding, F.; Wang, P.; Zhang, X.; Hou, J.; Luo, R.; Li, X.; et al. Identification of candidate genes that regulate the trade-off between seedling cold tolerance and fruit quality in melon (Cucumis melo L.). Hortic. Res. 2023, 10, uhad093. [Google Scholar] [CrossRef]
- Weng, J.; Rehman, A.; Li, P.; Chang, L.; Zhang, Y.; Niu, Q. Physiological and Transcriptomic analysis reveals the responses and difference to high temperature and humidity stress in two melon genotypes. Int. J. Mol. Sci. 2022, 23, 734. [Google Scholar] [CrossRef]
- Lang, X.; Zhao, X.; Zhao, J.; Ren, T.; Nie, L.; Zhao, W. MicroRNA Profiling Revealed the Mechanism of Enhanced Cold Resistance by Grafting in Melon (Cucumis melo L.). Plants 2024, 13, 1016. [Google Scholar] [CrossRef]
- Wang, H.; Xie, Y.; Yang, L.; Yan, W.; He, Z. Study on cold tolerance of different rootstocks of melon seedlings. IOP Conf. Ser. Earth Environ. Sci. 2019, 310, 052032. [Google Scholar] [CrossRef]
- Li, M.; Duan, X.; Wang, Q.; Chen, W.; Qi, H. A new morphological method to identify cold tolerance of melon at seedling stage. Funct. Plant Biol. 2019, 47, 80–90. [Google Scholar] [CrossRef]
- Liu, T.; Shi, J.; Li, M.; Ye, X.; Qi, H. Trehalose triggers hydrogen peroxide and nitric oxide to participate in melon seedlings oxidative stress tolerance under cold stress. Environ. Exp. Bot. 2021, 184, 104379. [Google Scholar] [CrossRef]
- Liu, T.; Han, Y.; Shi, J.; Liang, A.; Xu, D.; Ye, X.; Qi, H. Abscisic acid involved in trehalose improved melon photosynthesis via regulating oxidative stress tolerance and cell morphology structure under cold stress. Environ. Exp. Bot. 2022, 202, 105042. [Google Scholar] [CrossRef]
- Li, L.; Zhang, X.; Ding, F.; Hou, J.; Wang, J.; Luo, R.; Hu, J. Genome-wide identification of the melon (Cucumis melo L.) response regulator gene family and functional analysis of CmRR6 and CmPRR3 in response to cold stress. J. Plant Physiol. 2024, 292, 154160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Diao, Q.; Cao, Y.; Shi, M.; Zhang, H.; Jin, H.; Jiang, S.; Shen, H. Effect of sodium nitroprusside on the growth and enzymes related to nitrogen metabolism of melon seedlings under low temperature stress. Mol. Plant Breed. 2023, 14, 1–8. [Google Scholar] [CrossRef]
- Ma, M.; Liu, S.; Wang, Z.; Shao, R.; Ye, J.; Yan, W.; Che, G. Genome-wide identification of the SUN gene family in melon (Cucumis melo) and functional characterization of Two CmSUN genes in regulating fruit shape variation. Int. J. Mol. Sci. 2022, 23, 16047. [Google Scholar] [CrossRef]
- Yu, J.; Wu, S.; Sun, H.; Wang, X.; Tang, X.; Guo, S.; Fei, Z. CuGenDBv2: An updated database for cucurbit genomics. Nucleic Acids Res. 2023, 51, D1457–D1464. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Nogués, I.; Sekula, B.; Angelaccio, S.; Grzechowiak, M.; Tramonti, A.; Contestabile, R.; Ruszkowski, M. Arabidopsis thaliana serine hydroxymethyltransferases: Functions, structures, and perspectives. Plant Physiol. Biochem. 2022, 187, 37–49. [Google Scholar] [CrossRef]
- Pan, T.; Jin, H.; Zhou, C.; Yan, M. Rice Serine Hydroxymethyltransferases: Evolution, Subcellular Localization, Function and Perspectives. Plants 2024, 13, 1116. [Google Scholar] [CrossRef]
- Santatiwongchai, J.; Gleeson, D.; Gleeson, M.P. Theoretical Evaluation of the Reaction Mechanism of Serine Hydroxymethyltransferase. J. Phys. Chem. B 2018, 123, 407–418. [Google Scholar] [CrossRef]
- Wei, Z.; Sun, K.; Sandoval, F.J.; Cross, J.M.; Gordon, C.; Kang, C.; Roje, S. Folate polyglutamylation eliminates dependence of activity on enzyme concentration in mitochondrial serine hydroxymethyltransferases from Arabidopsis thaliana. Arch. Biochem. Biophys. 2013, 536, 87–96. [Google Scholar] [CrossRef]
- Yu, J.; Xie, Q.; Li, C.; Dong, Y.; Zhu, S.; Chen, J. Comprehensive characterization and gene expression patterns of LBD gene family in Gossypium. Planta 2020, 251, 81. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Hu, P.; Tao, Y.; Song, P.; Gao, H.; Guan, Y. Genome-wide identification and characterization of the Lateral Organ Boundaries Domain (LBD) gene family in polyploid wheat and related species. PeerJ 2021, 9, e11811. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Xue, H.; Xing, Z.; Xin, W.; Guo, L.; Bo, L.; Yan, C.; Chun, Y.; Jian, Z.; Hui, W. Identification and Expression Analysis of GASA Gene Family in Crape Myrtle. Mol. Plant Breed. 2024, 1–19. Available online: https://link.cnki.net/urlid/46.1068.S.20241111.1605.015 (accessed on 11 November 2024). (In Chinese).
- Su, L.; Wan, S.; Zhou, J.; Shao, Q.S.; Xing, B. Transcriptional regulation of plant seed development. Physiol. Plant. 2021, 173, 2013–2025. [Google Scholar] [CrossRef]
- Riemann, M.; Dhakarey, R.; Hazman, M.; Miro, B.; Kohli, A.; Nick, P. Exploring Jasmonates in the Hormonal Network of Drought and Salinity Responses. Front. Plant Sci. 2015, 6, 1077. [Google Scholar] [CrossRef]
- Wasternack, C. Jasmonates: An update on Biosynthesis, Signal Transduction and Action in Plant Stress Response, Growth and Development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef]
- Balbi, V.; Devoto, A. Jasmonate Signalling Network in Arabidopsis thaliana: Crucial regulatory nodes and new physiological scenarios. New Phytol. 2008, 177, 301–318. [Google Scholar] [CrossRef]
- Gusta, L.V.; Trischuk, R.; Weiser, C.J. Plant cold acclimation: The role of abscisic acid. J. Plant Growth Regul. 2005, 24, 308–318. [Google Scholar] [CrossRef]
- Sharma, R.; Sharma, P. Role of Abscisic Acid in Plant Stress. In New Insights into Phytohormones; IntechOpen: London, UK, 2023. [Google Scholar]
- McClung, C.R.; Hsu, M.; Painter, J.E.; Gagne, J.M.; Karlsberg, S.D.; Salomé, P.A. Integrated temporal regulation of the photorespiratory pathway. Circadian regulation of two Arabidopsis genes encoding serine hydroxymethyltransferase. Plant Physiol. 2000, 123, 381–392. [Google Scholar] [CrossRef]
- Voll, L.M.; Jamai, A.; Renné, P.; Voll, H.; McClung, C.R.; Weber, A.P. The photorespiratory Arabidopsis shm1 mutant is deficient in SHM1. Plant Physiol. 2006, 140, 59–66. [Google Scholar] [CrossRef]
- Bing, G.; Jia, Q.; Na, L.; Meng, S.; Li, W.; Jun, L.; Xiao, M. Genome-wide identification and expression analysis of SHMT gene family in foxtail millet (Setaria italica L.). Acta Agron. Sin. 2024, 1–14. Available online: https://link.cnki.net/urlid/11.1809.s.20241030.0852.002 (accessed on 11 November 2024). (In Chinese).
- Kito, K.; Tsutsumi, K.; Rai, V.; Theerawitaya, C.; Cha-Um, S.; Yamada-Kato, N.; Takabe, T. Isolation and functional characterization of 3-phosphoglycerate dehydrogenase involved in salt responses in sugar beet. Protoplasma 2017, 254, 2305–2313. [Google Scholar] [CrossRef]
- Liu, H.; Li, N.; Zhao, Y.; Kang, G.Z.; Zhao, Y.H.; Xu, H.W. Serine Hydroxymethyltransferase (SHMT) Gene Family in Wheat (Triticum aestivum L.): Identification, Evolution, and Expression Analysis. Agronomy 2022, 12, 1346. [Google Scholar] [CrossRef]
| Target Genes | Primer | Sequences | Amplicon Size (bp) |
|---|---|---|---|
| CmSHMT1 | MELO3C011107.1-F | ACATAAGTGGTTTAGTTGCCGC | 110 |
| MELO3C011107.1-R | AATCATTGCCCCCCGTG | ||
| CmSHMT2 | MELO3C020429.1-F | CATTAGTGGACTCGTTGCTGCT | 752 |
| MELO3C020429.1-R | GCTCTTCGATTTCCTTGTTGTT | ||
| CmSHMT3 | MELO3C019443.1-F | CTTGCCTTTTGCCCTAAACTAC | 116 |
| MELO3C019443.1-R | ATATCACACATCAGAACTGCTCCA | ||
| CmSHMT4 | MELO3C019450.1-F | TGTCTGTGTTATGGGTGGTGTG | 513 |
| MELO3C019450.1-R | GATTTGCCTCTGTTTCTCTTTCTC | ||
| CmSHMT5 | MELO3C010431.1-F | ATCTTTCCTTCCGCTTAAGTTCT | 179 |
| MELO3C010431.1-R | GTCGTCGCTATCATCGTCCA | ||
| CmSHMT6 | MELO3C016197.1-F | GTGAGAGAGAGTTTGCTGCGG | 196 |
| MELO3C016197.1-R | GGTATTGGGAATTGAGTGGTAAGAG | ||
| CmSHMT7 | MELO3C016200.1-F | TGTGACAACTTCTGATTTCCCTTTA | 181 |
| MELO3C016200.1-R | ATTTTCTCTGACCTTATCCCCC | ||
| CmSHMT8 | MELO3C007698.1-F | CCTGAGTGGAGTGGATAAAACG | 501 |
| MELO3C007698.1-R | GGATGGAGAGCCAGATAAAGATT | ||
| CmEFIa | EFIa-F | ACTGTGCTGTCCTCATTATTG | 98 |
| EFIa-R | AGGGTGAAAGCAAGAAGAGC |
| Gene Name | Gene ID | Amino Acid Sequence (aa) | Molecular Mass (kD) | Theoretical Isoelectric Point (pI) | Coefficient of Instability | Grand Average of Hydropathicity (GRAVY) | Prediction of Subcellular Localization |
|---|---|---|---|---|---|---|---|
| CmSHMT1 | MELO3C011107.1 | 516 | 57.16 | 8.14 | 35.46 | −0.276 | Chloroplast |
| CmSHMT2 | MELO3C020429.1 | 527 | 58.27 | 6.21 | 49.26 | −0.267 | Chloroplast |
| CmSHMT3 | MELO3C019443.1 | 167 | 19.06 | 9.44 | 43.49 | −0.334 | Chloroplast |
| CmSHMT4 | MELO3C019450.1 | 585 | 64.98 | 7.5 | 51.91 | −0.397 | Nucleus |
| CmSHMT5 | MELO3C010431.1 | 582 | 64.63 | 8.34 | 42.21 | −0.359 | Nucleus |
| CmSHMT6 | MELO3C016197.1 | 220 | 23.79 | 8.55 | 35.42 | 0.061 | Cytosol |
| CmSHMT7 | MELO3C016200.1 | 528 | 57.15 | 8.47 | 34.96 | −0.097 | Chloroplast |
| CmSHMT8 | MELO3C007698.1 | 549 | 61.20 | 8.39 | 39.99 | −0.287 | Mitochondrion |
| Gene Pairs | Rates of Non-Synonymous Substitution (Ka) | Rates of Synonymous Substitution (Ks) | Ka/Ks |
|---|---|---|---|
| CmSHMT1/CmSHMT8 | 0.104069 | 1.94726 | 0.05344364 |
| Element | Sequence | Description |
|---|---|---|
| AuxRE | AACGAC | Auxin-responsive elements |
| TC-rich repeats | GTTTTCTTAC | Cis-acting elements associated with defense mechanisms and stress responsiveness |
| GARE | TCTGTTG | Gibberellin-responsive elements |
| LTR | CCGAAA | Cis-acting elements associated with low-temperature responsiveness |
| TCARE | CCATCTTTTT | Cis-acting elements associated with salicylic acid responsiveness |
| ABRE | ACGTG/CACGTG | Cis-acting elements associated with the abscisic acid responsiveness |
| ARE | AAACCA | Cis-acting regulatory elements essential for the anaerobic induction |
| G-box | CACGTC/CACGAC | Cis-acting regulatory elements associated with light responsiveness |
| Box 4 | ATTAAT | Part of a conserved DNA module associated with light responsiveness |
| MeJARE | TGACG/CGTCA | Cis-acting regulatory elements associated with the MeJA-responsiveness |
| MRE | AACCTAA | MYB binding site associated with light responsiveness |
| MBS | CAACTG | MYB binding site associated with drought-inducibility |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; He, D.; Wu, Y.; Zhao, K.; Yang, C.; Zhong, Y.; Yang, L.; Niu, H.; Liu, S. Identification and Analysis of Melon (Cucumis melo L.) SHMT Gene Family Members and Their Functional Studies on Tolerance to Low-Temperature Stress. Agronomy 2025, 15, 203. https://doi.org/10.3390/agronomy15010203
Liu Y, He D, Wu Y, Zhao K, Yang C, Zhong Y, Yang L, Niu H, Liu S. Identification and Analysis of Melon (Cucumis melo L.) SHMT Gene Family Members and Their Functional Studies on Tolerance to Low-Temperature Stress. Agronomy. 2025; 15(1):203. https://doi.org/10.3390/agronomy15010203
Chicago/Turabian StyleLiu, Yanmin, Dandan He, Yizhou Wu, Kangqi Zhao, Changyi Yang, Yulu Zhong, Liuyang Yang, Haiyue Niu, and Sushuang Liu. 2025. "Identification and Analysis of Melon (Cucumis melo L.) SHMT Gene Family Members and Their Functional Studies on Tolerance to Low-Temperature Stress" Agronomy 15, no. 1: 203. https://doi.org/10.3390/agronomy15010203
APA StyleLiu, Y., He, D., Wu, Y., Zhao, K., Yang, C., Zhong, Y., Yang, L., Niu, H., & Liu, S. (2025). Identification and Analysis of Melon (Cucumis melo L.) SHMT Gene Family Members and Their Functional Studies on Tolerance to Low-Temperature Stress. Agronomy, 15(1), 203. https://doi.org/10.3390/agronomy15010203







