Activation of ABA Signaling Pathway and Up-Regulation of Salt-Responsive Genes Confer Salt Stress Tolerance of Wheat (Triticum aestivum L.) Seedlings
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Treatment
2.2. Protein Extraction and Alkylation
- A sample (0.4 g) of fresh leaves or roots was ground in liquid nitrogen using a pre-cooled mortar. For total protein extraction, 5 mL of a trichloroacetic acid/acetone solution containing 1 mM PMSF was added to the powdered tissue. Control treatments included three biological replicates, while stress treatments also had three biological replicates.
- The extracted protein was incubated in the dark at −20 °C for three hours. After incubation, the protein pellet was obtained by centrifugation at 13,000× g for 30 min at 4 °C. The pellet was then washed with 1 mL of acetone and rinsed [27].
- The pellet was finally resuspended in 1.5 mL of Buffer A, which contained 8 M urea, 4% CHAPS, 30 mM HEPES (pH 8.2), 2 mM Na2EDTA, 10 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride (PMSF) [27].
- A total of 15.8 µL of 200 mM DTT was added to 0.3 mL of the supernatant from step 3 after centrifugation (13,000× g, 4 °C for 30 min). The protein samples were then incubated at 56 °C for 1 h and immediately treated with 35 µL of 1 M iodoacetamide (IAM). Subsequently, 1 mL of cold acetone was added to precipitate the proteins in the sample. The pellet from this step was finally resuspended in 700 µL of 50 mM NH4HCO3. Insoluble materials were removed, and the total protein concentration was measured using the Coomassie Brilliant Blue G250 method [28].
2.3. Protein Digestion and Peptide Purification
2.4. LC-MS/MS Analysis of the Purified Peptides
2.5. Label-Free Quantification Analysis
2.6. Biochemical Analysis
2.7. qRT-PCR
2.8. Statistical Analysis
3. Results
3.1. Ion and Organic Solutes
3.2. Root Phytohormone
3.3. Gene Expression Analysis Involved in Salinity Tolerance and Carbohydrate Metabolism
3.4. Proteomic Profiling
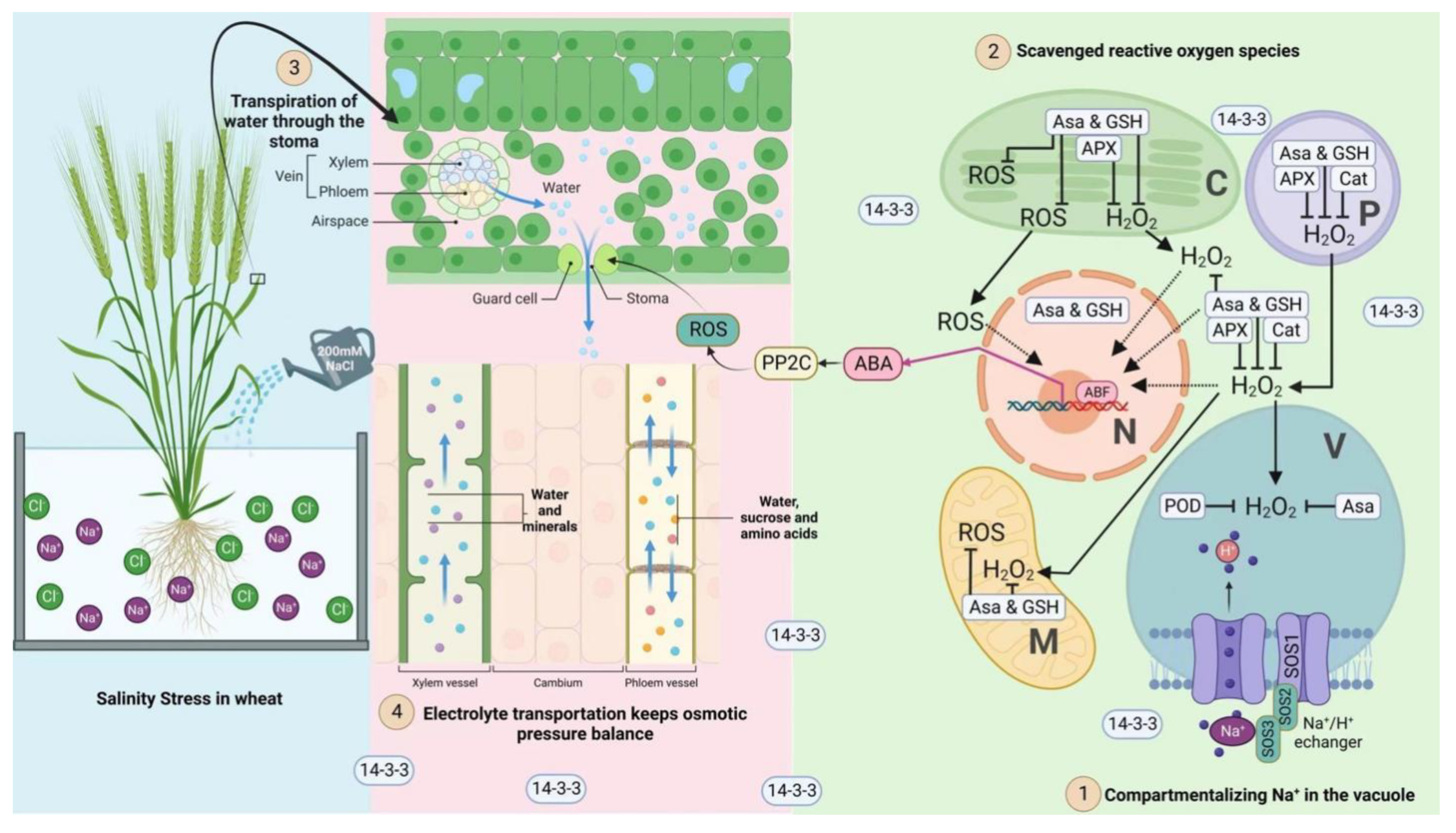
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shewry, P.R.; Hey, S.J. The contribution of wheat to human diet and health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef]
- Dubcovsky, J.; Dvorak, J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 2007, 316, 1862–1866. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Yang, Z.; Zhao, L.; Sun, F.; Liu, B. A newly formed hexaploid wheat exhibits immediate higher tolerance to nitrogen-deficiency than its parental lines. BMC Plant Biol. 2018, 18, 113. [Google Scholar] [CrossRef]
- Wu, D.; Cai, S.; Chen, M.; Ye, L.; Chen, Z.; Zhang, H.; Dai, F.; Wu, F.; Zhang, G. Tissue metabolic responses to salt stress in wild and cultivated barley. PLoS ONE 2013, 8, e55431. [Google Scholar] [CrossRef]
- Kosová, K.; Prášil, I.T.; Vítámvás, P. Protein contribution to plant salinity response and tolerance acquisition. Int. J. Mol. Sci. 2013, 14, 6757–6789. [Google Scholar]
- Pitman, M.G.; Läuchli, A. Global impact of salinity and agricultural ecosystems. In Salinity: Environment-Plants-Molecules; Springer: Berlin/Heidelberg, Germany, 2002; pp. 3–20. [Google Scholar]
- Jamil, A.; Riaz, S.; Ashraf, M.; Foolad, M. Gene expression profiling of plants under salt stress. Crit. Rev. Plant Sci. 2011, 30, 435–458. [Google Scholar] [CrossRef]
- Yang, J.; Cao, Y.; Yang, Z.; Zhang, W.; Sun, L.; Lu, C. Morphological, physiological and biochemical responses of biofuel plant Euphorbia lathyris to salt stress. Acta Agric. Scand. Sect. B–Soil Plant Sci. 2013, 63, 330–340. [Google Scholar]
- Foster, T.; Brozović, N.; Butler, A.; Neale, C.; Raes, D.; Steduto, P.; Fereres, E.; Hsiao, T.C. AquaCrop-OS: An open source version of FAO’s crop water productivity model. Agric. Water Manag. 2017, 181, 18–22. [Google Scholar] [CrossRef]
- Rahnama, A.; James, R.A.; Poustini, K.; Munns, R. Stomatal conductance as a screen for osmotic stress tolerance in durum wheat growing in saline soil. Funct. Plant Biol. 2010, 37, 255–263. [Google Scholar] [CrossRef]
- James, R.A.; Blake, C.; Byrt, C.S.; Munns, R. Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1; 4 and HKT1; 5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. J. Exp. Bot. 2011, 62, 2939–2947. [Google Scholar] [CrossRef] [PubMed]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Rao, S.A.; McNeilly, T. Assessment of salinity tolerance based upon seedling root growth response functions in maize (Zea mays L.). Euphytica 2003, 131, 81–89. [Google Scholar] [CrossRef]
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef]
- Dubcovsky, J.; María, G.S.; Epstein, E.; Luo, M.-C.; Dvořák, J. Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theor. Appl. Genet. 1996, 92, 448–454. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Läuchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef] [PubMed]
- Baghbani, A.; Namdari, A.; Kadkhodaie, A. Effect of salinity and nitrogen supply on nitrogen fixation nodules and nitrogen, sodium and potassium concentration of alfalfa cultivars. J. Basic Appl. Sci. Res. 2012, 2, 9978–9984. [Google Scholar]
- Kafi, M.; Stewart, W.; Borland, A. Carbohydrate and proline contents in leaves, roots, and apices of salt-tolerant and salt-sensitive wheat cultivars1. Russ. J. Plant Physiol. 2003, 50, 155–162. [Google Scholar] [CrossRef]
- Zhu, J.-K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Greenway, H.; Munns, R. Mechanisms of salt tolerance in nonhalophytes. Annu. Rev. Plant Physiol. 1980, 31, 149–190. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Millar, A.H.; Taylor, N.L. Wheat mitochondrial proteomes provide new links between antioxidant defense and plant salinity tolerance. J. Proteome Res. 2010, 9, 6595–6604. [Google Scholar] [CrossRef] [PubMed]
- Sobhanian, H.; Razavizadeh, R.; Nanjo, Y.; Ehsanpour, A.A.; Jazii, F.R.; Motamed, N.; Komatsu, S. Proteome analysis of soybean leaves, hypocotyls and roots under salt stress. Proteome Sci. 2010, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Zörb, C.; Schmitt, S.; Mühling, K.H. Proteomic changes in maize roots after short-term adjustment to saline growth conditions. Proteomics 2010, 10, 4441–4449. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yang, R.; Han, Y.; Gu, Z. Comparative proteomic and physiological analyses reveal the protective effect of exogenous calcium on the germinating soybean response to salt stress. J. Proteom. 2015, 113, 110–126. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.; Flowers, T.J.; Moore, A.L.; Harpham, N.V. An accurate and reproducible method for proteome profiling of the effects of salt stress in the rice leaf lamina. J. Exp. Bot. 2006, 57, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, Y.; Pareek, A.; Pareek, S.L.S. OsLdh7, a rice lactate dehydrogenase, confers stress resilience in rice under cadmium stress through NAD+/NADH regulation. Plant Physiol. Biochem. 2024, 215, 109009. [Google Scholar] [CrossRef]
- Sun, W.; Xiang, X.; Zhai, L.; Zhang, D.; Cao, Z.; Liu, L.; Zhang, Z. AGO_18b negatively regulates determinacy of spikelet meristems on the tassel central spike in maize. J. Integr. Plant Biol. 2018, 60, 65–78. [Google Scholar] [CrossRef]
- Zhao, T.; Wang, L.; Yang, J. Synergistic effects of combined application of biochar and arbuscular mycorrhizal fungi on the safe production of rice in cadmium contaminated soil. Sci. Total Environ. 2024, 951, 175499. [Google Scholar] [CrossRef]
- Baoxiang, W.; Yan, L.; Yifeng, W.; Jingfang, L.; Zhiguang, S.; Ming, C.; Yungao, X.; Bo, X.; Bo, Y.; Jian, L.; et al. OsbZIP72 Is Involved in Transcriptional Gene-Regulation Pathway of Abscisic Acid Signal Transduction by Activating Rice High-Affinity Potassium Transporter OsHKT1;1. Rice Sci. 2021, 28, 257–267. [Google Scholar] [CrossRef]
- Aslam, M.M.; Rashid, M.A.R.; Siddiqui, M.A.; Khan, M.T.; Farhat, F.; Yasmeen, S.; Khan, I.A.; Raja, S.; Rasool, F.; Sial, M.A.; et al. Recent Insights into Signaling Responses to Cope Drought Stress in Rice. Rice Sci. 2022, 29, 105–117. [Google Scholar] [CrossRef]
- Kartika, K.; Sakagami, J.-I.; Lakitan, B.; Yabuta, S.; Wijaya, A.; Kadir, S.; Widuri, L.I.; Siaga, E.; Nakao, Y. Morpho-Physiological Response of Oryza glaberrima to Gradual Soil Drying. Rice Sci. 2020, 27, 67–74. [Google Scholar] [CrossRef]
- Ahmed, S.; Rashid, M.A.R.; Zafar, S.A.; Azhar, M.T.; Waqas, M.; Uzair, M.; Rana, I.A.; Azeem, F.; Chung, G.; Ali, Z.; et al. Genome-wide investigation and expression analysis of APETALA-2 transcription factor subfamily reveals its evolution, expansion and regulatory role in abiotic stress responses in Indica Rice (Oryza sativa L. ssp. indica). Genomics 2021, 113, 1029–1043. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Xiao, C.; Xiao, B.; Wang, M.; Liu, J.; Bhanbhro, N.; Khan, A.; Wang, H.; Wang, H.; Yang, C. Proteomic profiling sheds light on alkali tolerance of common wheat (Triticum Aestivum L.). Plant Physiol. Biochem. 2019, 138, 58–64. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Xiao, C.; Cui, X.; Lu, H.; Han, L.; Liu, S.; Zheng, Y.; Wang, H.; Wang, H.; Yang, C. Comparative adaptive strategies of old and young leaves to alkali-stress in hexaploid wheat. Environ. Exp. Bot. 2020, 171, 103955. [Google Scholar] [CrossRef]
- Bremberger, C.; Lüttge, U. Dynamics of tonoplast proton pumps and other tonoplast proteins of Mesembryanthemum crystallinum L. during the induction of Crassulacean acid metabolism. Planta 1992, 188, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Giménez, M.J.; Pistón, F.; Atienza, S.G. Identification of suitable reference genes for normalization of qPCR data in comparative transcriptomics analyses in the Triticeae. Planta 2011, 233, 163–173. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ashraf, M.; Rauf, H. Inducing salt tolerance in maize (Zea mays L.) through seed priming with chloride salts: Growth and ion transport at early growth stages. Acta Physiol. Plant. 2001, 23, 407–414. [Google Scholar] [CrossRef]
- Patterson, J.H.; Newbigin, E.; Tester, M.; Bacic, A.; Roessner, U. Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. J. Exp. Bot. 2009, 60, 4089–4103. [Google Scholar]
- Rahman, H.; Jagadeeshselvam, N.; Valarmathi, R.; Sachin, B.; Sasikala, R.; Senthil, N.; Sudhakar, D.; Robin, S.; Muthurajan, R. Transcriptome analysis of salinity responsiveness in contrasting genotypes of finger millet (Eleusine coracana L.) through RNA-sequencing. Plant Mol. Biol. 2014, 85, 485–503. [Google Scholar] [CrossRef] [PubMed]
- Hussain, K.; Majeed, A.; Nawaz, K.; Nisar, M.F. Effect of different levels of salinity on growth and ion contents of black seeds (Nigella sativa L.). Curr. Res. J. Biol. Sci. 2009, 1, 135–138. [Google Scholar]
- Sheen, J.; Zhou, L.; Jang, J.-C. Sugars as signaling molecules. Curr. Opin. Plant Biol. 1999, 2, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Alamgir, A.; Ali, M.Y. Effect of salinity on leaf pigments, sugar and protein concentrations and chloroplast ATPase activity of rice (Oryza sativa L.). Bangladesh J. Bot. 1999, 28, 145–149. [Google Scholar]
- Less, H.; Galili, G. Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol. 2008, 147, 316–330. [Google Scholar] [CrossRef]
- Xiong, L.; Zhu, J.K. Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ. 2002, 25, 131–139. [Google Scholar] [CrossRef]
- Hartzendorf, T.; Rolletschek, H. Effects of NaCl-salinity on amino acid and carbohydrate contents of Phragmites australis. Aquat. Bot. 2001, 69, 195–208. [Google Scholar] [CrossRef]
- Costa, R.; Lobato, A.; Oliveira Neto, C.; Maia, P.; Alves, G.; Laughinghouse IV, H. Biochemical and physiological responses in two Vigna unguiculata (L.) Walp. cultivars under water stress. J. Agron. 2008, 7, 98–101. [Google Scholar] [CrossRef]
- Colombo, R.; Cerana, R. Enhanced activity of tonoplast pyrophosphatase in NaCl-grown cells of Daucus carota. J. Plant Physiol. 1993, 142, 226–229. [Google Scholar] [CrossRef]
- Silva, P.; Façanha, A.R.; Tavares, R.M.; Gerós, H. Role of tonoplast proton pumps and Na+/H+ antiport system in salt tolerance of Populus euphratica Oliv. J. Plant Growth Regul. 2010, 29, 23–34. [Google Scholar] [CrossRef]
- Verslues, P.E. ABA and cytokinins: Challenge and opportunity for plant stress research. Plant Mol. Biol. 2016, 91, 629–640. [Google Scholar] [CrossRef] [PubMed]

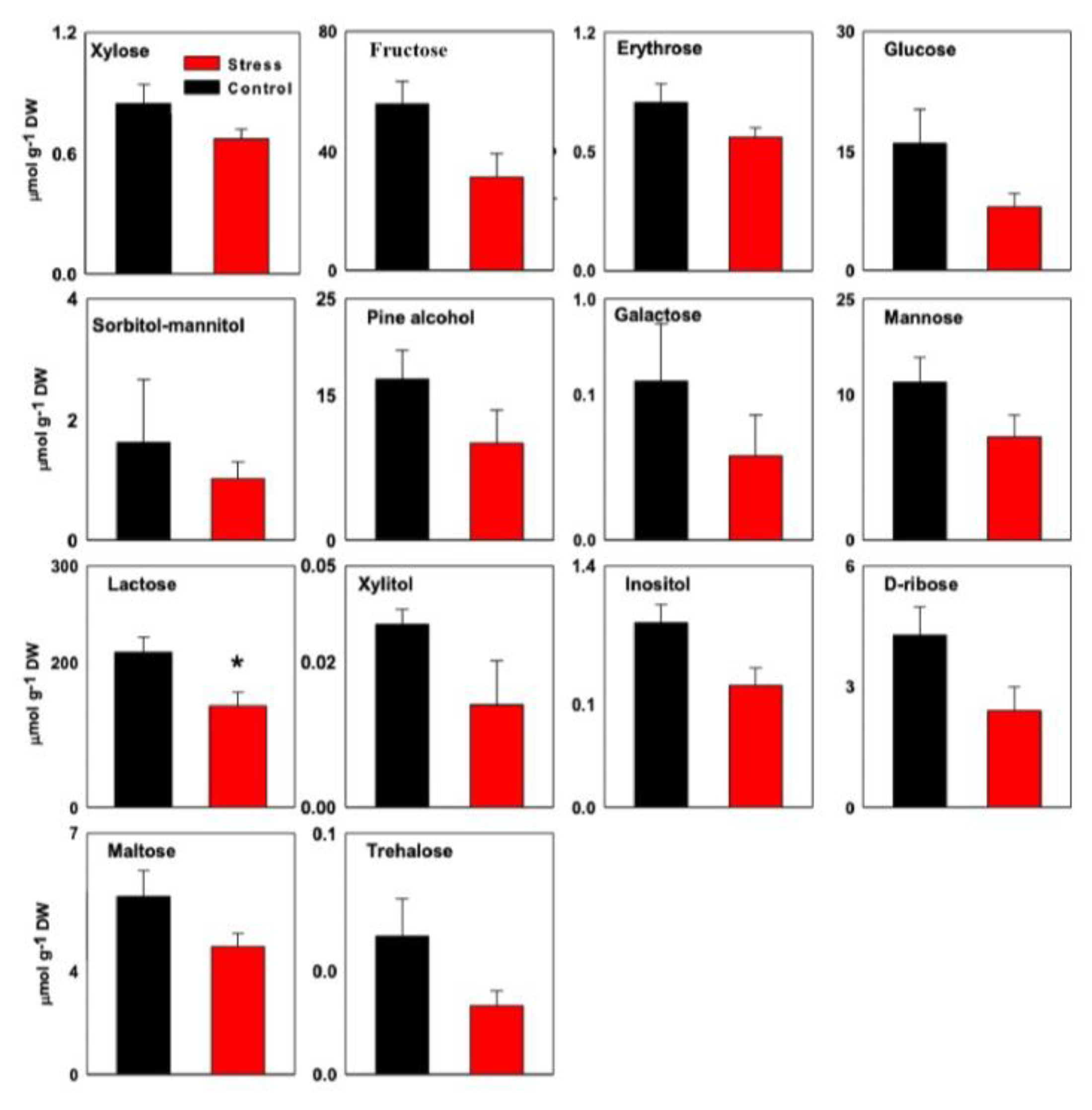

| Name | Control Plant (µg g−1 DW) | Stressed Plant (µg g−1 DW) | Fold Change (Stress/Control) | p-Value (t-Test) |
|---|---|---|---|---|
| GA3 | 0.0183 ± 0.0051 | 0.0138 ± 0.0018 | 0.7501 | 0.295 |
| ABA | 0.0028 ± 0.0002 | 0.0063 ± 0.0007 | 2.2959 | 0.0079 |
| IAA | 0.000 ± 0.0000 | 0.0164 ± 0.0232 | --- | 0.3739 |
| Trans-zeatin | 0.016 ± 0.0021 | 0.0169 ± 0.001 | 1.0536 | 0.06308 |
| JA | 0.3204 ± 0.0207 | 0.5687 ± 0.0379 | 1.7754 | 0.0012 |
| SA | 0.1337 ± 0.0196 | 0.2098 ± 0.0249 | 1.5693 | 0.0275 |
| Dihydrozeatin | 0.0005 ± 0.0005 | 0.0006 ± 0.0004 | 1.207 | 0.8131 |
| GA1 | 0.0247 ± 0.0067 | 0.1024 ± 0.0154 | 4.1522 | 0.0026 |
| Gene ID | Gene Name | Fold Change | p-Value (t-Test) |
|---|---|---|---|
| (Stress/Control) | |||
| TraesCS6B01G080000 | Glutathione S-transferase | 424.977 | 0.321 |
| TraesCS1D01G094700 | Glutathione S-transferase | 1.69 | 0.228 |
| TraesCS7B01G370000 | Glutathione S-transferase | 2.464 | 0.048 |
| TraesCS5B01G426800 | Dehydrin | 6.503 | 0.019 |
| TraesCS2B01G613900 | Peroxidase | 1.08 | 0.005 |
| TraesCS1A01G141100 | LEA | 0.811 | 0.261 |
| TraesCS2B01G125800 | Peroxidase | 3.998 | 0.092 |
| TraesCS2A01G016500 | Pectate lyase | 1.692 | 0.107 |
| TraesCS6A01G350600 | Dehydrin | 0.993 | 0.981 |
| TraesCS7A01G560000 | Dehydrin | 0.853 | 0.001 |
| TraesCS6A01G350700 | Dehydrin | 0.418 | 0.012 |
| TraesCS3B01G266200 | PP2C | 0.306 | 0.013 |
| TraesCS3B01G411300 | ABF | 2.682 | 0.005 |
| TraesCS3D01G250900 | V-H+ ATPase-b2 | 0.776 | 0.82 |
| TraesCS2A01G289300 | HAK1 | 0.789 | 0.036 |
| TraesCS3A01G446600 | Potassium transporter | 1.737 | 0.043 |
| TraesCS2D01G419000 | Potassium transporter | 0.146 | 0.074 |
| TraesCS4D01G361300.1 | HKT1;5 | 1.145 | 0.003 |
| Gene Name | Fold Change (Stress/Control) | p-Value (t-Test) |
|---|---|---|
| TaSPSV | 0.58 | 0.365 |
| TaSPSIV | 40.48 | 0.194 |
| TaSPSIII | 0.88 | 0.79 |
| TaSPSIIa | 2.01 | 0.121 |
| TaSPSIa | 1.92 | 0.492 |
| TaFK4b | 0.53 | 0.478 |
| TaFK4a | 0.64 | 0.033 |
| TaCesA10 | 0.52 | 0.473 |
| TaCesA4 | 1.93 | 0.228 |
| TaCesA1b | 0.47 | 0.332 |
| TaUGDC1 | 1.06 | 0.949 |
| TaUGDH1 | 0.55 | 0.364 |
| TaPDH-E3-1 | 0.69 | 0.4304 |
| TaPDH-E1-B1 | 1.06 | 0.002 |
| Ta6-SFT2 | 1.06 | 0.83 |
| Ta6-SFT1 | 1.4 | 0.001 |
| Ta1-SST2 | 1.7 | 0.013 |
| Ta1-SST1 | 1.94 | 0.092 |
| TaSPSVI | 1.79 | 0.013 |
| TaSuS3 | 2.29 | 0.039 |
| TaSuS4 | 1.68 | 0.174 |
| TaSuS5 | 0.37 | 0.375 |
| TaSuS7 | 1.01 | 0.966 |
| TaSuS9 | 2.07 | 0.104 |
| TaSuS11 | 2.3 | 0.026 |
| TaSAInv1 | 0.72 | 0.482 |
| TaSAInv2 | 2.11 | 0.014 |
| TaRP36 | 0.56 | 0.209 |
| TaRP15 | 0.78 | 0.479 |
| S.no | Protein Name | Protein Number |
|---|---|---|
| 1 | Ascorbate | 3 |
| 2 | Citrate synthase | 6 |
| 3 | Glutathione S-transferase | 7 |
| 4 | Dehydrin | 8 |
| 5 | Peroxidase | 4 |
| 6 | Peroxidase | 2 |
| 7 | Heat shock factor-binding protein 1 | 4 |
| 8 | Dehydrin, conserved site | 1 |
| 9 | V-type proton ATPase subunit G | 5 |
| 10 | Superoxide dismutase [Cu-Zn] | 8 |
| 11 | V-type proton ATPase subunit E | 1 |
| 12 | 14-3-3 protein | 5 |
| 13 | Peroxidase | 1 |
| 14 | Thioredoxin | 2 |
| 15 | Malate dehydrogenase | 1 |
| 16 | Superoxide dismutase | 3 |
| 17 | Cold shock protein | 1 |
| 18 | Plasma membrane-associated cation-binding protein 1 | 3 |
| 19 | Hsp70-Hsp90 organizing protein 1 | 5 |
| 20 | V-type proton ATPase subunit d | 1 |
| 21 | ATPase family AAA domain-containing protein 3 | 2 |
| 22 | Heat-inducible transcription repressor | 6 |
| 23 | Photosystem I reaction center subunit II | 4 |
| Protein ID | Protein Name | Fold Change (Stress/Control) | Adj. p-Value |
|---|---|---|---|
| TraesCS3B01G147100.1 | Glutaredoxin | 100 | 2.68892 × 10−16 |
| TraesCS3B01G248200.1 | Receptor-like kinase | 100 | 2.68892 × 10−16 |
| TraesCS1D01G207400.1 | Glutathione S-transferase | 100 | 2.68892 × 10−16 |
| TraesCS6D01G234700.1 | Dehydrin | 100 | 2.68892 × 10−16 |
| TraesCS4A01G209400.1 | Cytochrome b-c1 complex subunit 6 | 100 | 2.68892 × 10−16 |
| TraesCS6D01G378400.1 | Potassium channel beta subunit | 100 | 2.68892 × 10−16 |
| TraesCS5B01G195600.1 | Heat shock factor-binding protein 1 | 100 | 2.68892 × 10−16 |
| TraesCS5A01G369800.1 | Dehydrin, conserved site | 100 | 2.68892 × 10−16 |
| TraesCS2B01G457800.1 | V-type proton ATPase subunit G | 7.856 | 8.69717 × 10−6 |
| TraesCS2D01G123300.1 | Superoxide dismutase [Cu-Zn] | 5.785 | 5.0951 × 10−5 |
| TraesCS3A01G238700.1 | V-type proton ATPase subunit E | 24.96 | 9.77338 × 10−5 |
| TraesCS4B01G159900.1 | 14-3-3 protein | 5.155 | 0.000695605 |
| TraesCS7B01G375600.1 | Peroxidase | 9.257 | 0.001057598 |
| TraesCS2B01G389500.1 | Thioredoxin | 5.432 | 0.005073489 |
| TraesCS3A01G234800.2 | Acyl-CoA-binding protein | 4.724 | 0.007562613 |
| TraesCS2A01G537100.1 | Superoxide dismutase | 3.961 | 0.010151094 |
| TraesCS1B01G273000.2 | Cold shock protein | 3.692 | 0.020824126 |
| TraesCS3B01G183300.2 | Plasma membrane-associated cation-binding protein 1 | 3.284 | 0.038020934 |
| TraesCS2A01G386800.1 | Hsp70-Hsp90 organizing protein 1 | 3.64 | 0.04285163 |
| TraesCS3A01G210400.2 | V-type proton ATPase subunit d | 7.228 | 0.043992867 |
| TraesCS1D01G445100.1 | Heat shock family protein | 0.01 | 2.68892 × 10−16 |
| TraesCS4B01G269500.1 | Heat-inducible transcription repressor | 0.01 | 2.68892 × 10−16 |
| TraesCS5A01G457500.1 | Photosystem I reaction center subunit II | 0.033 | 1.56163 × 10−5 |
| TraesCS6A01G198500.1 | Prohibitin | 8.078 | 0.038129986 |
| TraesCS5A01G138700.1 | Cysteine proteinase | 4.276 | 0.032385383 |
| TraesCS3D01G529500.1 | Cytochrome b5 | 3.632 | 0.026628283 |
| TraesCS4D01G018500.1 | RNA-binding protein | 3.336 | 0.015415861 |
| TraesCS3A01G104600.1 | Late embryogenesis abundant (LEA) | 12.616 | 0.005187525 |
| TraesCS4A01G139600.1 | 40S ribosomal protein S21 | 5.398 | 0.00510172 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, Z.; Khan, A.; Khan, A.; Tao, Z.; Zhang, S.; Long, Q.; Lin, J.; Luo, S. Activation of ABA Signaling Pathway and Up-Regulation of Salt-Responsive Genes Confer Salt Stress Tolerance of Wheat (Triticum aestivum L.) Seedlings. Agronomy 2024, 14, 2095. https://doi.org/10.3390/agronomy14092095
Zou Z, Khan A, Khan A, Tao Z, Zhang S, Long Q, Lin J, Luo S. Activation of ABA Signaling Pathway and Up-Regulation of Salt-Responsive Genes Confer Salt Stress Tolerance of Wheat (Triticum aestivum L.) Seedlings. Agronomy. 2024; 14(9):2095. https://doi.org/10.3390/agronomy14092095
Chicago/Turabian StyleZou, Zhiyou, Aziz Khan, Adnan Khan, Zhongyi Tao, Sheng Zhang, Qiteng Long, Jinfu Lin, and Shunshe Luo. 2024. "Activation of ABA Signaling Pathway and Up-Regulation of Salt-Responsive Genes Confer Salt Stress Tolerance of Wheat (Triticum aestivum L.) Seedlings" Agronomy 14, no. 9: 2095. https://doi.org/10.3390/agronomy14092095
APA StyleZou, Z., Khan, A., Khan, A., Tao, Z., Zhang, S., Long, Q., Lin, J., & Luo, S. (2024). Activation of ABA Signaling Pathway and Up-Regulation of Salt-Responsive Genes Confer Salt Stress Tolerance of Wheat (Triticum aestivum L.) Seedlings. Agronomy, 14(9), 2095. https://doi.org/10.3390/agronomy14092095






