The Roles of Glutaredoxins in Wheat (Triticum aestivum L.) under Biotic and Abiotic Stress Conditions, including Fungal and Hormone Treatments
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Sequence Analysis of TaGRXs
2.2. Phylogenetic Analysis of TaGRXs
2.3. Physicochemical Properties of TaGRXs
2.4. Gene Structure and Motif Analysis of TaGRXs
2.5. Homology Modeling and Chromosome Mapping of TaGRXs
2.6. Analysis of Cis-Acting Elements of TaGRXs
2.7. Gene Expression Analysis of TaGRXs
2.8. Wheat Hormone Treatment
2.9. Fungal Pathogenic Stress Treatment
2.10. Real-Time Quantitative PCR (RT-qPCR) Analysis
3. Results
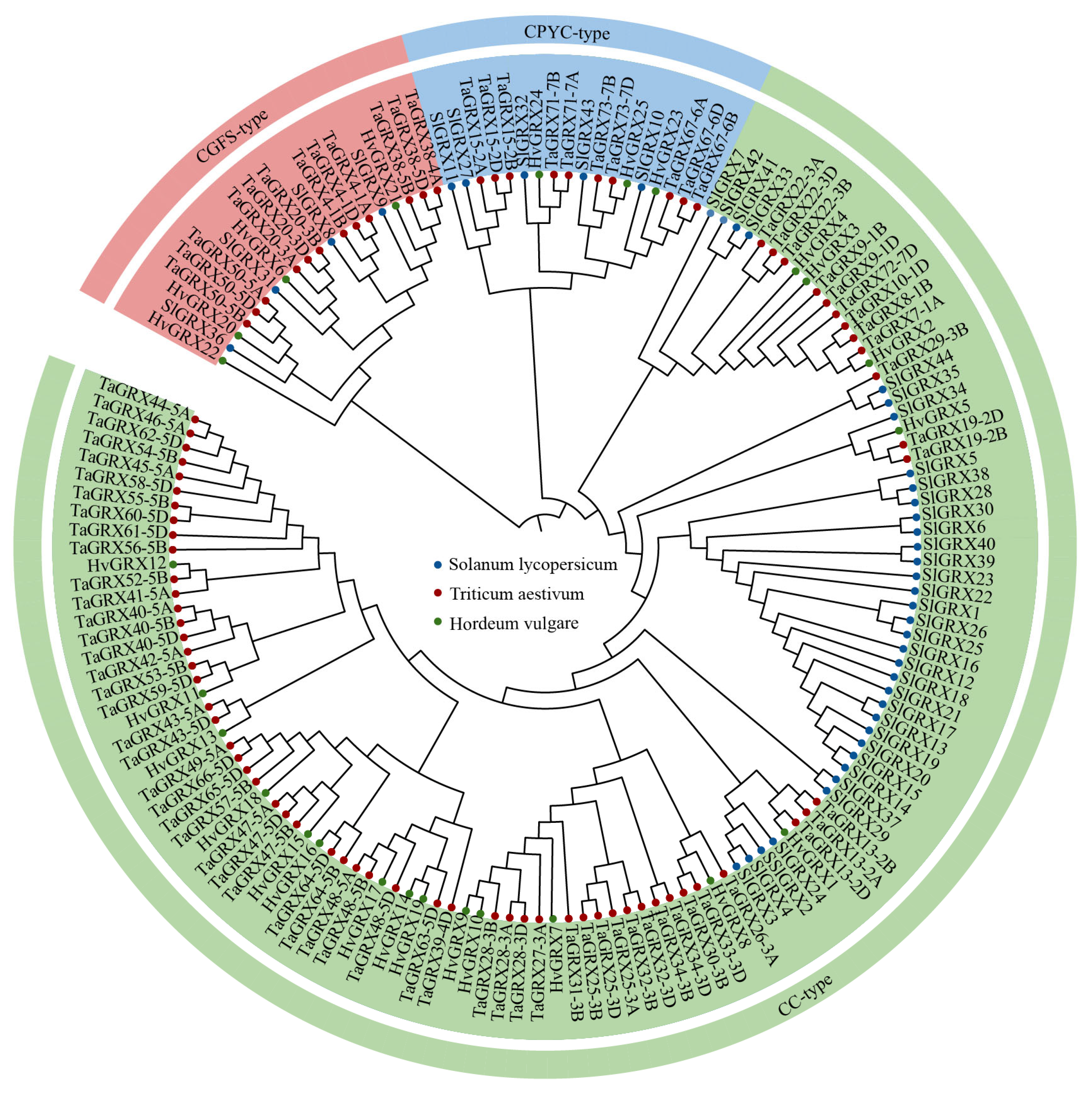
3.1. Identification and Phylogenetic Analysis of TaGRXs
3.2. Protein Features of TaGRXs
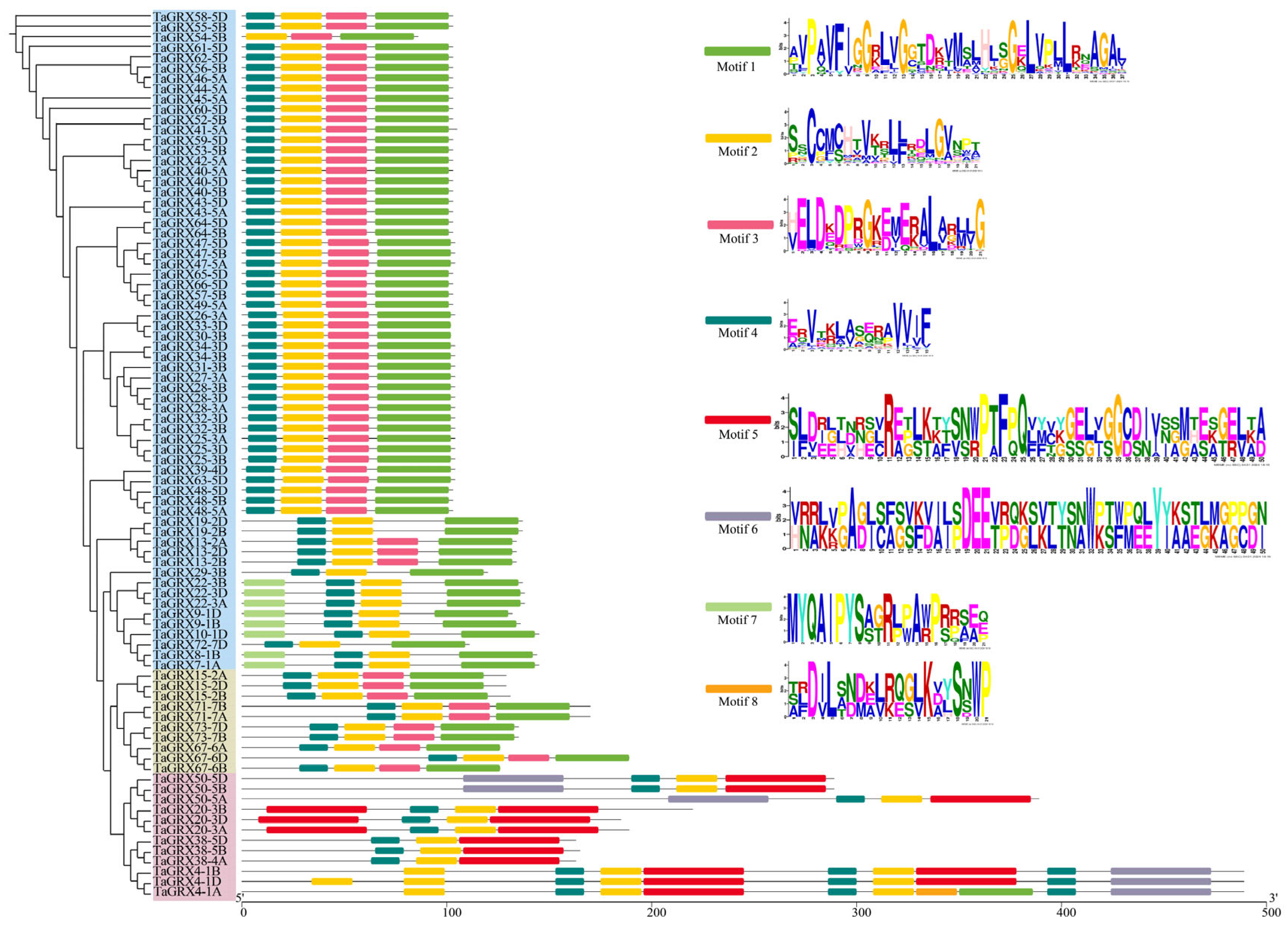
3.3. Gene Structure and Motif of TaGRXs
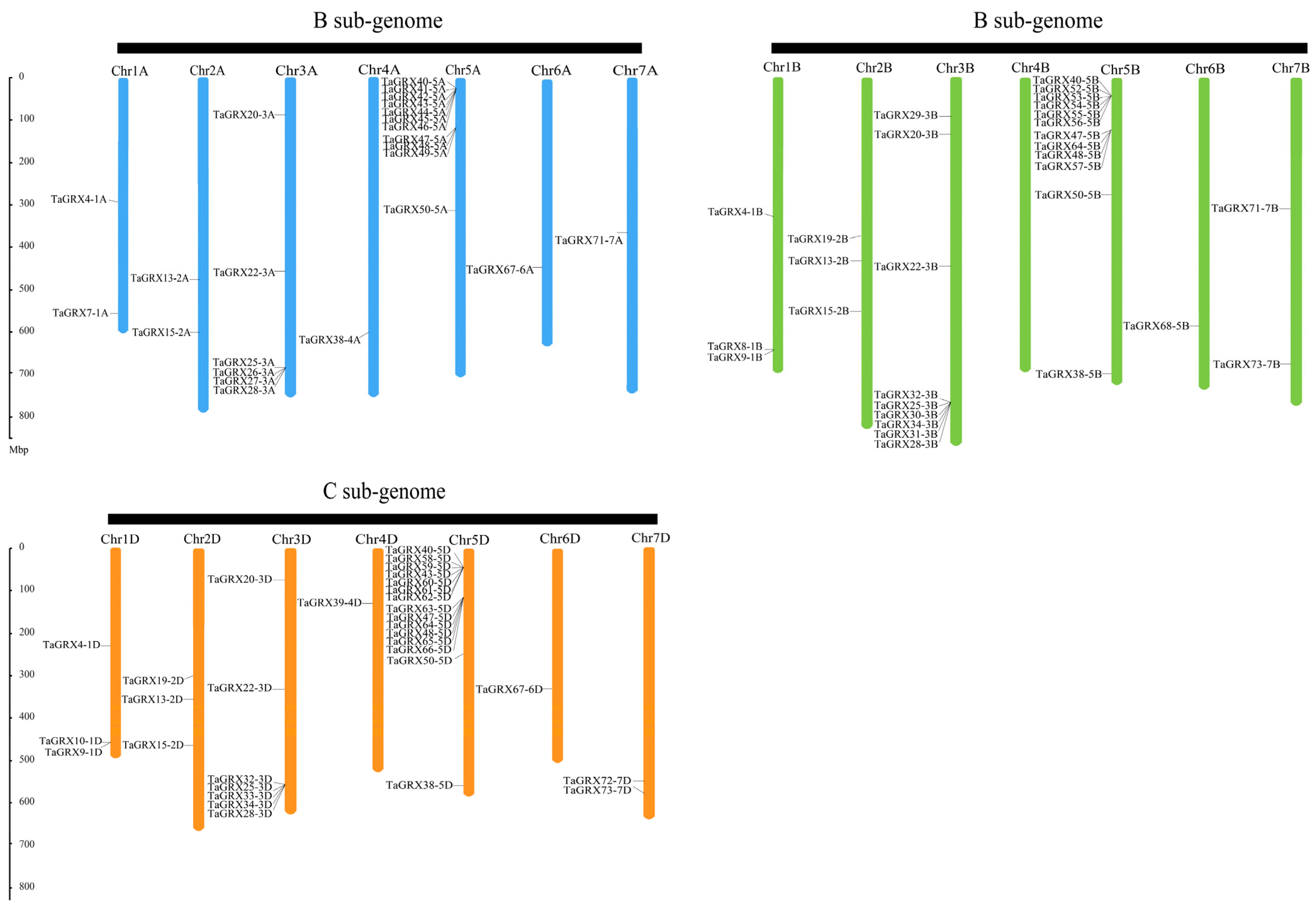
3.4. Structural Modeling and Chromosomal Location of TaGRXs
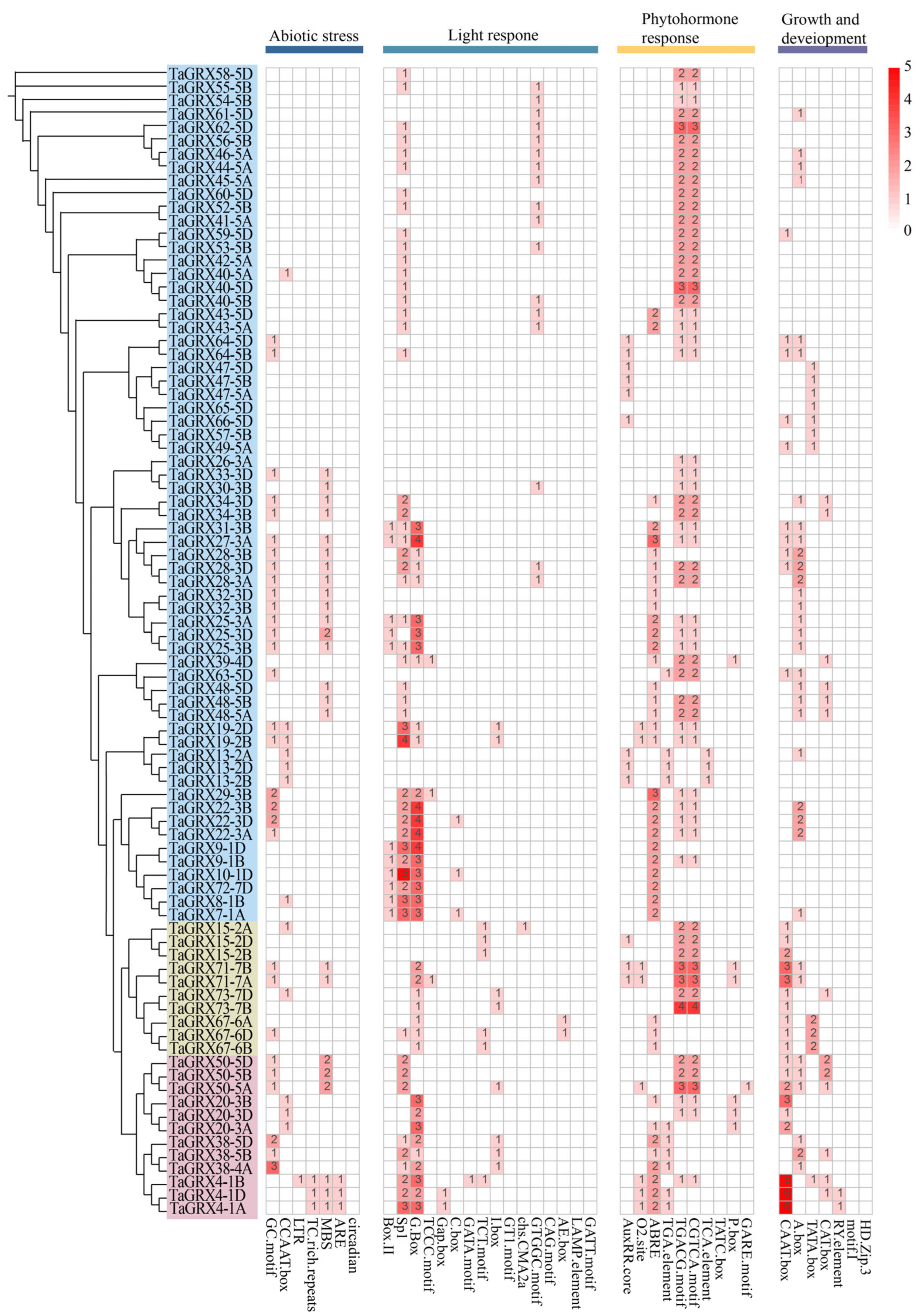
3.5. Conserved Cis-Acting Elements of TaGRXs
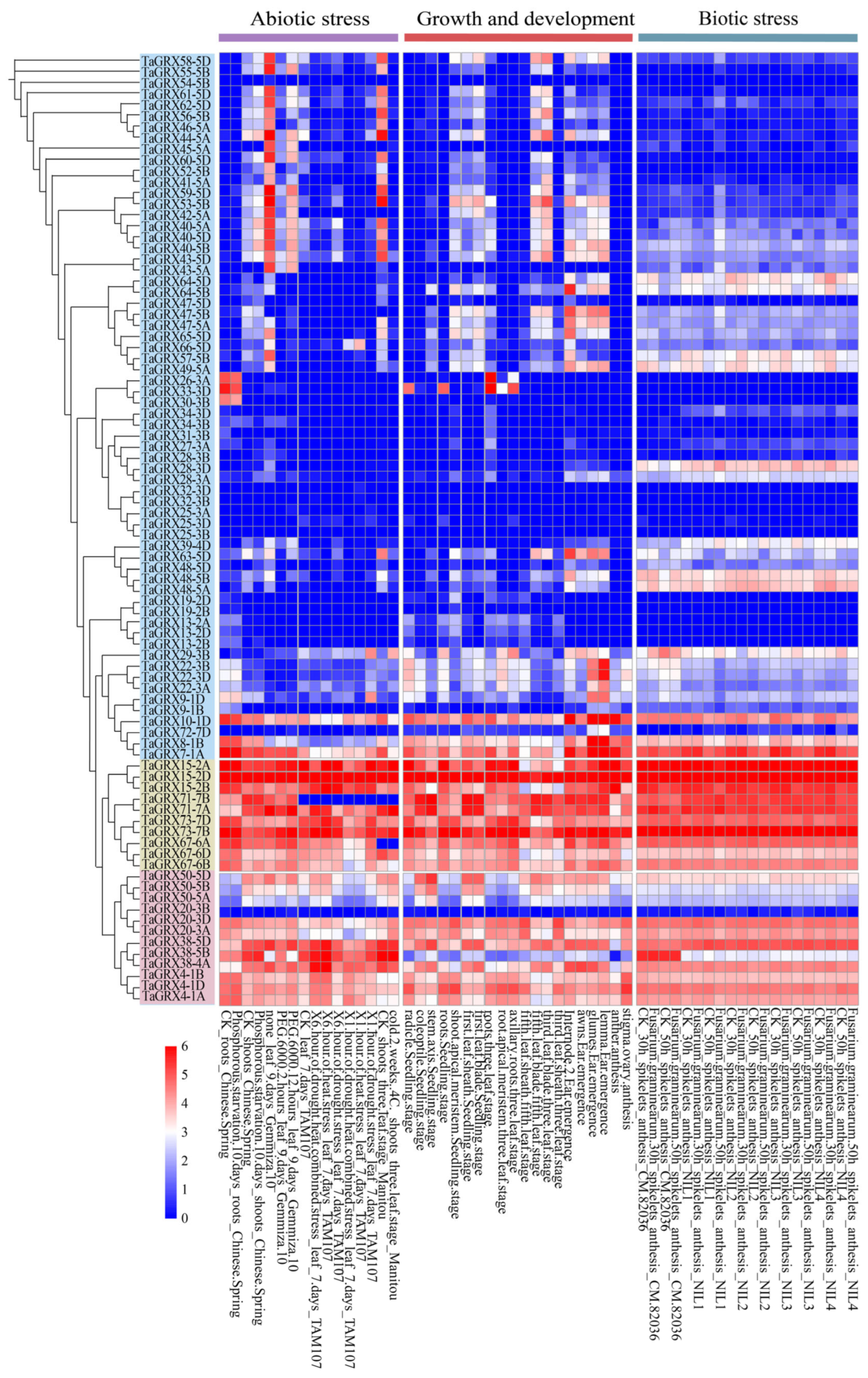
3.6. Expression Analysis of TaGRXs
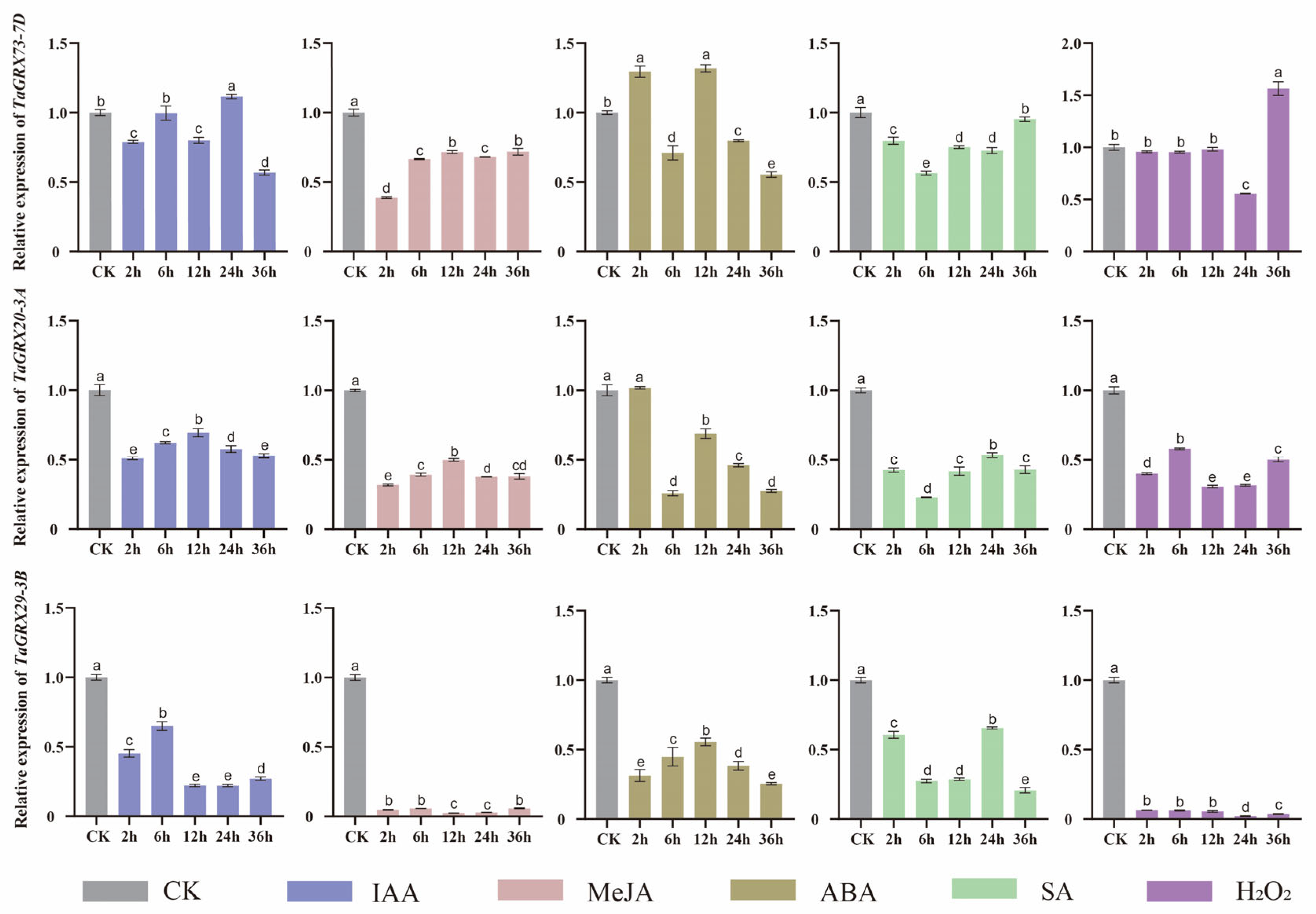
3.7. Hormonal Response Expression Analysis
3.8. Fungal Pathogenic Stress Response Expression Analysis
3.9. Tissue-Specific Expression Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lamers, J.; van der Meer, T.; Testerink, C. How Plants Sense and Respond to Stressful Environments. Plant Physiol. 2020, 182, 1624–1635. [Google Scholar] [CrossRef] [PubMed]
- Sevilla, F.; Martí, M.C.; De Brasi-Velasco, S.; Jiménez, A. Redox regulation, thioredoxins, and glutaredoxins in retrograde signalling and gene transcription. J. Exp. Bot. 2023, 74, 5955–5969. [Google Scholar] [CrossRef] [PubMed]
- Snezhkina, A.V.; Kudryavtseva, A.V.; Kardymon, O.L.; Savvateeva, M.V.; Melnikova, N.V.; Krasnov, G.S.; Dmitriev, A.A. ROS Generation and Antioxidant Defense Systems in Normal and Malignant Cells. Oxidative Med. Cell. Longev. 2019, 2019, 6175804. [Google Scholar] [CrossRef] [PubMed]
- Lillig, C.H.; Berndt, C.; Holmgren, A. Glutaredoxin systems. Biochim. Biophys. Acta 2019, 1780, 1304–1317. [Google Scholar] [CrossRef]
- Ogata, F.T.; Branco, V.; Vale, F.F.; Coppo, L. Glutaredoxin: Discovery, redox defense and much more. Redox Biol. 2021, 43, 101975. [Google Scholar] [CrossRef]
- Gutsche, N.; Thurow, C.; Zachgo, S.; Gatz, C. Plant-specific CC-type glutaredoxins: Functions in developmental processes and stress responses. Biol. Chem. 2021, 396, 495–509. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, X.; Zhang, L.; Chen, S. CC-type glutaredoxin gene CsGRX4 in cucumber responds to Botrytis cinerea via JA signaling pathway. Sci. Hortic. 2022, 306, 111440. [Google Scholar] [CrossRef]
- Yang, Y.; Xue, W.; Chen, P.; Yuan, X.; Li, X.; Zhang, T.; Chen, S. Identification and expression analyzes of CC-type glutaredoxin in cucumber (Cucumis sativus L.) under abiotic stress. Sci. Hortic. 2021, 289, 110417. [Google Scholar] [CrossRef]
- Guo, X.; Yu, X.; Xu, Z.; Zhao, P.; Zou, L.; Li, W.; Geng, M.; Zhang, P.; Peng, M.; Ruan, M. CC-type glutaredoxin, MeGRXC3, associates with catalases and negatively regulates drought tolerance in cassava (Manihot esculenta Crantz). Plant Biotechnol. J. 2022, 20, 2389–2405. [Google Scholar] [CrossRef]
- Son, S.; Kim, H.; Lee, K.S.; Kim, S.; Park, S.R. Rice glutaredoxin GRXS15 confers broad-spectrum resistance to Xanthomonas oryzae pv. oryzae and Fusarium fujikuroi. Biochem. Biophys. Res. Commun. 2020, 533, 1385–1392. [Google Scholar] [CrossRef]
- Guo, Y.S.; Huang, C.J.; Xie, Y.; Song, F.M.; Zhou, X.P. A tomato glutaredoxin gene SlGRX1 regulates plant responses to oxidative, drought and salt stresses. Planta 2010, 232, 1499–1509. [Google Scholar] [CrossRef] [PubMed]
- Laporte, D.; Olate, E.; Salinas, P.; Salazar, M.; Jordana, X.; Holuigue, L. Glutaredoxin GRXS13 plays a key role in protection against photooxidative stress in Arabidopsis. J. Exp. Bot. 2012, 63, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I. Reactive oxygen species (ROS): An introduction. In Reactive Oxygen Species in Plants: The Right Balance; Springer Nature: Singapore, 2023; pp. 1–22. [Google Scholar]
- Liedgens, L.; Zimmermann, J.; Wäschenbach, L.; Geissel, F.; Laporte, H.; Gohlke, H.; Morgan, B.; Deponte, M. Quantitative assessment of the determinant structural differences between redox-active and inactive glutaredoxins. Nat. Commun. 2004, 11, 1725. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Tang, J.; Gao, S.; Cheng, X.; Du, L.; Chu, C. Control of rice pre-harvest sprouting by glutaredoxin-mediated abscisic acid signaling. Plant J. 2019, 100, 1036–1051. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, C.; Zhu, Y.; Zhang, L.; Chen, T.; Zhou, F.; Chen, H.; Lin, Y. The calcium-dependent kinase OsCPK24 functions in cold stress responses in rice. J. Integr. Plant Biol. 2018, 60, 173–188. [Google Scholar] [CrossRef]
- Shiferaw, B.; Smale, M.; Braun, H.J.; Duveiller, E.; Reynolds, M.; Muricho, G. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 2013, 5, 291–317. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2014. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 2 December 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Xiao, Y.; Wen, Y.; Li, K.; Ma, Z.; Yin, J. Genome-wide characterization and function analysis uncovered roles of wheat LIMs in responding to adverse stresses and TaLIM8-4D function as a susceptible gene. Plant Genome 2022, 15, e20246. [Google Scholar] [CrossRef]
- Jiang, W.; Geng, Y.; Liu, Y.; Chen, S.; Cao, S.; Li, W.; Chen, H.; Ma, D.; Yin, J. Genome-wide identification and characterization of SRO gene family in wheat: Molecular evolution and expression profiles during different stresses. Plant Physiol. Biochem. 2020, 154, 590–611. [Google Scholar] [CrossRef]
- Jiang, B.; Su, C.; Wang, Y.; Xu, X.; Li, Y.; Ma, D. Genome-wide identification of Glutathione peroxidase (GPX) family genes and silencing TaGPX3.2A reduced disease resistance in wheat. Plant Physiol. Biochem. 2023, 204, 108139. [Google Scholar] [CrossRef]
- Hu, Y.; Su, C.; Zhang, Y.; Li, Y.; Chen, X.; Shang, H.; Hu, X. A Puccinia striiformis f. sp. tritici effector inhibits high-temperature seedling-plant resistance in wheat. Plant J. 2022, 112, 249–267. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Wang, Y.; Ma, Z.; Xu, X.; Ma, D.; Yang, L. Genome-wide identification of long intergenic non-coding RNAs of responsive to powdery mildew stress in wheat (Triticum aestivum). Front. Plant Sci. 2023, 14, 1297580. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.K.; Pandey, A.; Hamurcu, M.; Rajpal, V.R.; Vyhnanek, T.; Topal, A.; Raina, S.N.; Gezgin, S. Insight into the Boron Toxicity Stress-Responsive Genes in Boron-Tolerant Triticum dicoccum Shoots Using RNA Sequencing. Agronomy 2023, 13, 631. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chai, Y.C.; Mieyal, J.J. Glutathione and Glutaredoxin-Key Players in Cellular Redox Homeostasis and Signaling. Antioxidants 2023, 12, 1553. [Google Scholar] [CrossRef] [PubMed]
- Elsanosi, H.A.; Zhang, J.; Mostafa, S.; Geng, X.; Zhou, G.; Awdelseid, A.H.M.; Song, L. Genome-wide identification, structural and gene expression analysis of BTB gene family in soybean. BMC Plant Biol. 2024, 24, 663. [Google Scholar] [CrossRef]
- Zhai, R.; Ye, S.; Ye, J.; Wu, M.; Zhu, G.; Yu, F.; Wang, X.; Feng, Y.; Zhang, X. Glutaredoxin in Rice Growth, Development, and Stress Resistance: Mechanisms and Research Advances. Int. J. Mol. Sci. 2024, 24, 16968. [Google Scholar] [CrossRef]
- Liu, H.; Lyu, H.M.; Zhu, K.; Van de Peer, Y.; Max Cheng, Z.M. The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. Plant J. Cell Mol. Biol. 2021, 105, 1072–1082. [Google Scholar] [CrossRef]
- Jiang, D.; Yang, W.; Pi, J.; Yang, G.; Luo, Y.; Du, S.; Huang, L.J. Genome-Wide Identification, Characterization, and Expression Profiling of the Glutaredoxin Gene Family in Tea Plant (Camellia sinensis). Forests 2023, 14, 1647. [Google Scholar] [CrossRef]
- Garg, R.; Jhanwar, S.; Tyagi, A.K.; Jain, M. Genome-wide survey and expression analysis suggest diverse roles of glutaredoxin gene family members during development and response to various stimuli in rice. DNA Res. 2010, 17, 353–367. [Google Scholar] [CrossRef]
- Malik, W.A.; Wang, X.; Wang, X.; Shu, N.; Cui, R.; Chen, X.; Wang, D.; Lu, X.; Yin, Z.; Wang, J.; et al. Genome-wide expression analysis suggests glutaredoxin genes response to various stresses in cotton. Int. J. Biol. Macromol. 2020, 153, 470–491. [Google Scholar] [CrossRef] [PubMed]
- Shiade, S.R.; Zand-Silakhoor, A.; Fathi, A.; Rahimi, R.; Minkina, T.M.; Rajput, V.D.; Zulfiqar, U.; Chaudhary, T. Plant Metabolites and Signaling Pathways in Response to Biotic and Abiotic Stresses: Exploring Bio stimulant Applications. Plant Stress 2024, 12, 100454. [Google Scholar] [CrossRef]
- Liu, F.; Xi, M.; Liu, T.; Wu, X.; Ju, L.; Wang, D. The central role of transcription factors in bridging biotic and abiotic stress responses for plants’ resilience. New Crops 2023, 1, 100005. [Google Scholar] [CrossRef]
- Hu, Y.; Wu, Q.; Peng, Z.; Sprague, S.A.; Wang, W.; Park, J.; Akhunov, E.; Jagadish, K.S.V.; Nakata, P.A.; Cheng, N.; et al. Silencing of OsGRX17 in rice improves drought stress tolerance by modulating ROS accumulation and stomatal closure. Sci. Rep. 2017, 7, 15950. [Google Scholar] [CrossRef]
- Hou, J.; Sun, Q.; Li, J.; Ahammed, G.J.; Yu, J.; Fang, H.; Xia, X. Glutaredoxin S25 and its interacting TGACG motif-binding factor TGA2 mediate brassinosteroid-induced chlorothalonil metabolism in tomato plants. Environ. Pollut. 2019, 255 Pt 2, 113256. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.K.; Verma, S.; Tripathi, R.D.; Chakrabarty, D. A rice glutaredoxin regulate the expression of aquaporin genes and modulate root responses to provide arsenic tolerance. Ecotoxicol. Environ. Saf. 2020, 195, 110471. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Jin, L.; Sun, Y.; Zhan, C.; Tang, S.; Qin, T.; Liu, N.; Huang, J. OsNAC120 balances plant growth and drought tolerance by integrating GA and ABA signaling in rice. Plant Commun. 2024, 5, 100782. [Google Scholar] [CrossRef]
- Huang, L.J.; Yang, W.; Chen, J.; Yu, P.; Wang, Y.; Li, N. Molecular identification and functional characterization of an environmental stress responsive glutaredoxin gene ROXY1 in Quercus glauca. Plant Physiol. Biochem. 2024, 207, 108367. [Google Scholar] [CrossRef]
- Ning, X.; Sun, Y.; Wang, C.; Zhang, W.; Sun, M.; Hu, H.; Liu, J.; Yang, L. A Rice CPYC-Type Glutaredoxin OsGRX 20 in Protection against Bacterial Blight, Methyl Viologen and Salt Stresses. Front. Plant Sci. 2018, 9, 111. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, M.; Xu, X.; Dong, Y.; Bimpong, D.; Liu, L.; Li, Y.; Shen, H.; Wang, Y. The Roles of Glutaredoxins in Wheat (Triticum aestivum L.) under Biotic and Abiotic Stress Conditions, including Fungal and Hormone Treatments. Agronomy 2024, 14, 2057. https://doi.org/10.3390/agronomy14092057
Song M, Xu X, Dong Y, Bimpong D, Liu L, Li Y, Shen H, Wang Y. The Roles of Glutaredoxins in Wheat (Triticum aestivum L.) under Biotic and Abiotic Stress Conditions, including Fungal and Hormone Treatments. Agronomy. 2024; 14(9):2057. https://doi.org/10.3390/agronomy14092057
Chicago/Turabian StyleSong, Mengyuan, Xiao Xu, Ye Dong, Daniel Bimpong, Lijun Liu, Yanli Li, Huiquan Shen, and Youning Wang. 2024. "The Roles of Glutaredoxins in Wheat (Triticum aestivum L.) under Biotic and Abiotic Stress Conditions, including Fungal and Hormone Treatments" Agronomy 14, no. 9: 2057. https://doi.org/10.3390/agronomy14092057
APA StyleSong, M., Xu, X., Dong, Y., Bimpong, D., Liu, L., Li, Y., Shen, H., & Wang, Y. (2024). The Roles of Glutaredoxins in Wheat (Triticum aestivum L.) under Biotic and Abiotic Stress Conditions, including Fungal and Hormone Treatments. Agronomy, 14(9), 2057. https://doi.org/10.3390/agronomy14092057






