Genome-Wide Identification and Analysis of the Aux/IAA Gene Family in Rosa hybrida—“The Fairy”: Evidence for the Role of RhIAA25 in Adventitious Root Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification of the Aux/IAA Gene Family in R. hybrida
2.3. Analysis of the Phylogenetic Tree, Conserved Domain, Motif, and Gene Structure of RhAux/IAA
2.4. Chromosomal Localization and Collinearity Analysis of RhAux/IAA
2.5. Analysis of Cis-Acting Elements in RhAux/IAA Genes
2.6. Analysis of Expression of RhAux/IAA Genes during Rooting Based on RNA-Seq
2.7. Quantitative Real-Time PCR
2.8. Subcellular Localization
2.9. Yeast Self-Activation Analysis of RhIAA25
2.10. Genetic Transformation and Identification of Transgenic RhIAA25 in A. thaliana
2.11. Statistical Analyses
3. Results
3.1. Identification of RhAux/IAA Genes
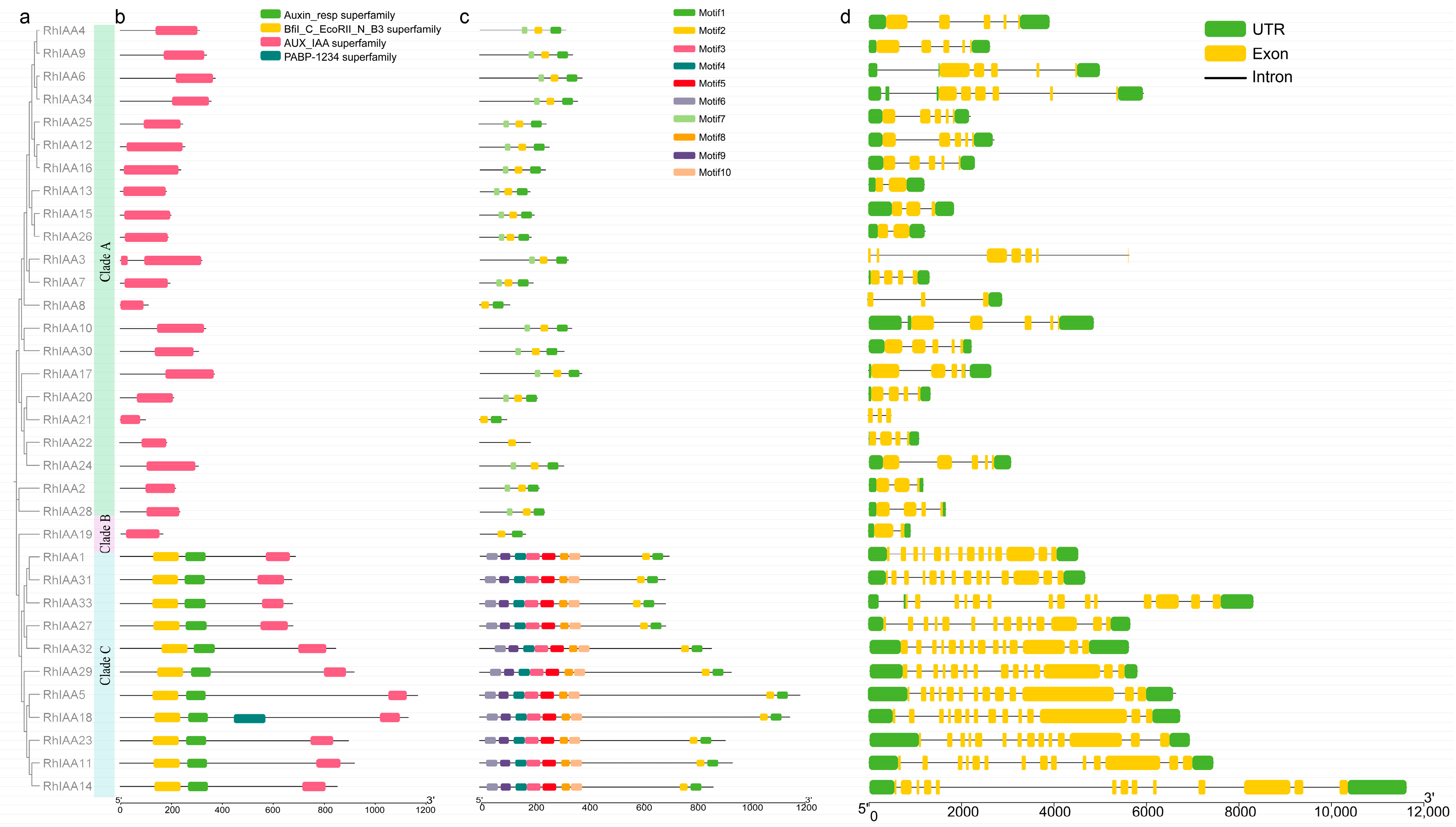
3.2. Phylogenetic Analysis of RhAux/IAA Genes
3.3. Gene Structure Analysis of RhAux/IAA Family Members
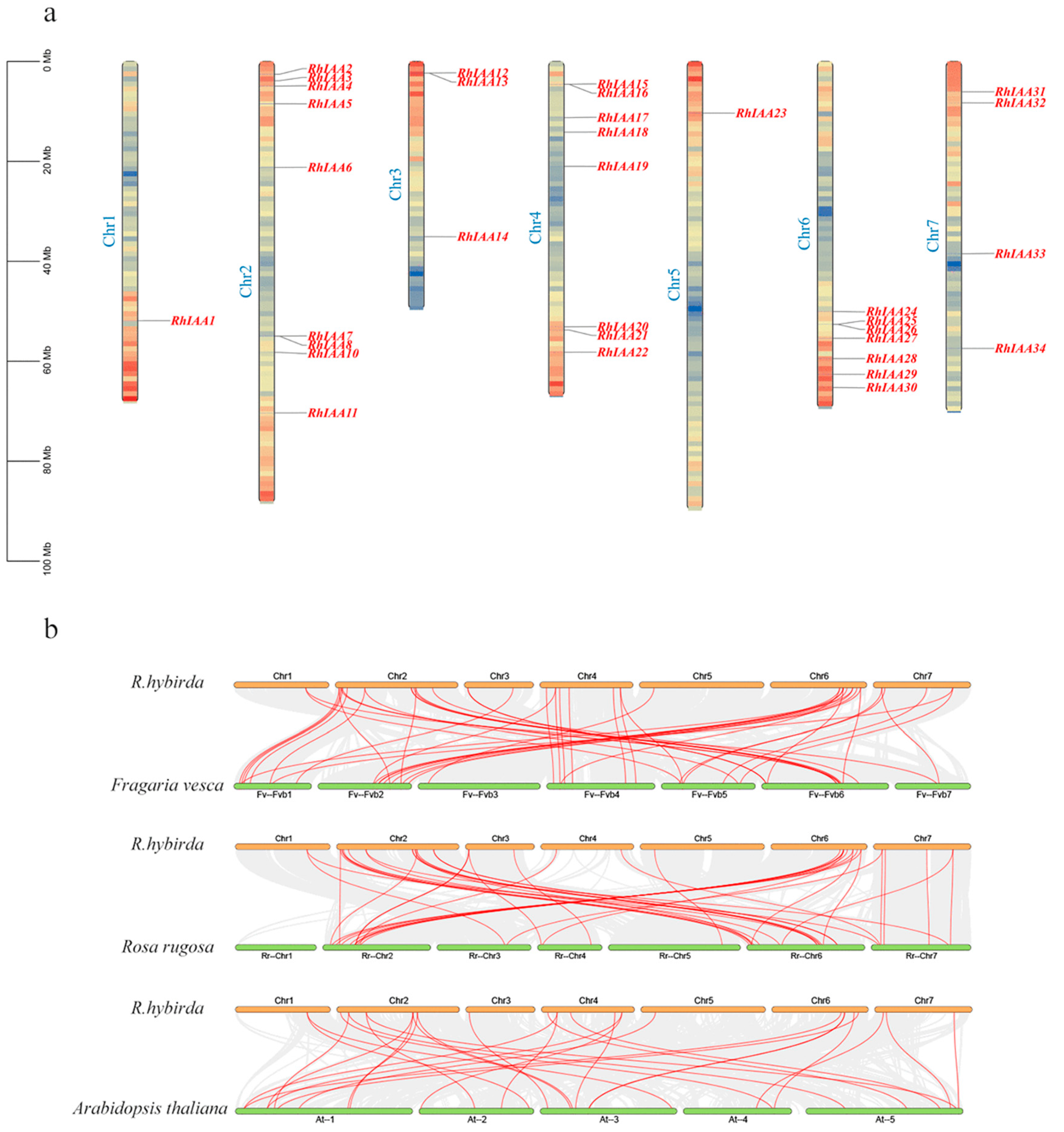
3.4. The Chromosomal Localization and Collinearity Analysis of the RhAux/IAA Genes
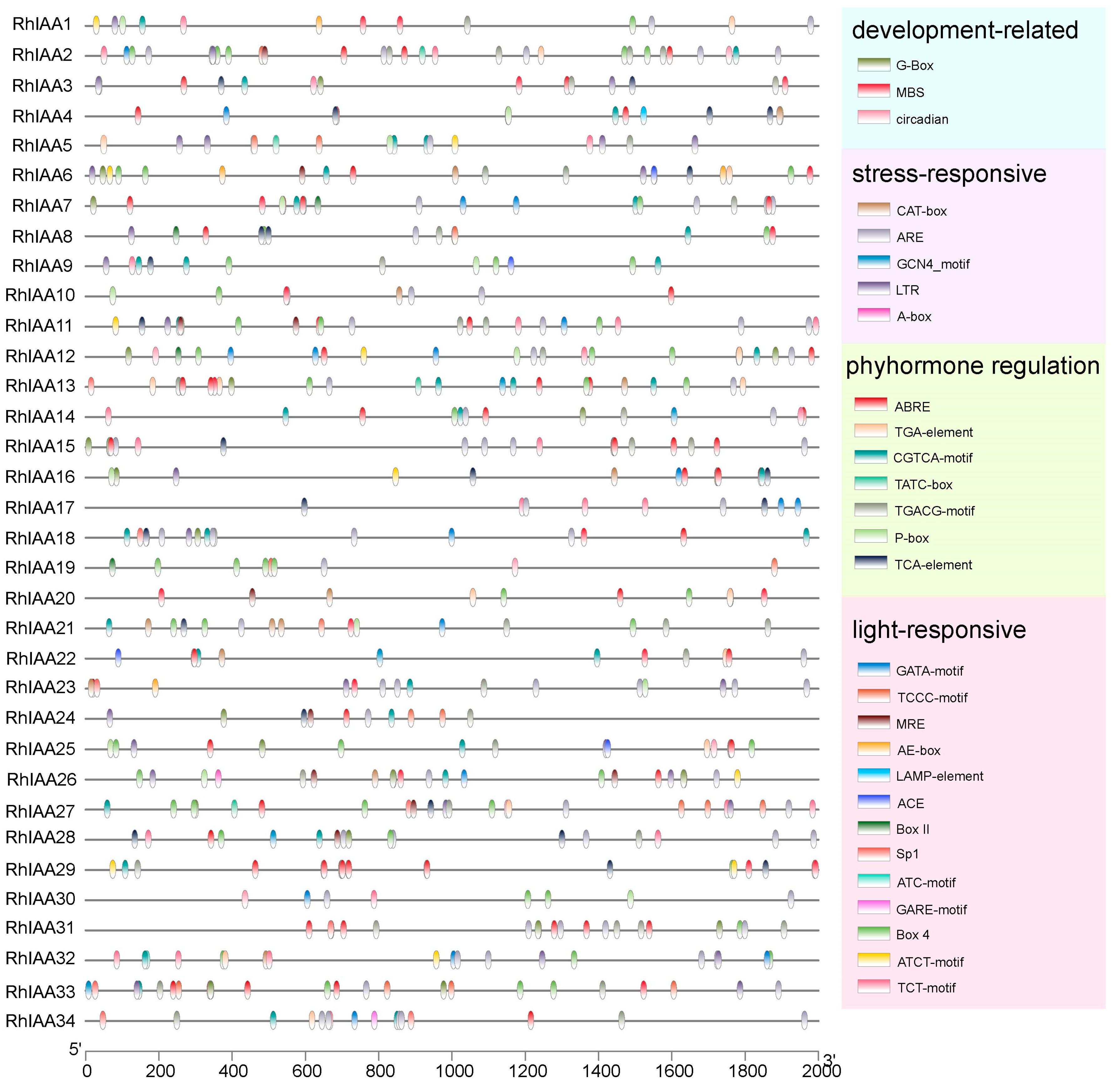
3.5. Cis-Elements in the Promoters of RhAux/IAA Genes
3.6. The Expression Pattern of RhAux/IAA Genes during Adventitious Root Development
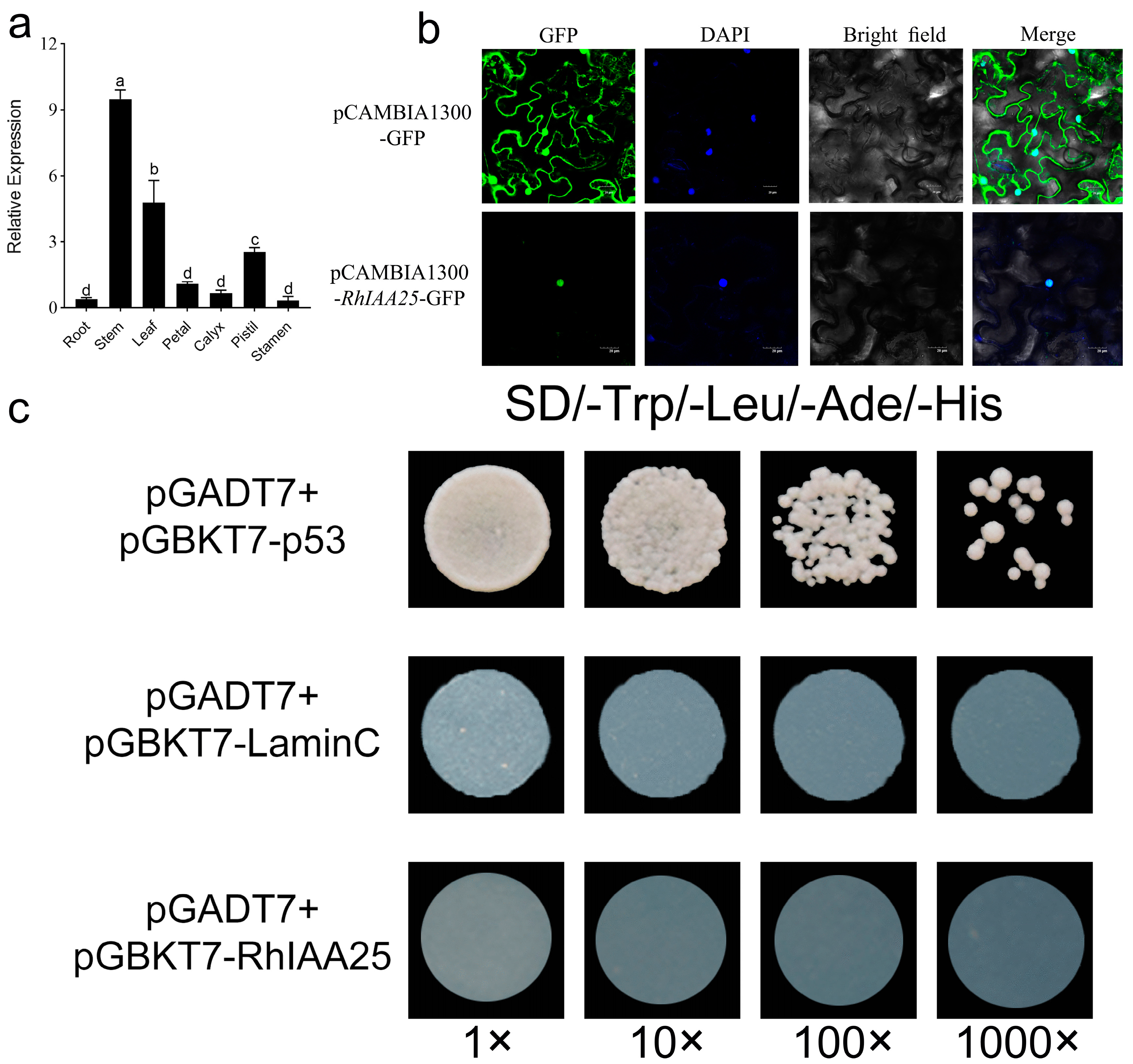
3.7. The Expression Pattern and Molecular Characteristics of RhIAA25
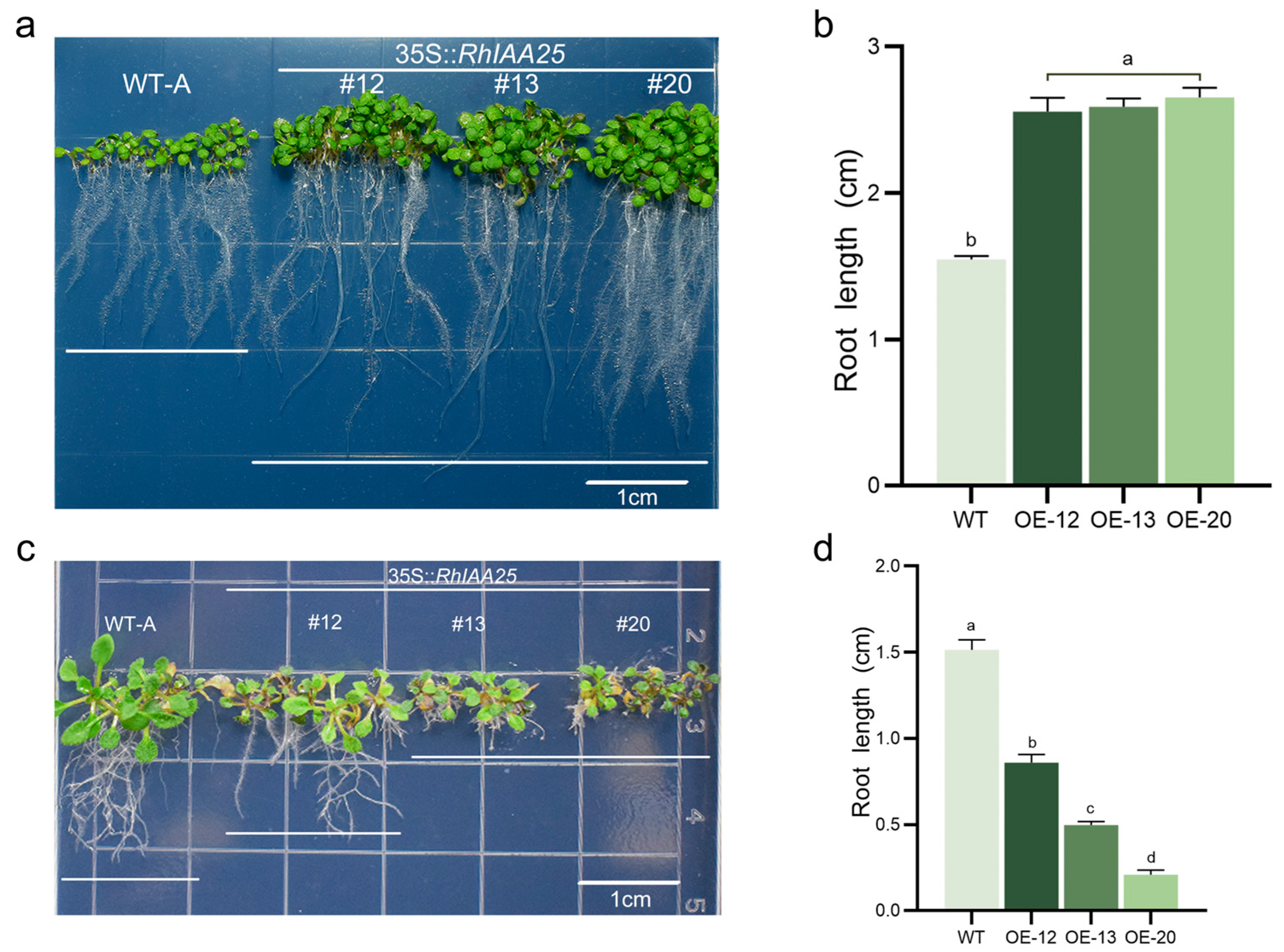
3.8. The Effect of Overexpressing RhIAA25 on Arabidopsis Root Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Wu, X.; Wang, X.; Dai, M.; Peng, Y. Crop root system architecture in drought response. J. Genet. Genom. 2024, in press. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zeng, R.; Liao, H. Improving crop nutrient efficiency through root architecture modifications. J. Integr. Plant Biol. 2016, 58, 193–202. [Google Scholar] [CrossRef]
- Bellini, C.; Pacurar, D.I.; Perrone, I. Adventitious roots and lateral roots: Similarities and differences. Annu. Rev. Plant Biol. 2014, 65, 639–666. [Google Scholar] [CrossRef]
- Mhimdi, M.; Pérez-Pérez, J.M. Understanding of Adventitious Root Formation: What Can We Learn From Comparative Genetics? Front. Plant Sci. 2020, 11, 582020. [Google Scholar] [CrossRef]
- Ermel, F.F.; Vizoso, S.; Charpentier, J.P.; Jay-Allemand, C.; Catesson, A.M.; Couée, I. Mechanisms of primordium formation during adventitious root development from walnut cotyledon explants. Planta 2000, 211, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Urquhart, S.; Foo, E.; Reid, J.B. The role of strigolactones in photomorphogenesis of pea is limited to adventitious rooting. Physiol. Plant 2015, 153, 392–402. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, A.; Gobin, B.; Van Huylenbroeck, J.; Van Labeke, M.-C. Adventitious rooting of Chrysanthemum is stimulated by a low red:far-red ratio. J. Plant Physiol. 2019, 236, 117–123. [Google Scholar] [CrossRef]
- King, J.J.; Stimart, D.P.; Fisher, R.H.; Bleecker, A.B. A Mutation Altering Auxin Homeostasis and Plant Morphology in Arabidopsis. Plant Cell 1995, 7, 2023–2037. [Google Scholar] [CrossRef]
- Bai, T.; Dong, Z.; Zheng, X.; Song, S.; Jiao, J.; Wang, M.; Song, C. Auxin and Its Interaction with Ethylene Control Adventitious Root Formation and Development in Apple Rootstock. Front. Plant Sci. 2020, 11, 574881. [Google Scholar] [CrossRef]
- Li, Y.; Han, S.; Qi, Y. Advances in structure and function of auxin response factor in plants. J. Integr. Plant Biol. 2023, 65, 617–632. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.-J.; Zhang, J.-Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef] [PubMed]
- Fukaki, H.; Tameda, S.; Masuda, H.; Tasaka, M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002, 29, 153–168. [Google Scholar] [CrossRef]
- Leyser, H.M.; Pickett, F.B.; Dharmasiri, S.; Estelle, M. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 1996, 10, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Rogg, L.E.; Lasswell, J.; Bartel, B. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 2001, 13, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Umemura, I.; Gomi, K.; Hasegawa, Y.; Kitano, H.; Sazuka, T.; Matsuoka, M. Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J. 2006, 46, 297–306. [Google Scholar] [CrossRef]
- Ni, J.; Wang, G.; Zhu, Z.; Zhang, H.; Wu, Y.; Wu, P. OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice. Plant J. 2011, 68, 433–442. [Google Scholar] [CrossRef]
- Deng, W.; Yan, F.; Liu, M.; Wang, X.; Li, Z. Down-regulation of SlIAA15 in tomato altered stem xylem development and production of volatile compounds in leaf exudates. Plant Signal. Behav. 2012, 7, 911–913. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Di, P.; Wang, Y. Genome-Wide Identification and Analysis of the Aux/IAA Gene Family in Panax ginseng: Evidence for the Role of PgIAA02 in Lateral Root Development. Int. J. Mol. Sci. 2024, 25, 3470. [Google Scholar] [CrossRef]
- Jung, H.; Lee, D.-K.; Choi, Y.D.; Kim, J.-K. OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci. 2015, 236, 304–312. [Google Scholar] [CrossRef]
- Zhang, A.; Yang, X.; Lu, J.; Song, F.; Sun, J.; Wang, C.; Lian, J.; Zhao, L.; Zhao, B. OsIAA20, an Aux/IAA protein, mediates abiotic stress tolerance in rice through an ABA pathway. Plant Sci. 2021, 308, 110903. [Google Scholar] [CrossRef]
- Raymond, O.; Gouzy, J.; Just, J.; Badouin, H.; Verdenaud, M.; Lemainque, A.; Vergne, P.; Moja, S.; Choisne, N.; Pont, C.; et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 2018, 50, 772–777. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Gao, P.; Zhou, T.; Pang, S.; Zhang, J.; Yang, T.; Zhang, W.; Dong, J.; Che, D. Genome-Wide Identification and Expression Analysis of the Trehalose-6-phosphate Synthase and Trehalose-6-phosphate Phosphatase Gene Families in Rose (Rosa hybrida cv ‘Carola’) under Different Light Conditions. Plants 2024, 13, 114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, J.; Fan, Y.; Dong, J.; Gao, P.; Jiang, W.; Yang, T.; Che, D. RNA sequencing analysis reveals PgbHLH28 as the key regulator in response to methyl jasmonate-induced saponin accumulation in Platycodon grandiflorus. Hortic. Res. 2024, 11, uhae058. [Google Scholar] [CrossRef]
- Tsuda, K.; Qi, Y.; Nguyen, L.V.; Bethke, G.; Tsuda, Y.; Glazebrook, J.; Katagiri, F. An efficient Agrobacterium-mediated transient transformation of Arabidopsis. Plant J. 2012, 69, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Guruprasad, K.; Reddy, B.V.; Pandit, M.W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990, 4, 155–161. [Google Scholar] [CrossRef]
- Lakehal, A.; Chaabouni, S.; Cavel, E.; Le Hir, R.; Ranjan, A.; Raneshan, Z.; Novák, O.; Păcurar, D.I.; Perrone, I.; Jobert, F.; et al. A Molecular Framework for the Control of Adventitious Rooting by TIR1/AFB2-Aux/IAA-Dependent Auxin Signaling in Arabidopsis. Mol. Plant 2019, 12, 1499–1514. [Google Scholar] [CrossRef]
- Okushima, Y.; Fukaki, H.; Onoda, M.; Theologis, A.; Tasaka, M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 2007, 19, 118–130. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Goldfarb, B.; Lanz-Garcia, C.; Lian, Z.; Whetten, R. Aux/IAA gene family is conserved in the gymnosperm, loblolly pine (Pinus taeda). Tree Physiol. 2003, 23, 1181–1192. [Google Scholar] [CrossRef] [PubMed]
- Rouse, D.; Mackay, P.; Stirnberg, P.; Estelle, M.; Leyser, O. Changes in auxin response from mutations in an Aux/IAA gene. Science 1998, 279, 1371–1373. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liu, Y.; Zhang, S.; Shui, D.; Xia, Z.; Sun, J. Genome-wide identification and expression analysis of the Aux/IAA gene family in turnip (Brassica rapa ssp. rapa). BMC Plant Biol. 2023, 23, 342. [Google Scholar] [CrossRef]
- Su, Y.; He, H.; Wang, P.; Ma, Z.; Mao, J.; Chen, B. Genome-wide characterization and expression analyses of the auxin/indole-3-acetic acid (Aux/IAA) gene family in apple (Malus domestica). Gene 2021, 768, 145302. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Feng, G.; Yang, Z.; Wu, J.; Liu, B.; Xu, X.; Nie, G.; Huang, L.; Zhang, X. Genome-Wide Characterization of the Aux/IAA Gene Family in Orchardgrass and a Functional Analysis of DgIAA21 in Responding to Drought Stress. Int. J. Mol. Sci. 2023, 24, 16184. [Google Scholar] [CrossRef]
- Huang, D.; Wang, Q.; Duan, D.; Dong, Q.; Zhao, S.; Zhang, M.; Jing, G.; Liu, C.; van Nocker, S.; Ma, F.; et al. Overexpression of MdIAA9 confers high tolerance to osmotic stress in transgenic tobacco. PeerJ 2019, 7, e7935. [Google Scholar] [CrossRef]
- Zeng, Y.; Verstraeten, I.; Trinh, H.K.; Lardon, R.; Schotte, S.; Olatunji, D.; Heugebaert, T.; Stevens, C.; Quareshy, M.; Napier, R.; et al. Chemical induction of hypocotyl rooting reveals extensive conservation of auxin signalling controlling lateral and adventitious root formation. New Phytol. 2023, 240, 1883–1899. [Google Scholar] [CrossRef]
- Yamauchi, T.; Tanaka, A.; Inahashi, H.; Nishizawa, N.K.; Tsutsumi, N.; Inukai, Y.; Nakazono, M. Fine control of aerenchyma and lateral root development through AUX/IAA- and ARF-dependent auxin signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 20770–20775. [Google Scholar] [CrossRef]
- Edelmann, H.G. Plant root development: Is the classical theory for auxin-regulated root growth false. Protoplasma 2022, 259, 823–832. [Google Scholar] [CrossRef]
- Luo, S.; Li, Q.; Liu, S.; Pinas, N.M.; Tian, H.; Wang, S. Constitutive Expression of OsIAA9 Affects Starch Granules Accumulation and Root Gravitropic Response in Arabidopsis. Front. Plant Sci. 2015, 6, 1156. [Google Scholar] [CrossRef]
- Si, C.; Zeng, D.; da Silva, J.A.T.; Qiu, S.; Duan, J.; Bai, S.; He, C. Genome-wide identification of Aux/IAA and ARF gene families reveal their potential roles in flower opening of Dendrobium officinale. BMC Genom. 2023, 24, 199. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Shad, M.A.; Shu, Y.; Nong, S.; Li, X.; Wu, S.; Yang, J.; Rao, M.J.; Aslam, M.Z.; Huang, X.; et al. Genome-Wide Analysis of the Auxin/Indoleacetic Acid (Aux/IAA) Gene Family in Autopolyploid Sugarcane (Saccharum spontaneum). Int. J. Mol. Sci. 2024, 25, 7473. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Zhang, L.; Zhu, R.; Jiang, X.; Yue, C.; Su, Y.; Ren, H.; Wang, M. A Gain-of-Function Mutant of IAA7 Inhibits Stem Elongation by Transcriptional Repression of EXPA5 Genes in Brassica napus. Int. J. Mol. Sci. 2021, 22, 9018. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | Isoelectric Point (pl) | Molecular Weight (Da) | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| RhIAA1 | Chr1g0360041 | 6.57 | 76,056.96 | 52.02 | 70.87 | −0.444 | Nucleus |
| RhIAA2 | Chr2g0087791 | 8.40 | 24,832.97 | 47.94 | 70.51 | −0.780 | Nucleus |
| RhIAA3 | Chr2g0089801 | 8.71 | 36,549.63 | 40.36 | 63.03 | −0.678 | Nucleus |
| RhIAA4 | Chr2g0091261 | 7.59 | 33,159.66 | 45.32 | 69.00 | −0.427 | Nucleus |
| RhIAA5 | Chr2g0095551 | 6,22 | 130,207.21 | 69,93 | 69.70 | −0.707 | Nucleus |
| RhIAA6 | Chr2g0109881 | 8.09 | 40,175.07 | 48.94 | 61.08 | −0.601 | Nucleus |
| RhIAA7 | Chr2g0137301 | 6.60 | 21,507.46 | 58.19 | 71.95 | −0.457 | Nucleus |
| RhIAA8 | Chr2g0137311 | 5.15 | 12,409.52 | 46.65 | 78.91 | −0.112 | Nucleus |
| RhIAA9 | Chr2g0139301 | 8.09 | 37,084.92 | 47.92 | 62.90 | −0.615 | Nucleus |
| RhIAA10 | Chr2g0140681 | 9.16 | 36,783.42 | 52.43 | 68.39 | −0.699 | Nucleus |
| RhIAA11 | Chr2g0152951 | 6.00 | 101,532.47 | 66.77 | 72.86 | −0.490 | Nucleus |
| RhIAA12 | Chr3g0451071 | 6.05 | 27,871.51 | 33.99 | 67.83 | −0.596 | Nucleus |
| RhIAA13 | Chr3g0451081 | 8.24 | 20,409.43 | 45.62 | 60.88 | −0.646 | Nucleus |
| RhIAA14 | Chr3g0487771 | 5.79 | 94,404.83 | 66.83 | 69.33 | −0.509 | Nucleus |
| RhIAA15 | Chr4g0389611 | 6.41 | 21,983.12 | 41.43 | 75.98 | −0.531 | Nucleus |
| RhIAA16 | Chr4g0389621 | 8.59 | 26,427.61 | 42.17 | 67.47 | −0.470 | Nucleus |
| RhIAA17 | Chr4g0395511 | 8.18 | 39,694.86 | 38.55 | 72.41 | −0.301 | Nucleus |
| RhIAA18 | Chr4g0397771 | 6.24 | 124,586.58 | 64.85 | 70.86 | −0.598 | Nucleus |
| RhIAA19 | Chr4g0402441 | 9.65 | 18,232.67 | 65.45 | 86.91 | −0.478 | Nucleus |
| RhIAA20 | Chr4g0428011 | 7.19 | 23,209.94 | 50.72 | 79.38 | −0.505 | Nucleus |
| RhIAA21 | Chr4g0428791 | 5.76 | 11,445.23 | 20.32 | 89.49 | −0.249 | Nucleus |
| RhIAA22 | Chr4g0434391 | 5.00 | 21,066.55 | 49.22 | 81.08 | −0.481 | Nucleus |
| RhIAA23 | Chr5g0014961 | 6.04 | 98,764.09 | 65.78 | 73.67 | −0.434 | Nucleus |
| RhIAA24 | Chr6g0286601 | 6.34 | 32,064.59 | 39.66 | 66.08 | −0.394 | Nucleus |
| RhIAA25 | Chr6g0289571 | 6.46 | 26,908.69 | 42.57 | 62.08 | −0.629 | Nucleus |
| RhIAA26 | Chr6g0289581 | 5.22 | 21,129.74 | 49.08 | 59.10 | −0.748 | Nucleus |
| RhIAA27 | Chr6g0292551 | 5.83 | 75,288.01 | 60.28 | 70.36 | −0.411 | Nucleus |
| RhIAA28 | Chr6g0297821 | 5.93 | 26,495.80 | 37.58 | 72.01 | −0.712 | Nucleus |
| RhIAA29 | Chr6g0302551 | 5.22 | 100,600.14 | 57.27 | 74.22 | −0.391 | Nucleus |
| RhIAA30 | Chr6g0306631 | 8.21 | 34,123.51 | 54.45 | 70.59 | −0.721 | Nucleus |
| RhIAA31 | Chr7g0186081 | 6.52 | 74,700.30 | 51.15 | 72.81 | −0.482 | Nucleus |
| RhIAA32 | Chr7g0188911 | 6.15 | 93,554.81 | 53.60 | 65.06 | −0.656 | Nucleus |
| RhIAA33 | Chr7g0219771 | 6.03 | 75,041.53 | 60.25 | 70.89 | −0.488 | Nucleus |
| RhIAA34 | Chr7g0233101 | 7.05 | 39,188.39 | 47.89 | 71.21 | −0.578 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Zhang, Y.; Huangfu, M.; Fan, Y.; Zhang, J.; Yang, T.; Che, D.; Dong, J. Genome-Wide Identification and Analysis of the Aux/IAA Gene Family in Rosa hybrida—“The Fairy”: Evidence for the Role of RhIAA25 in Adventitious Root Development. Agronomy 2024, 14, 2005. https://doi.org/10.3390/agronomy14092005
Zhang W, Zhang Y, Huangfu M, Fan Y, Zhang J, Yang T, Che D, Dong J. Genome-Wide Identification and Analysis of the Aux/IAA Gene Family in Rosa hybrida—“The Fairy”: Evidence for the Role of RhIAA25 in Adventitious Root Development. Agronomy. 2024; 14(9):2005. https://doi.org/10.3390/agronomy14092005
Chicago/Turabian StyleZhang, Wuhua, Yifei Zhang, Minge Huangfu, Yingdong Fan, Jinzhu Zhang, Tao Yang, Daidi Che, and Jie Dong. 2024. "Genome-Wide Identification and Analysis of the Aux/IAA Gene Family in Rosa hybrida—“The Fairy”: Evidence for the Role of RhIAA25 in Adventitious Root Development" Agronomy 14, no. 9: 2005. https://doi.org/10.3390/agronomy14092005
APA StyleZhang, W., Zhang, Y., Huangfu, M., Fan, Y., Zhang, J., Yang, T., Che, D., & Dong, J. (2024). Genome-Wide Identification and Analysis of the Aux/IAA Gene Family in Rosa hybrida—“The Fairy”: Evidence for the Role of RhIAA25 in Adventitious Root Development. Agronomy, 14(9), 2005. https://doi.org/10.3390/agronomy14092005





