Moving toward Automaticity: A Robust Synthetic Occlusion Image Method for High-Throughput Mushroom Cap Phenotype Extraction
Abstract
1. Introduction
- (1)
- A novel edible mushroom synthetic cap occlusion image dataset generation method was proposed, which could automatically generate synthetic occlusion images and corresponding annotation in the realm of image processing. The method could also be applied to generate other amodal instance segmentation datasets and could effectively solve the problem in which the amodal ground truth cannot be obtained and greatly reduce the time for data collection and data annotation.
- (2)
- Based on the method of generating a synthetic image dataset, an Oudemansiella raphanipes, Agrocybe cylindraceas and Pholiota nameko amodal instance segmentation dataset was proposed to simulate a real-world dataset. It is the first synthetic edible mushroom image dataset for cap amodal instance segmentation.
- (3)
- An amodal mask-based method was proposed for calculating the width and length of caps. To the best of our knowledge, this is the first work that applies amodal instance segmentation to measure the width and length of caps based on synthetic training.
2. Materials and Methods
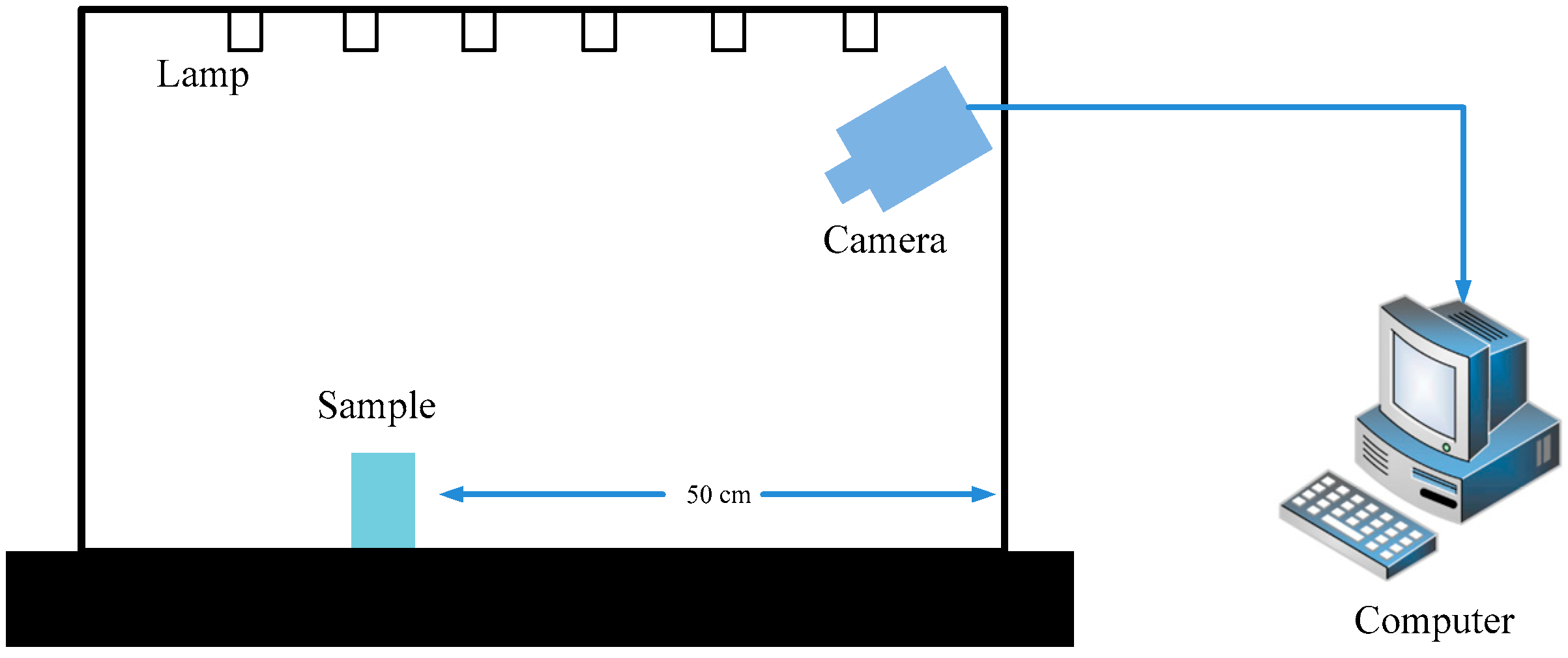
2.1. Raw Data Acquisition
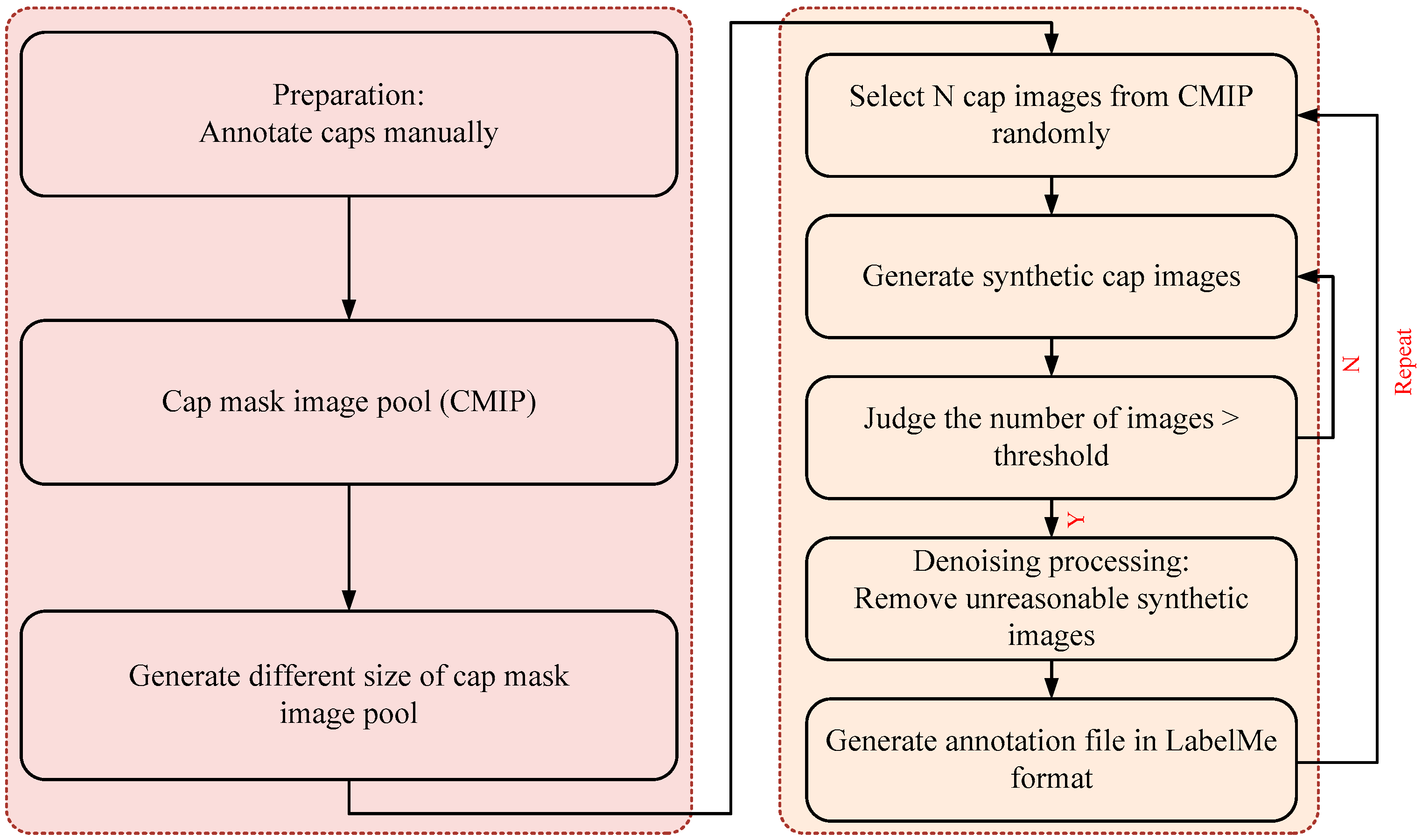
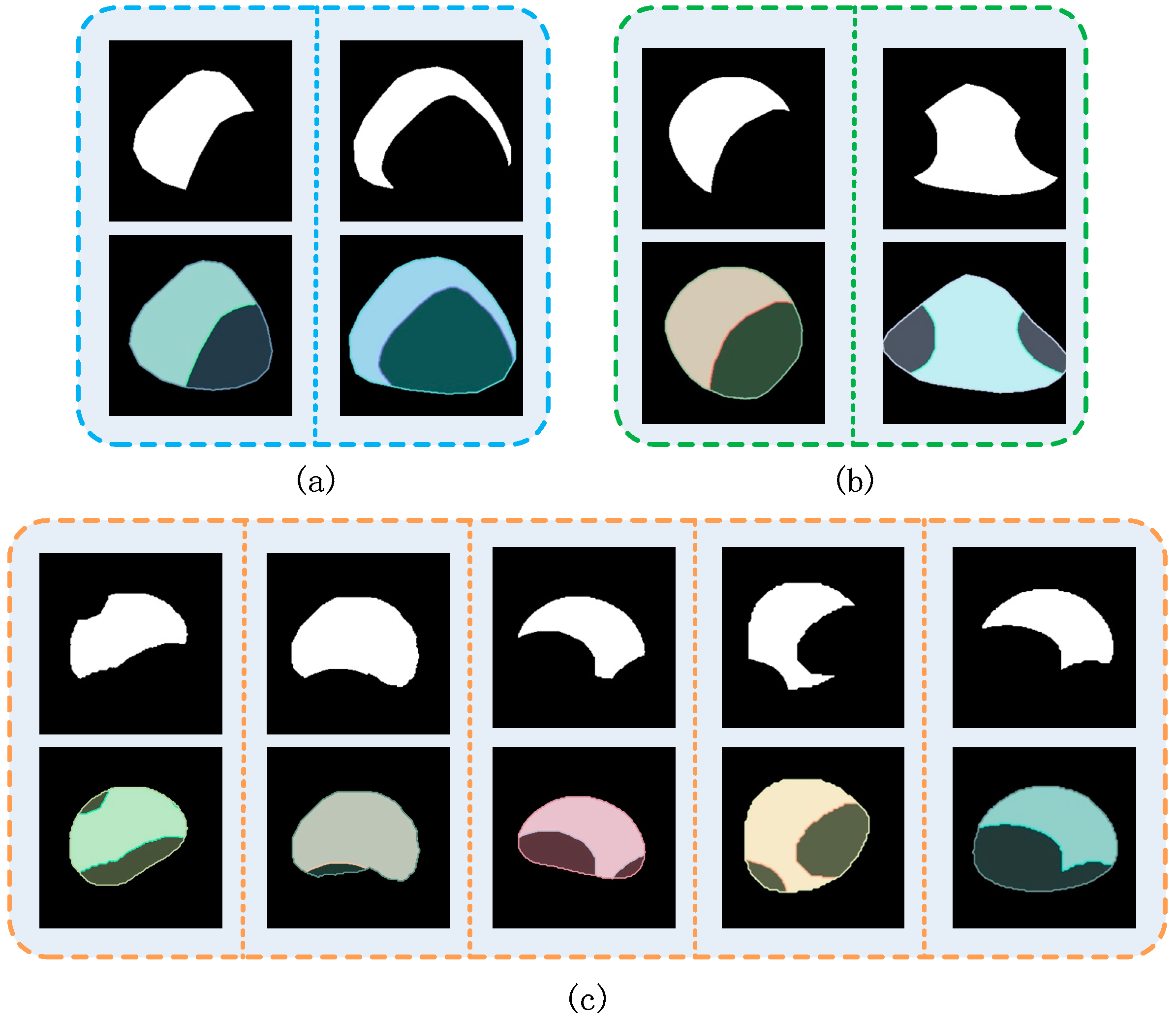
2.2. Synthetic Image Generation and Annotation Method
- (1)
- Select a cap mask image, , randomly as occluder and another . Images from CMIP were occluded.
- (2)
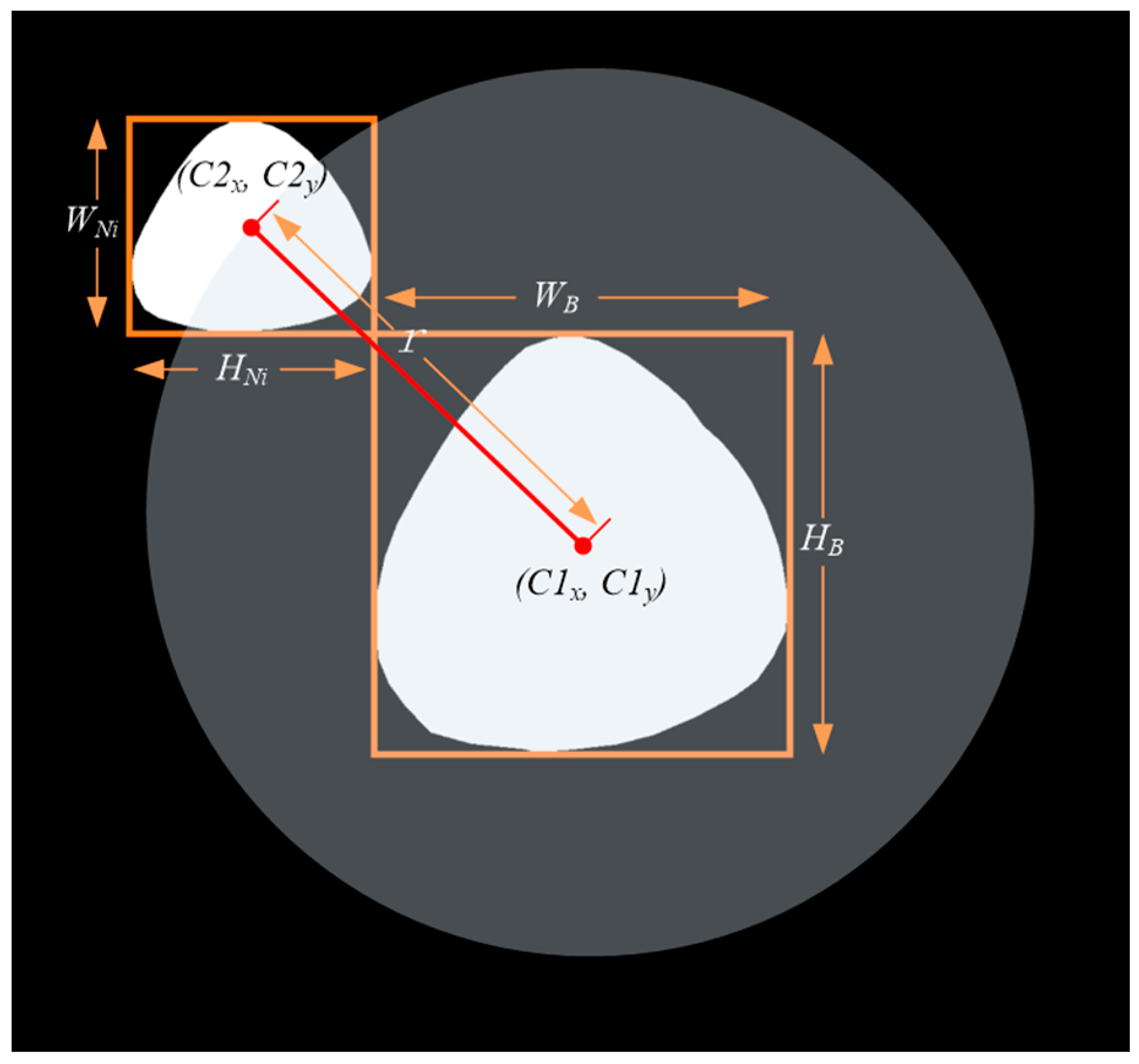
- As shown in Figure 4, calculate the position of the cap; we considered , , , , respectively, as the width and height of the bounding box of and . Furthermore, we set a movable area based on Equation (1) and generated a random point in the area of the circle with radius as the initial move position of the . The center of was mapped on according to the random point and then a mapped image was obtained.
- (3)
- Occlusion processing. Based on step (2), iterate over the point of and ; we considered that if the point belongs both to and , then it is the occluded point, and then we removed it.
- (4)
- Denoising processing. As the synthetic image after step (3) sometimes has more than one region, we only maintained the largest region. Finally, combine the synthetic images and the Json file when annotated manually to generate a new Json file, which is suitable for LabelMe.
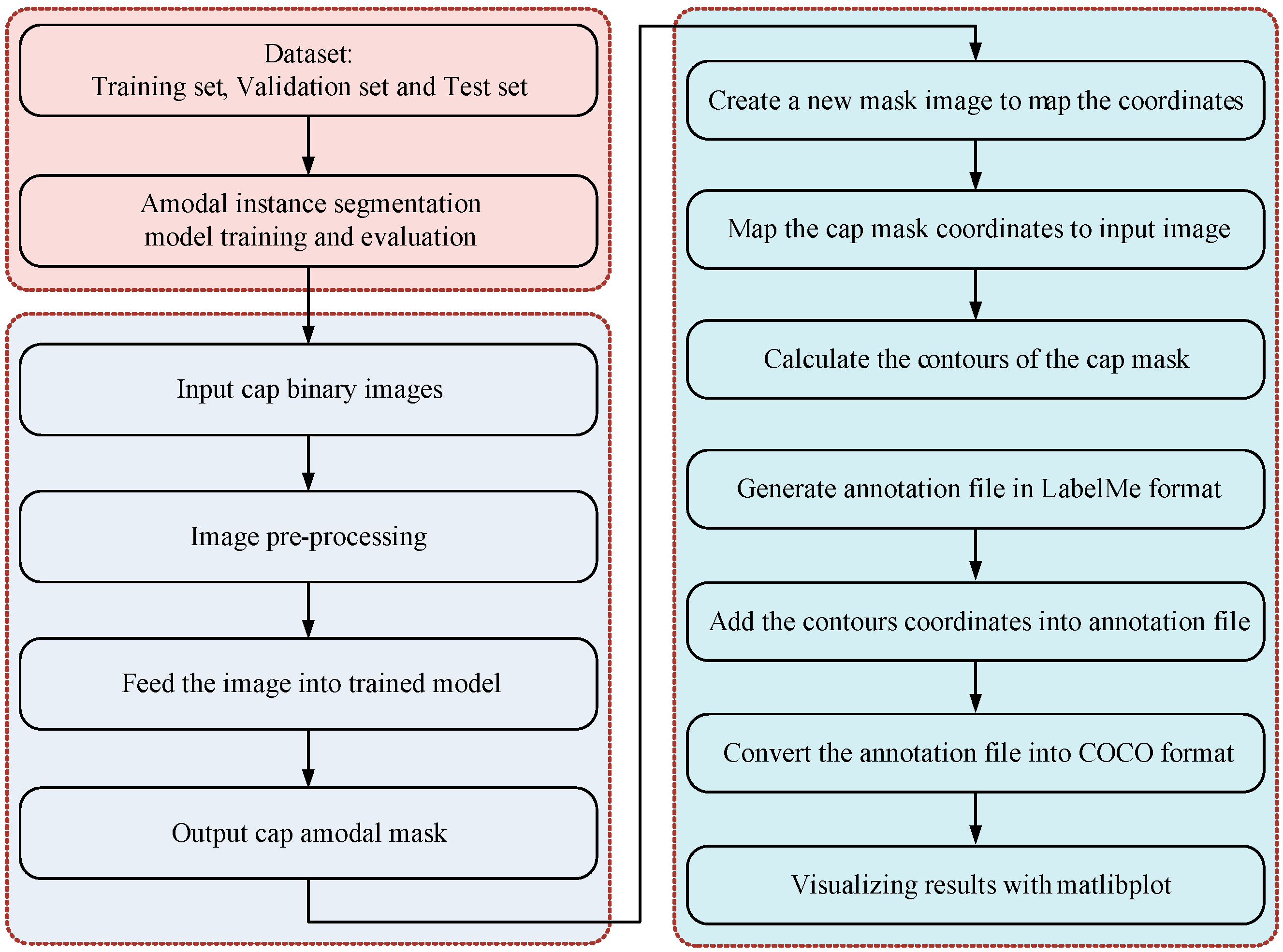
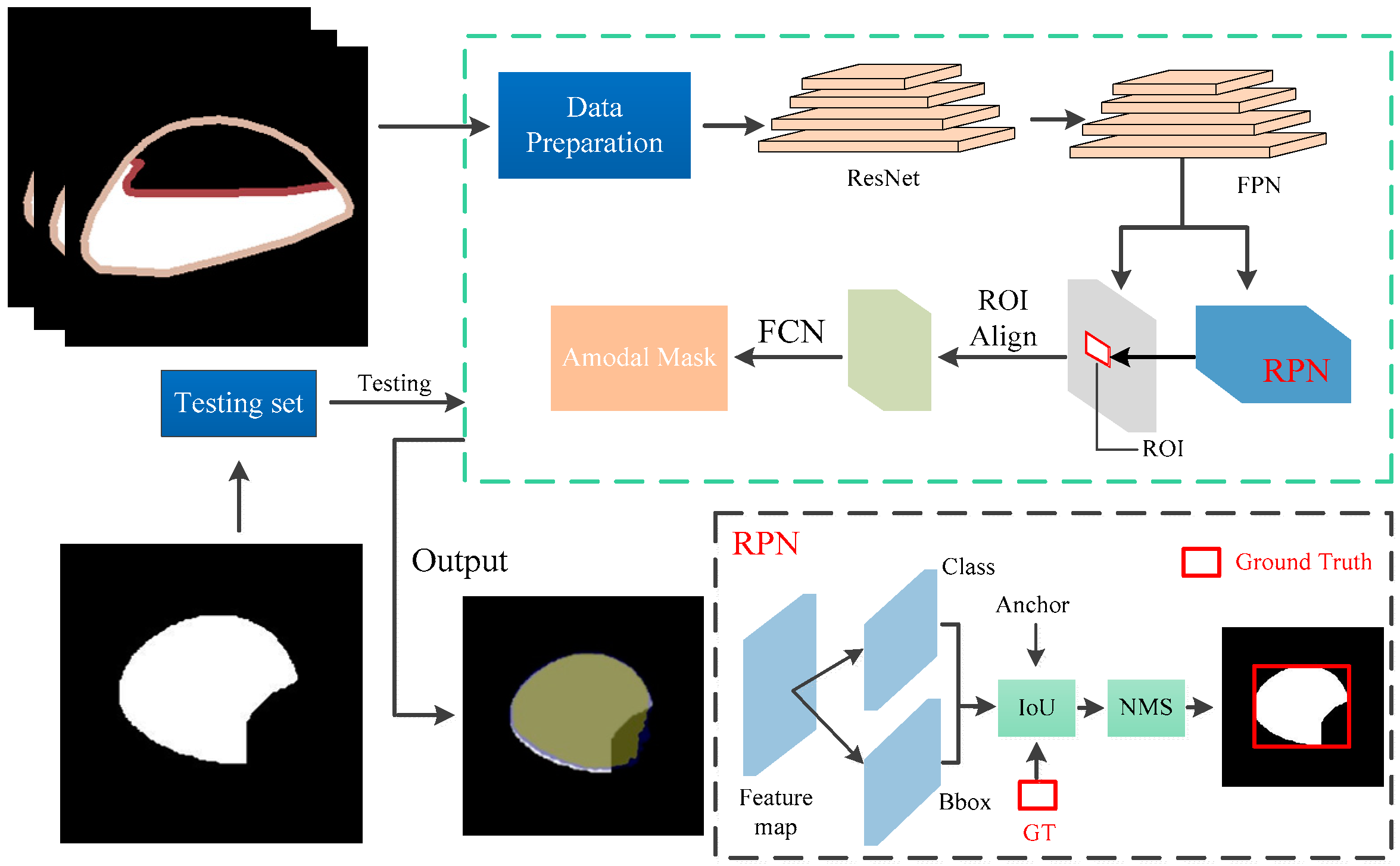
2.3. Amodal Instance Segmentation and Size Estimation
2.3.1. Occlusion R-CNN
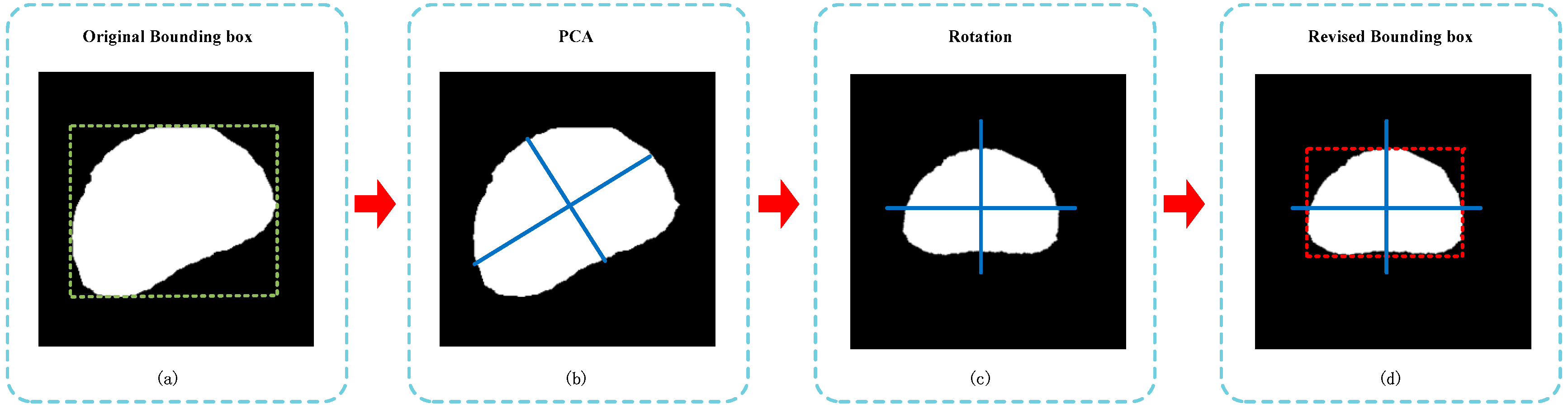
2.3.2. Size Estimation
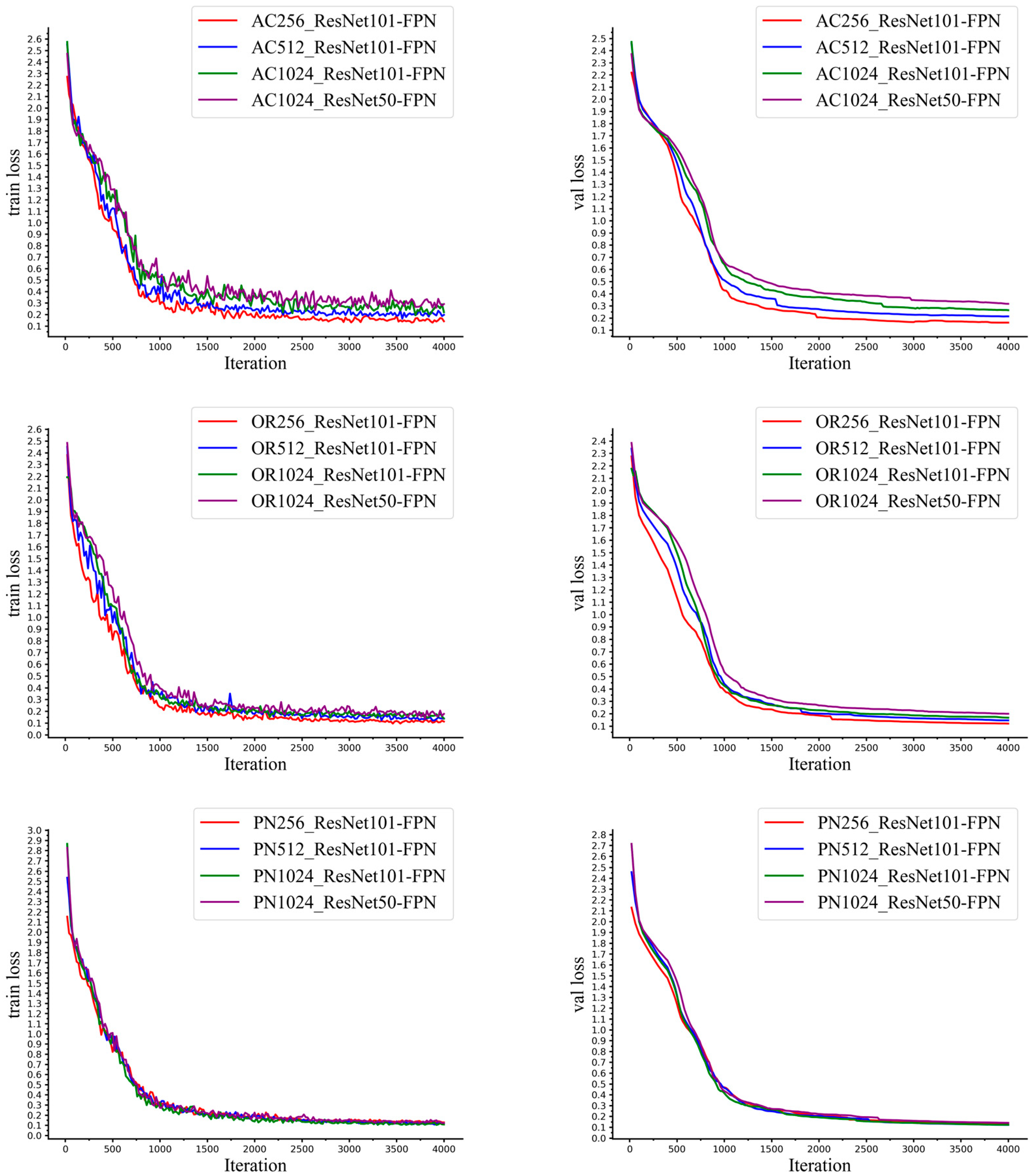
2.4. Implementation Details
2.5. Evaluation Metrics
3. Results
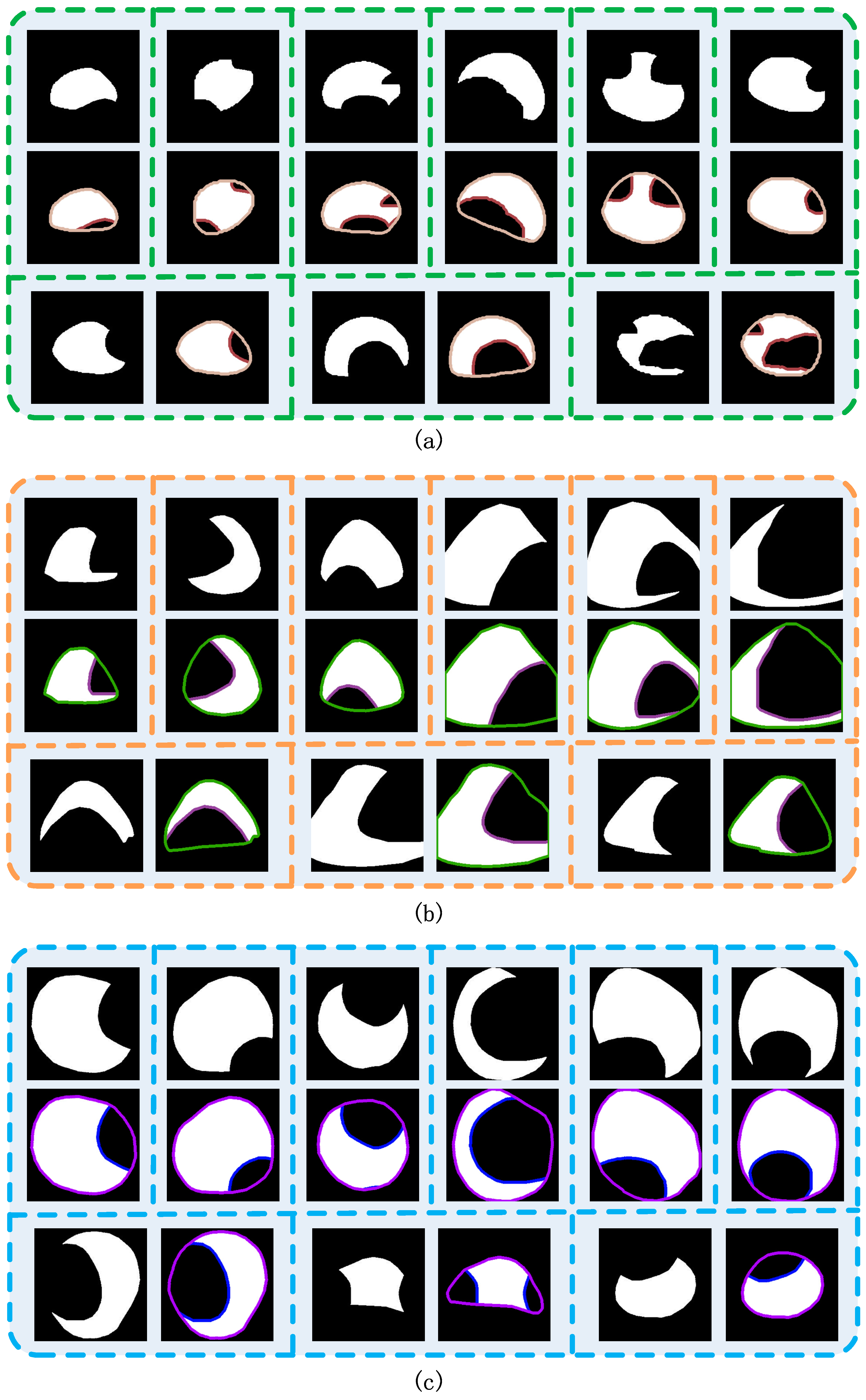
3.1. Synthetic Cap Occlusion Image Dataset
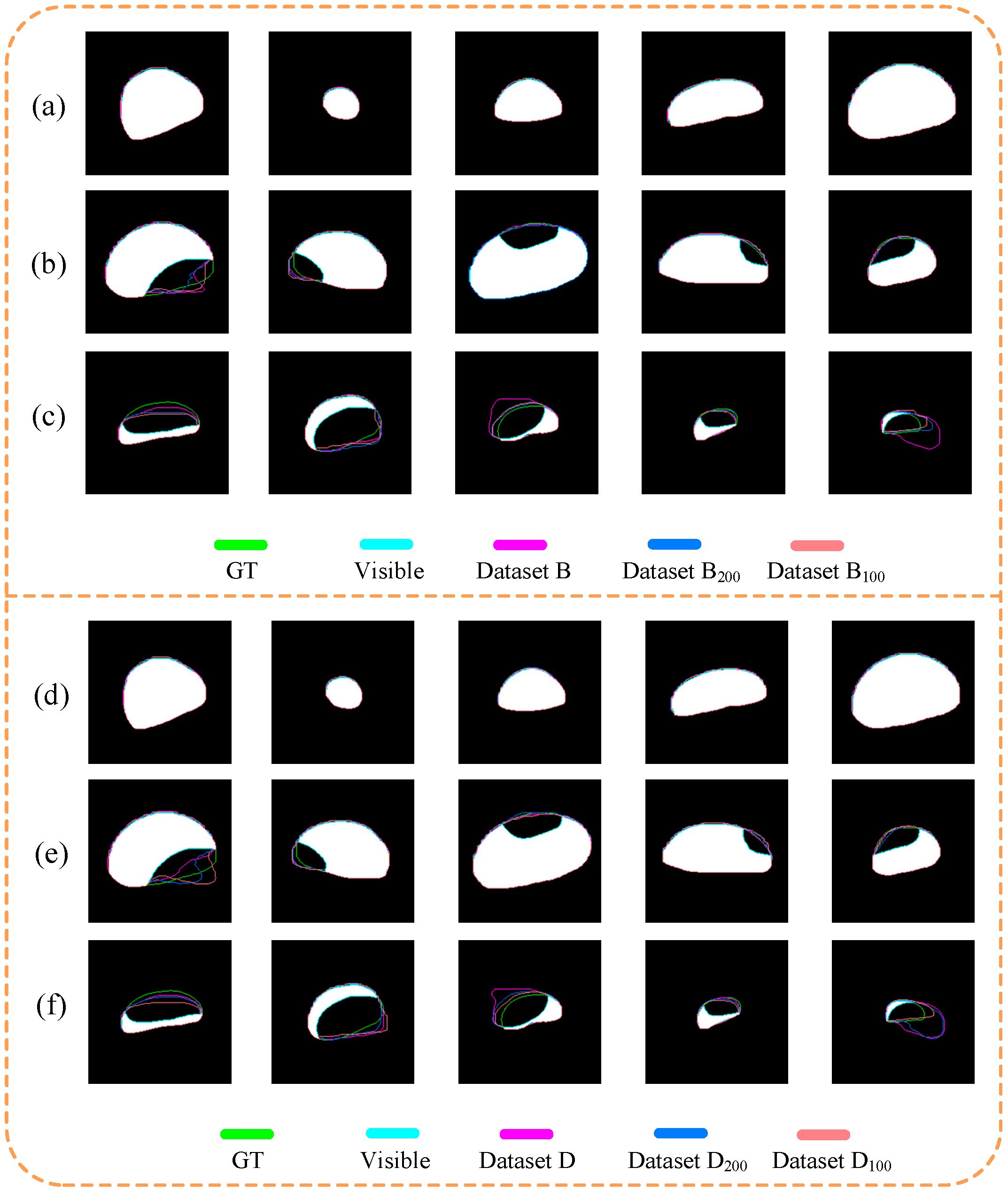
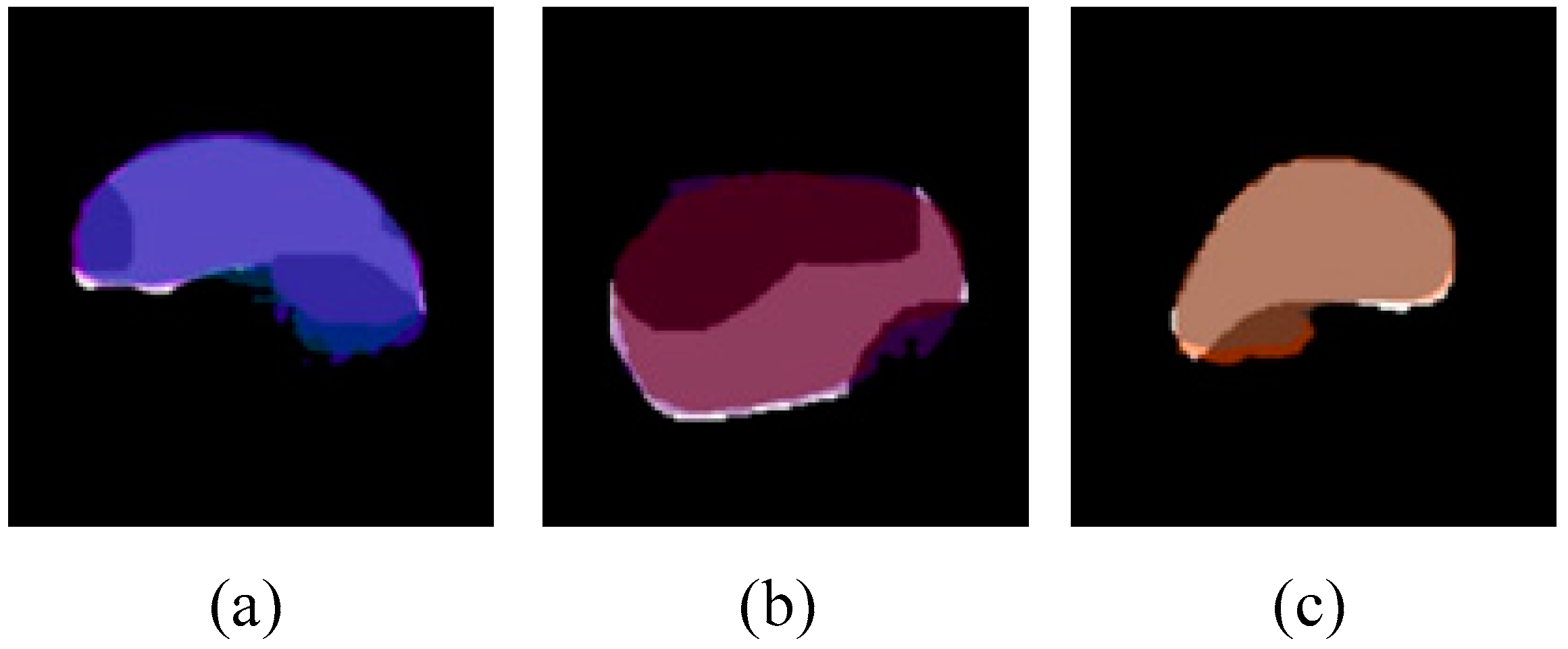
3.2. Amodal Instance Segmentation Results of Cap
3.3. Performance of the Models Trained on Different Datasets
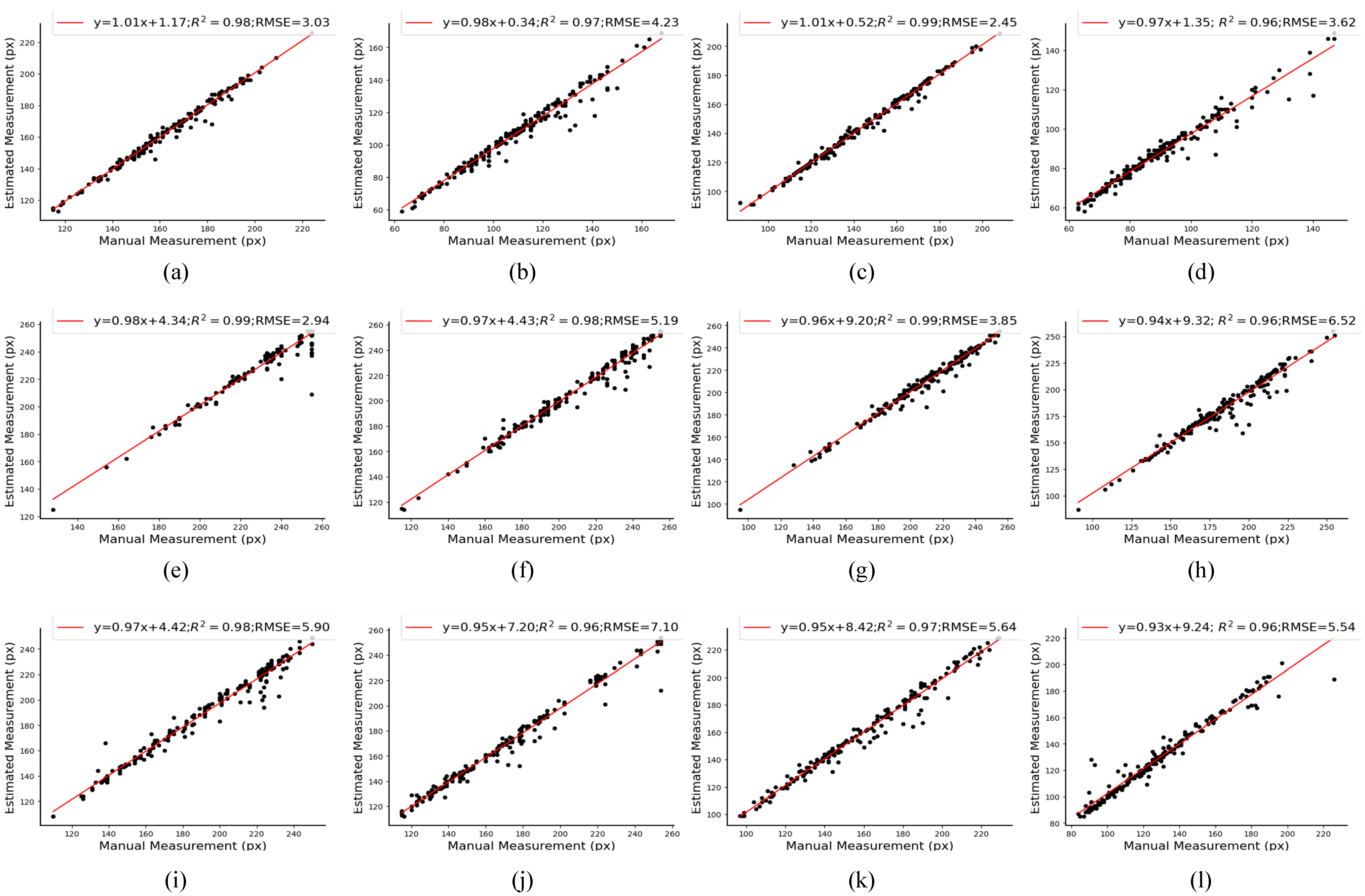
3.4. Size Estimation of Caps
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.X.; Zhang, T.; Huang, X.J.; Yin, J.Y.; Nie, S.P. Heteroglycans from the Fruiting Bodies of Agrocybe Cylindracea: Fractionation, Physicochemical Properties and Structural Characterization. Food Hydrocoll. 2021, 114, 106568. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, X.; Mu, X.; Hu, M.; Wang, J.; Huang, X.; Nie, S. Protective Effects of Flavonoids Isolated from Agrocybe Aegirita on Dextran Sodium Sulfate-Induced Colitis. eFood 2021, 2, 288–295. [Google Scholar] [CrossRef]
- Li, S.; Yan, Z.; Guo, Y.; Su, X.; Cao, Y.; Jiang, B.; Yang, F.; Zhang, Z.; Xin, D.; Chen, Q.; et al. SPM-IS: An Auto-Algorithm to Acquire a Mature Soybean Phenotype Based on Instance Segmentation. Crop J. 2022, 10, 1412–1423. [Google Scholar] [CrossRef]
- Van De Vooren, J.G.; Polder, G.; Van Der Heijden, G.W.A.M. Application of Image Analysis for Variety Testing of Mushroom. Euphytica 1991, 57, 245–250. [Google Scholar] [CrossRef]
- Van De Vooren, J.G.; Polder, G.; Van Der Heijden, G.W.A.M. Identification of Mushroom Cultivars Using Image Analysis. Trans. ASAE 1992, 35, 347–350. [Google Scholar] [CrossRef]
- Tanabata, T.; Shibaya, T.; Hori, K.; Ebana, K.; Yano, M. American Society of Plant Biologists (ASPB) Smart-Grain: High-Throughput Phenotyping Software for Measuring Seed Shape through Image Analysis. Physiology 2012, 160, 1871–1880. [Google Scholar] [CrossRef]
- Zhao, M.; Wu, S.; Li, Y. Improved YOLOv5s-Based Detection Method for Termitomyces Albuminosus. Trans. Chin. Soc. Agric. Eng. 2023, 39, 267–276. [Google Scholar] [CrossRef]
- Yin, H.; Xu, J.; Wang, Y.; Hu, D.; Yi, W. A Novel Method of Situ Measurement Algorithm for Oudemansiella Raphanipies Caps Based on YOLO v4 and Distance Filtering. Agronomy 2023, 13, 134. [Google Scholar] [CrossRef]
- Yang, S.; Zheng, L.; Yang, H.; Zhang, M.; Wu, T.; Sun, S.; Tomasetto, F.; Wang, M. A Synthetic Datasets Based Instance Segmentation Network for High-Throughput Soybean Pods Phenotype Investigation. Expert Syst. Appl. 2022, 192, 116403. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Lawrence Zitnick, C.; Irvine, U. Microsoft COCO: Common Objects in Context. In Proceedings of the Computer Vision—ECCV 2014: 13th European Conference, Zurich, Switzerland, 6–12 September 2014; Proceedings, Part IV. Springer: Cham, Switzerland, 2014. [Google Scholar]
- Xu, P.; Fang, N.; Liu, N.; Lin, F.; Yang, S.; Ning, J. Visual Recognition of Cherry Tomatoes in Plant Factory Based on Improved Deep Instance Segmentation. Comput. Electron. Agric 2022, 197, 106991. [Google Scholar] [CrossRef]
- Zhou, W.; Chen, Y.; Li, W.; Zhang, C.; Xiong, Y.; Zhan, W.; Huang, L.; Wang, J.; Qiu, L. SPP-Extractor: Automatic Phenotype Extraction for Densely Grown Soybean Plants. Crop J. 2023, 11, 1569–1578. [Google Scholar] [CrossRef]
- Kuznichov, D.; Zvirin, A.; Honen, Y.; Kimmel, R. Data Augmentation for Leaf Segmentation and Counting Tasks in Rosette Plants. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops, Long Beach, CA, USA, 16–17 June 2019; IEEE Computer Society: Washington, DC, USA, 2019; pp. 2580–2589. [Google Scholar]
- Toda, Y.; Okura, F.; Ito, J.; Okada, S.; Kinoshita, T.; Tsuji, H.; Saisho, D. Training Instance Segmentation Neural Network with Synthetic Datasets for Crop Seed Phenotyping. Commun. Biol. 2020, 3, 173. [Google Scholar] [CrossRef] [PubMed]
- Follmann, P.; König, R.; Härtinger, P.; Klostermann, M. Learning to See the Invisible: End-to-End Trainable Amodal Instance Segmentation. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision 2019, Waikoloa, HI, USA, 7–11 January 2019; pp. 1328–1336. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision 2017, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury Google, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. Adv. Neural Inf. Process. Syst. 2019, 32, 8026–8037. [Google Scholar] [CrossRef]
- Russell, B.C.; Torralba, A.; Murphy, K.P.; Freeman, W.T. LabelMe: A Database and Web-Based Tool for Image Annotation. Int. J. Comput. Vis. 2008, 77, 157–173. [Google Scholar] [CrossRef]
- Igathinathane, C.; Pordesimo, L.O.; Columbus, E.P.; Batchelor, W.D.; Methuku, S.R. Shape Identification and Particles Size Distribution from Basic Shape Parameters Using ImageJ. Comput. Electron. Agric. 2008, 63, 168–182. [Google Scholar] [CrossRef]
- Yin, H.; Wei, Q.; Gao, Y.; Hu, H.; Wang, Y. Moving toward Smart Breeding: A Robust Amodal Segmentation Method for Occluded Oudemansiella Raphanipes Cap Size Estimation. Comput. Electron. Agric. 2024, 220, 108895. [Google Scholar] [CrossRef]



| Dataset | Image Size | Dataset | Test Dataset | Generation Time/h |
|---|---|---|---|---|
| Train./Val. | ||||
| AC | 256 | 12,000/3000 | 2000 | 5.19 |
| 512 | 12,000/3000 | 2000 | 20.31 | |
| 1024 | 12,000/3000 | 2000 | 79.90 | |
| OR | 256 | 12,000/3000 | 2000 | 4.72 |
| 512 | 12,000/3000 | 2000 | 18.42 | |
| 1024 | 12,000/3000 | 2000 | 72.63 | |
| PN | 256 | 12,000/3000 | 2000 | 5.67 |
| 512 | 12,000/3000 | 2000 | 21.25 | |
| 1024 | 12,000/3000 | 2000 | 83.82 |
| Pre-trained model | Pre-trained ImageNet weights | |||||||||
| Iteration | 4000 | |||||||||
| Backbone | ResNet101-FPN | |||||||||
| Dataset | 256 × 256 | 512 × 512 | 1024 × 1024 | |||||||
| Test dataset | 256 | 512 | 1024 | 256 | 512 | 1024 | 256 | 512 | 1024 | |
| AC | R | 0.9 | 0.86 | 0.80 | 0.96 | 0.90 | 0.81 | 0.91 | 0.89 | 0.85 |
| AP0.5 | 1.0 | 0.99 | 0.99 | 1.0 | 1.0 | 0.99 | 1.0 | 1.0 | 1.0 | |
| AP0.75 | 0.98 | 0.95 | 0.88 | 0.99 | 0.98 | 0.90 | 0.99 | 0.99 | 0.96 | |
| AP@[0.5:0.95] | 0.93 | 0.85 | 0.76 | 0.93 | 0.88 | 0.79 | 0.88 | 0.86 | 0.82 | |
| OR | R | 0.97 | 0.94 | 0.89 | 0.97 | 0.95 | 0.92 | 0.96 | 0.95 | 0.94 |
| AP0.5 | 1.0 | 1.0 | 0.99 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| AP0.75 | 0.99 | 0.98 | 0.95 | 0.99 | 0.99 | 0.97 | 0.99 | 0.99 | 0.98 | |
| AP@[0.5:0.95] | 0.96 | 0.92 | 0.88 | 0.95 | 0.94 | 0.90 | 0.94 | 0.94 | 0.93 | |
| PN | R | 0.96 | 0.48 | 0.45 | 0.60 | 0.98 | 0.97 | 0.60 | 0.98 | 0.97 |
| AP0.5 | 1.0 | 0.79 | 0.73 | 0.94 | 0.99 | 0.99 | 0.94 | 0.99 | 0.99 | |
| AP0.75 | 0.99 | 0.36 | 0.33 | 0.47 | 0.99 | 0.99 | 0.46 | 0.99 | 0.99 | |
| AP@[0.5:0.95] | 0.95 | 0.40 | 0.37 | 0.51 | 0.96 | 0.96 | 0.51 | 0.96 | 0.96 | |
| Dataset | 1024 × 1024 | ||||||
| Pre-trained model | Pre-trained ImageNet weights | ||||||
| Iteration | 4000 | ||||||
| Backbone | ResNet50-FPN | ResNet101-FPN | |||||
| Image size | 256 | 512 | 1024 | 256 | 512 | 1024 | |
| AC | R | 0.90 | 0.87 | 0.84 | 0.91 | 0.89 | 0.85 |
| AP0.5 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| AP0.75 | 0.99 | 0.97 | 0.93 | 0.99 | 0.99 | 0.96 | |
| AP@[0.5:0.95] | 0.88 | 0.84 | 0.81 | 0.88 | 0.86 | 0.82 | |
| OR | R | 0.94 | 0.91 | 0.88 | 0.96 | 0.95 | 0.94 |
| AP0.5 | 1.0 | 1.0 | 0.99 | 1.0 | 1.0 | 1.0 | |
| AP0.75 | 0.99 | 0.99 | 0.98 | 0.99 | 0.99 | 0.98 | |
| AP@[0.5:0.95] | 0.92 | 0.89 | 0.86 | 0.94 | 0.94 | 0.93 | |
| PN | R | 0.6 | 0.97 | 0.97 | 0.60 | 0.98 | 0.97 |
| AP0.5 | 0.94 | 0.99 | 0.99 | 0.94 | 0.99 | 0.99 | |
| AP0.75 | 0.50 | 0.99 | 0.99 | 0.46 | 0.99 | 0.99 | |
| AP@[0.5:0.95] | 0.52 | 0.96 | 0.96 | 0.51 | 0.96 | 0.96 | |
| Training Set | Evaluated Set | AP |
|---|---|---|
| B | B | 0.80 |
| A | 0.79 | |
| C | C | 0.89 |
| A | 0.97 | |
| D | D | 0.81 |
| A | 0.79 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Q.; Wang, Y.; Yang, S.; Guo, C.; Wu, L.; Yin, H. Moving toward Automaticity: A Robust Synthetic Occlusion Image Method for High-Throughput Mushroom Cap Phenotype Extraction. Agronomy 2024, 14, 1337. https://doi.org/10.3390/agronomy14061337
Wei Q, Wang Y, Yang S, Guo C, Wu L, Yin H. Moving toward Automaticity: A Robust Synthetic Occlusion Image Method for High-Throughput Mushroom Cap Phenotype Extraction. Agronomy. 2024; 14(6):1337. https://doi.org/10.3390/agronomy14061337
Chicago/Turabian StyleWei, Quan, Yinglong Wang, Shenglan Yang, Chaohui Guo, Lisi Wu, and Hua Yin. 2024. "Moving toward Automaticity: A Robust Synthetic Occlusion Image Method for High-Throughput Mushroom Cap Phenotype Extraction" Agronomy 14, no. 6: 1337. https://doi.org/10.3390/agronomy14061337
APA StyleWei, Q., Wang, Y., Yang, S., Guo, C., Wu, L., & Yin, H. (2024). Moving toward Automaticity: A Robust Synthetic Occlusion Image Method for High-Throughput Mushroom Cap Phenotype Extraction. Agronomy, 14(6), 1337. https://doi.org/10.3390/agronomy14061337






