Genome-Wide Identification of Phytochrome-Interacting Factor (PIF) Gene Family in Potatoes and Functional Characterization of StPIF3 in Regulating Shade-Avoidance Syndrome
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Processing Method
2.2. Genome-Wide Identification and Annotation of Potato PIF Genes
2.3. Bioinformatics Analysis of StPIF Genes
2.4. Gene Expression Analysis of StPIFs
2.5. Tissue-Specific Analysis of Expression Levels
2.6. Subcellular Localization of StPIF3
2.7. Plasmid Construction and Generation of the Solanum tuberosum Transgenic Plant
2.8. Shade-Avoidance Syndrome Analysis of Transgenic Potato Plants
3. Results
3.1. Identification of Potato PIFs
3.2. Subcellular Localization, Chromosome Localization, and Gene Structure Analysis of Potato PIFs
3.3. Multiple Sequence Alignment and Phylogenetic Analysis
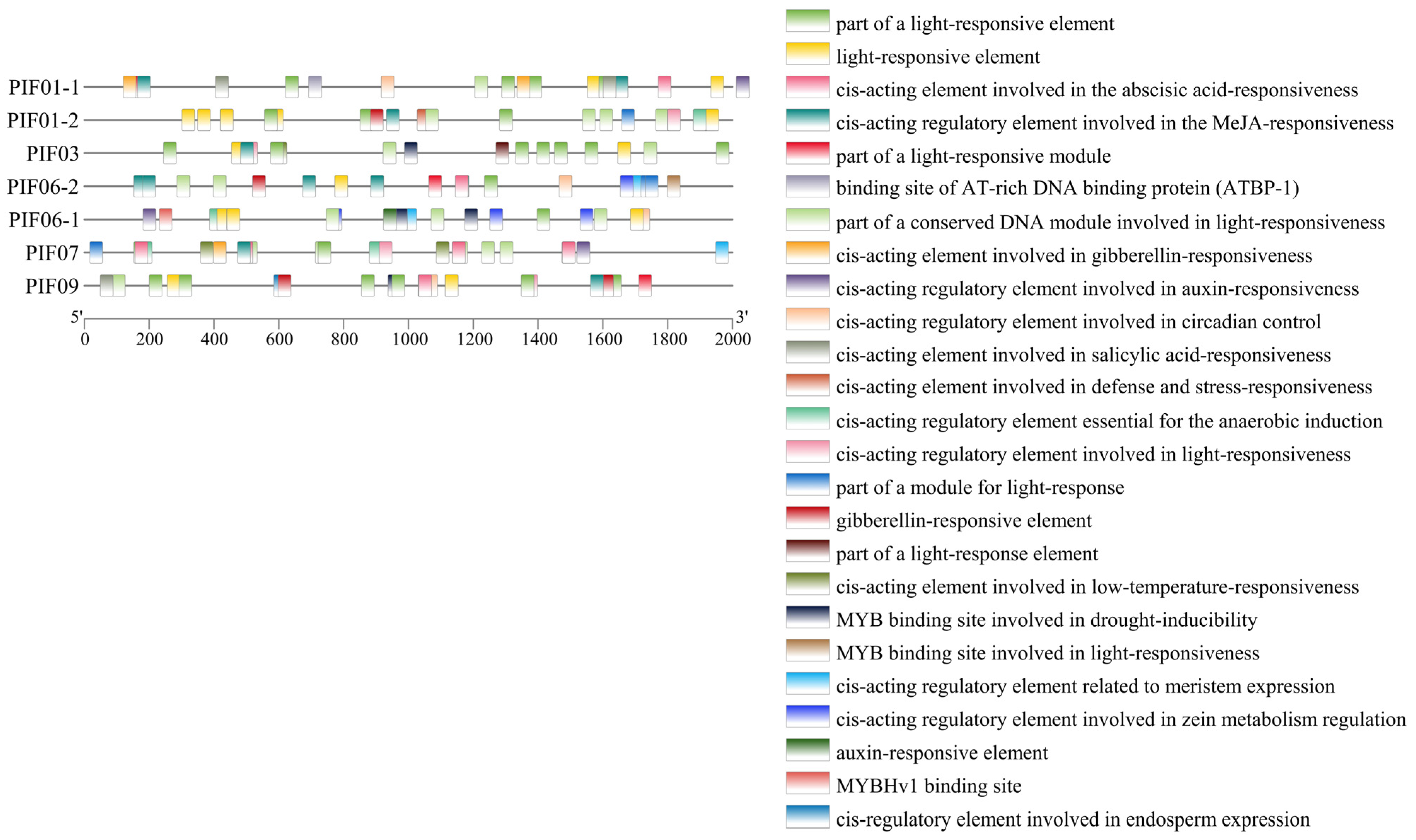
3.4. Conserved Motif and Cis-Acting Element Analysis of PIF Family
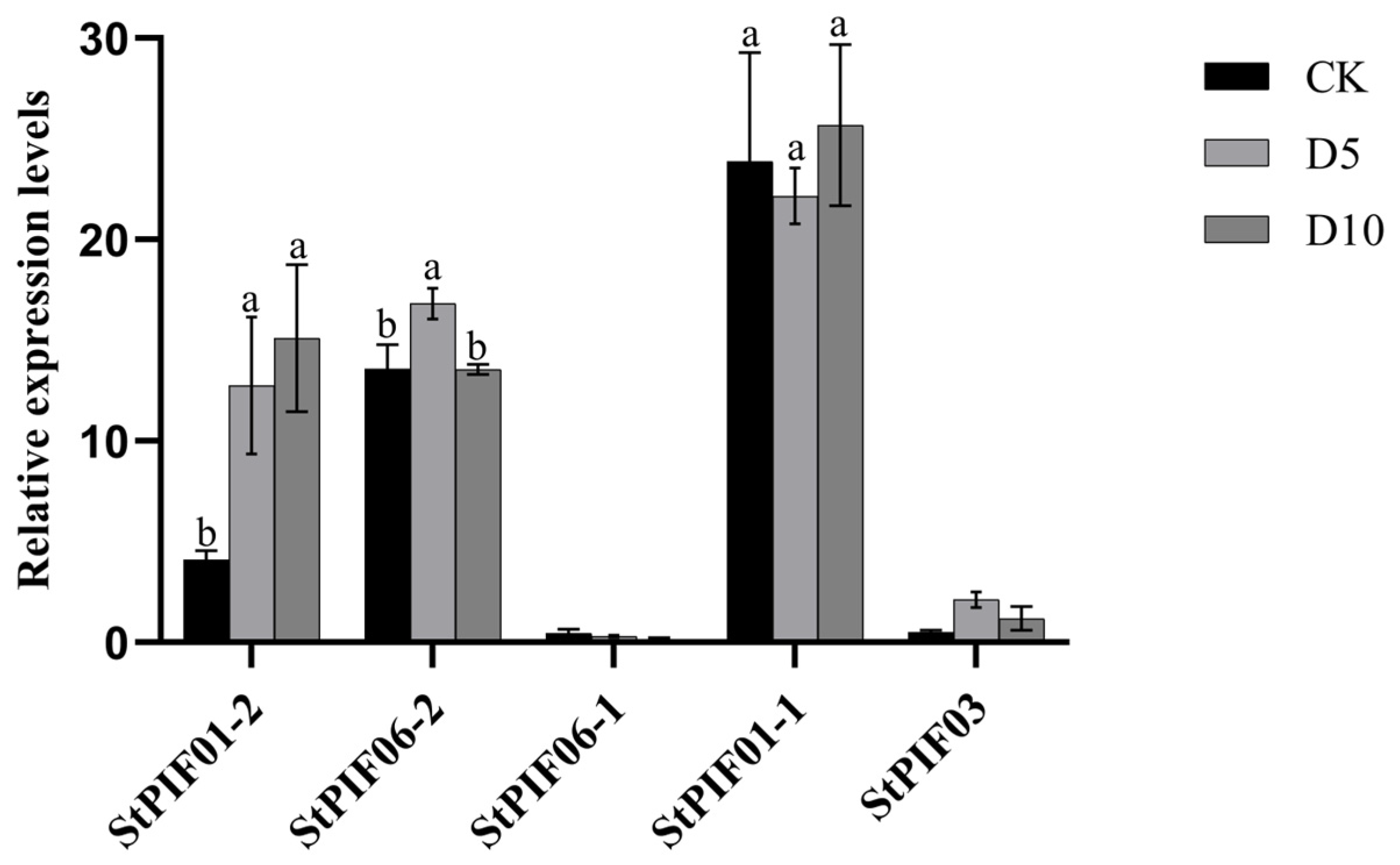
3.5. QRT-PCR Analysis of StPIF Genes under Shaded Stress
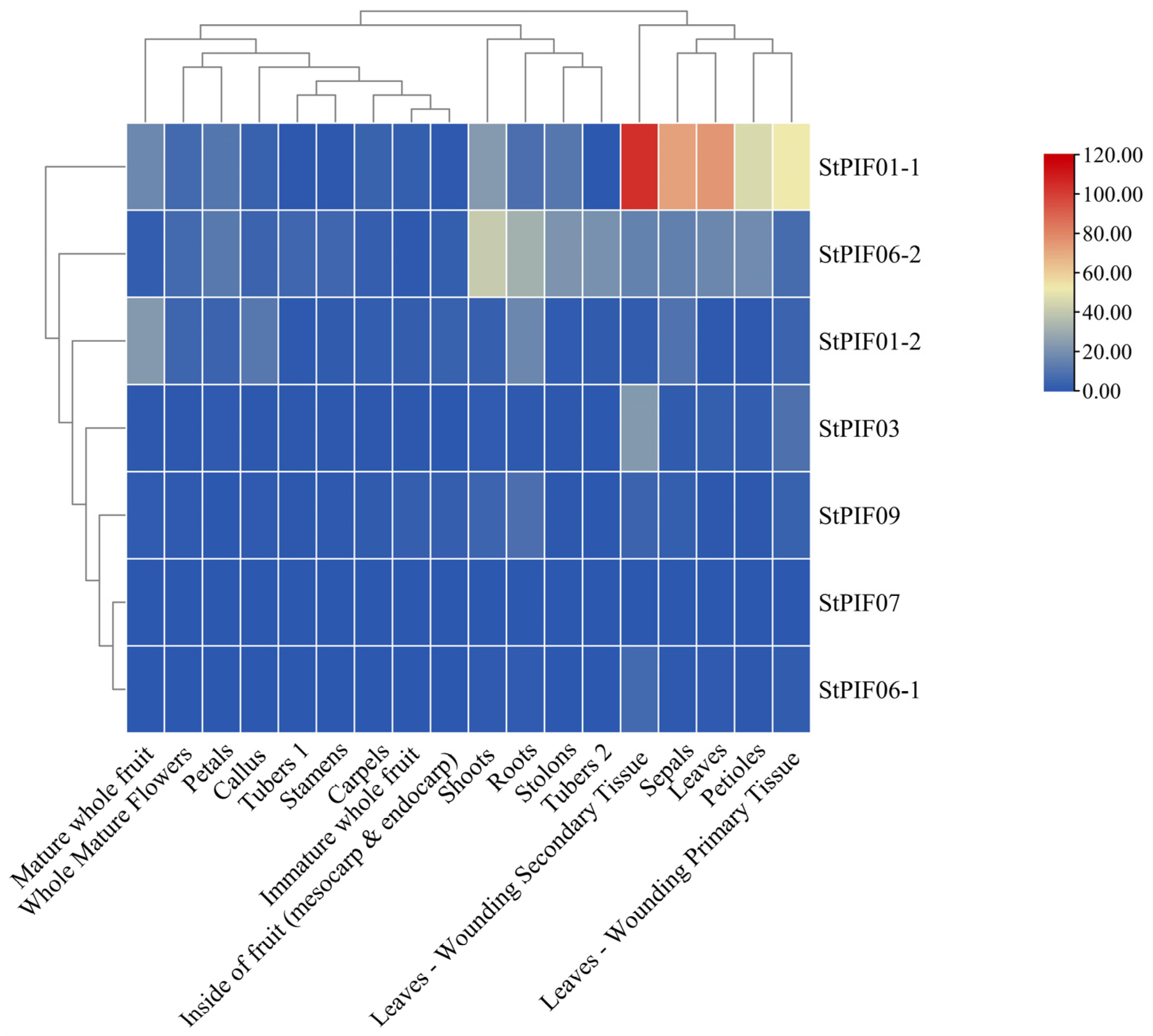
3.6. Tissue-Specific Analysis of Expression Levels of StPIFs in Potatoes
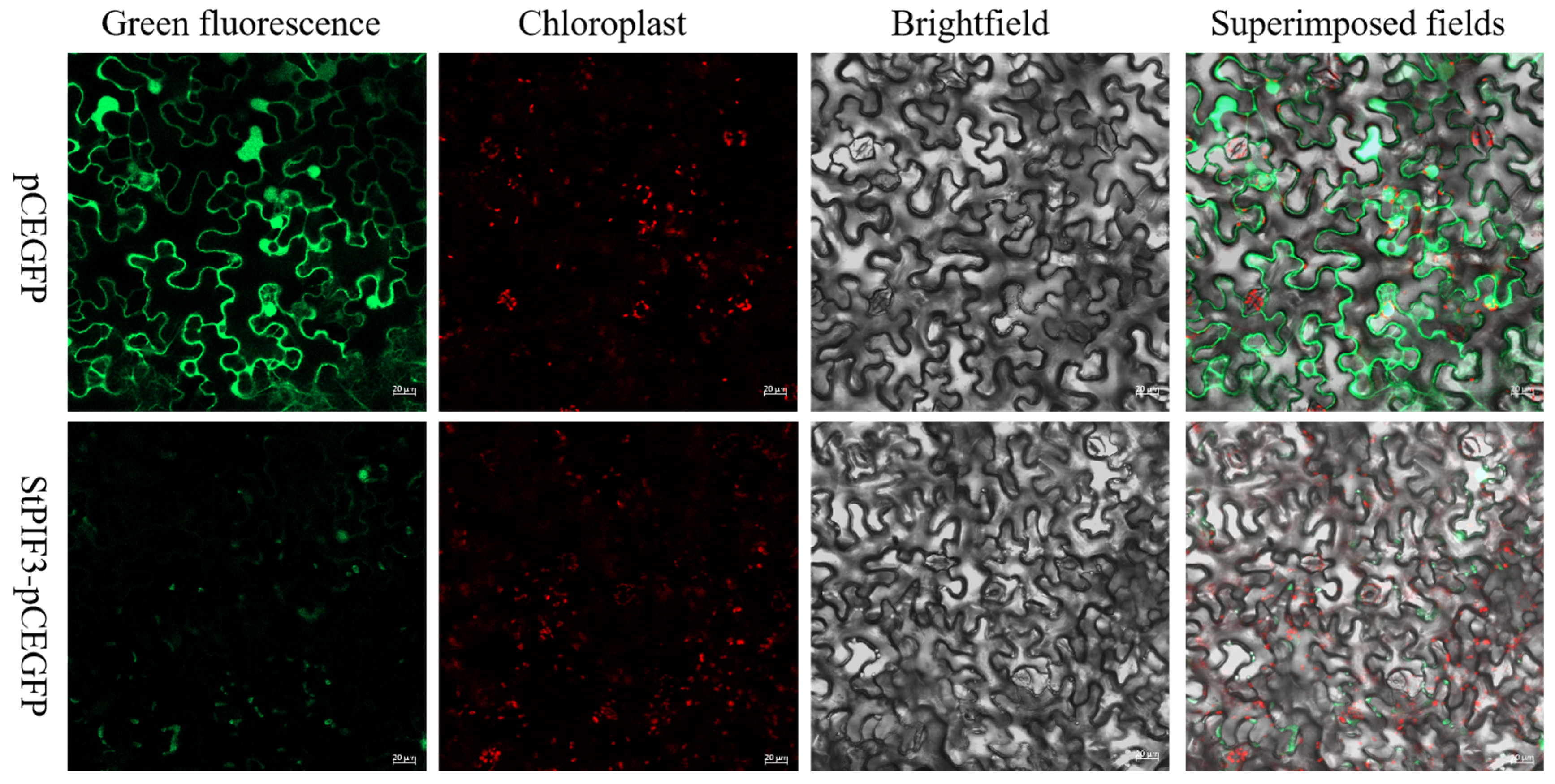
3.7. Subcellular Localization of StPIF3
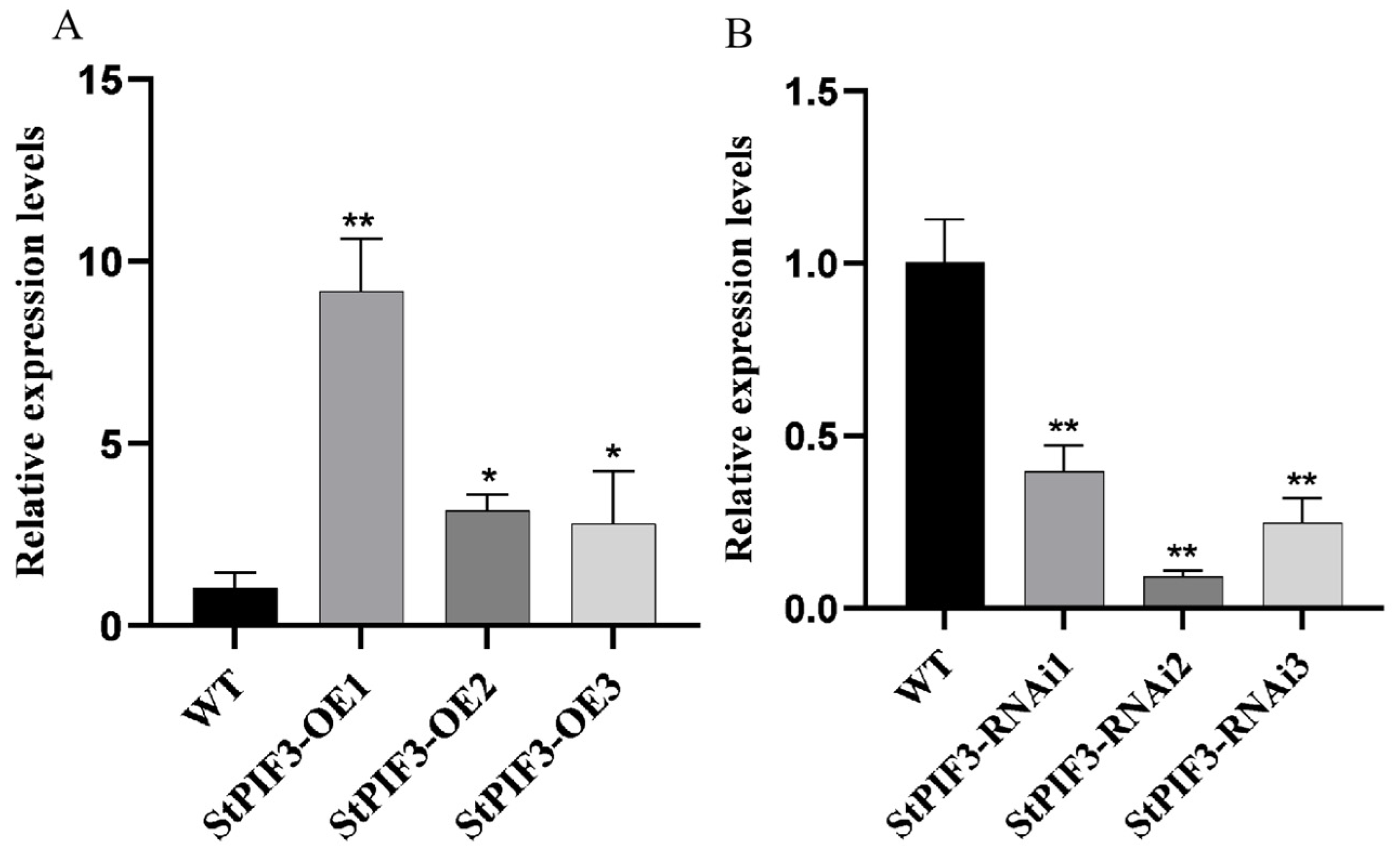
3.8. Potato Transformation and Identification
3.9. StPIF3 Regulates Shade-Avoidance Syndrome by Coordinating Chlorophyll Accumulation
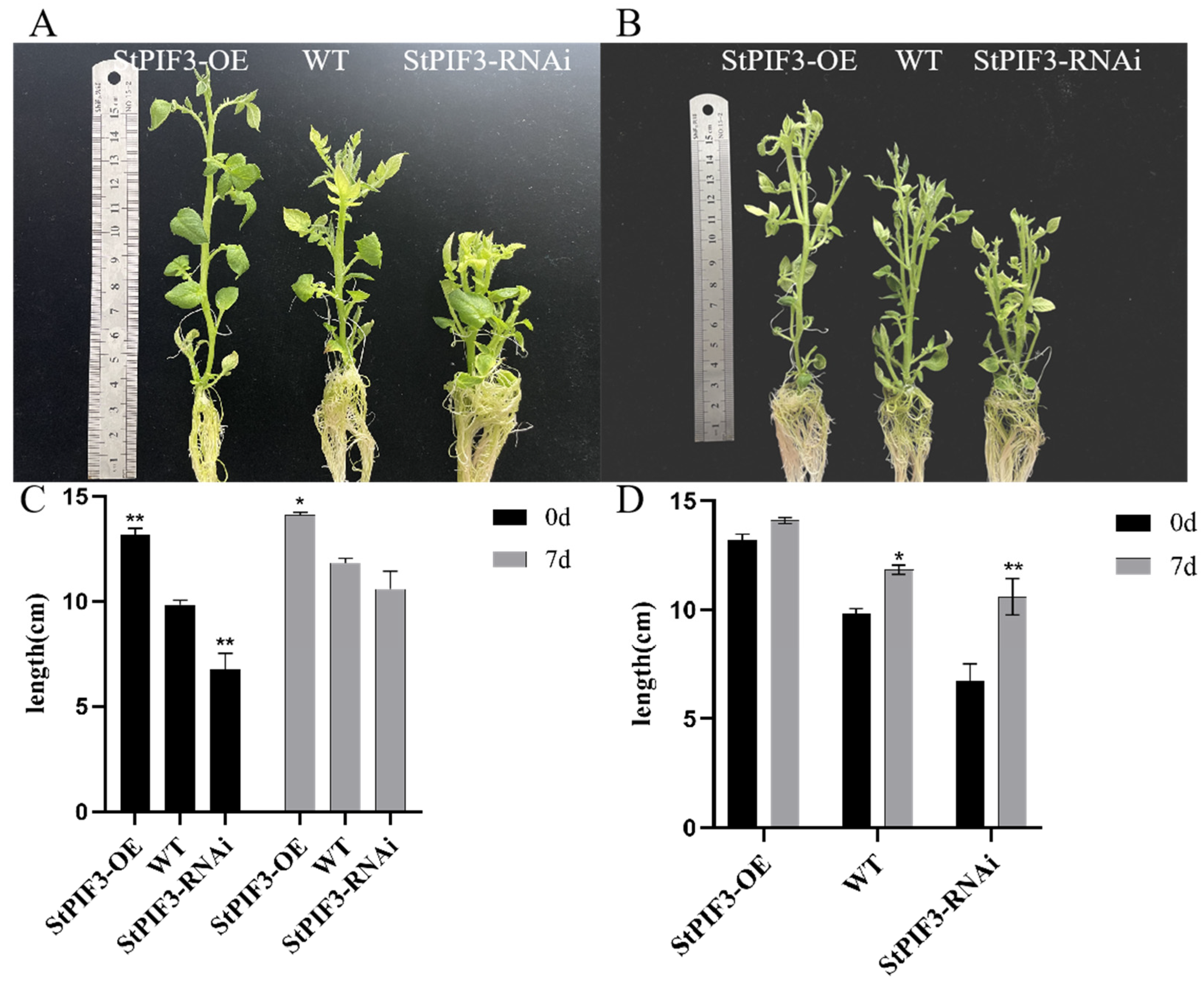
3.10. StPIF3 Regulates Shade-Avoidance Syndrome by Regulating Plant Heights
4. Discussion
4.1. StPIF Identification and Evolution in Potatoes
4.2. PIF Transcription Factors Exhibit Phylogenetic Conservation across Plantae
4.3. PIFs Function as Key Nodes Conjoining Environmental and Hormonal Signals
4.4. StPIF3 Regulates Shade-Avoidance Syndrome in Potatoes
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bae, G.; Choi, G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 2008, 59, 281–311. [Google Scholar] [CrossRef] [PubMed]
- Roeber, V.M.; Bajaj, I.; Rohde, M.; Schmulling, T.; Cortleven, A. Light acts as a stressor and influences abiotic and biotic stress responses in plants. Plant Cell Environ. 2021, 44, 645–664. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Larner, V.S.; Whitelam, G.C. The signal transducing photoreceptors of plants. Int. J. Dev. Biol. 2005, 49, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chory, J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011, 21, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.F.; Campos, M.L.; Azevedo, R.A. The role of phytochrome in stress tolerance. J. Integr. Plant Biol. 2011, 53, 920–929. [Google Scholar] [CrossRef] [PubMed]
- Ballare, C.L. Light regulation of plant defense. Annu. Rev. Plant Biol. 2014, 65, 335–363. [Google Scholar] [CrossRef] [PubMed]
- Castillon, A.; Shen, H.; Huq, E. Phytochrome interacting factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007, 12, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Leivar, P.; Quail, P.H. PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 2011, 16, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Choi, G. Phytochrome-interacting factor from Arabidopsis to liverwort. Curr. Opin. Plant Biol. 2017, 35, 54–60. [Google Scholar] [CrossRef]
- Nakamura, Y.; Kato, T.; Yamashino, T.; Murakami, M.; Mizuno, T. Characterization of a set of phytochrome-interacting factor-like bHLH proteins in Oryza sativa. Biosci. Biotechnol. Biochem. 2007, 71, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Lv, W.; Zhang, H.; Ma, L.; Li, P.; Ge, L. Genome-wide analysis of the basic Helix-Loop-Helix (bHLH) transcription factor family in maize. BMC Plant Biol. 2018, 18, 235. [Google Scholar] [CrossRef]
- Rosado, D.; Gramegna, G.; Cruz, A.; Lira, B.S.; Freschi, L.; de Setta, N. Phytochrome interacting factors (PIFs) in Solanum lycopersicum: Diversity, evolutionary history and expression profiling during different developmental processes. PLoS ONE 2016, 11, e0165929. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.F.; Wang, X.; Yang, Y.Y.; You, C.X.; Zhang, Z.L.; Hao, Y.J. Identification of phytochrome-interacting factor family members and functional analysis of MdPIF4 in Malus domestica. Int. J. Mol. Sci. 2020, 21, 7350. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.K.; Zheng, T.; Zhu, X.D.; Jiu, S.T.; Liu, Z.J.; Guan, L.; Jia, H.F.; Fang, J.G. Genome-wide identification of PIFs in Grapes (Vitis vinifera L.) and their transcriptional analysis under lighting/shading conditions. Genes 2018, 9, 451. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Wen, G.; Zhao, C. Phylogeny and evolution of plant Phytochrome Interacting Factors (PIFs) gene family and functional analyses of PIFs in Brachypodium distachyon. Plant Cell Rep. 2022, 41, 1209–1227. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Nishihama, R.; Kataoka, H.; Hosaka, M.; Manabe, R.; Nomoto, M. Phytochrome signaling is mediated by phytochrome interacting factor in the liverwort Marchantia polymorpha. Plant Cell 2016, 28, 1406–1421. [Google Scholar] [CrossRef] [PubMed]
- Possart, A.; Xu, T.; Paik, I.; Hanke, S.; Keim, S.; Hermann, H.M.; Wolf, L.; Hiß, M.; Becker, C.; Huq, E.; et al. Characterization of phytochrome interacting factors from the moss Physcomitrella patens illustrates conservation of phytochrome signaling modules in land plants. Plant Cell 2017, 29, 310–330. [Google Scholar] [CrossRef] [PubMed]
- Leivar, P.; Monte, E. PIFs: Systems integrators in plant development. Plant Cell 2014, 26, 56–78. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Y.; Wang, J.; Li, P.; Zhao, C.; Chen, Y.; Bi, Y. Phytochrome-interacting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light in Arabidopsis seedlings. Plant Sci. 2015, 238, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Josse, E.M.; Halliday, K.J. A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol. Biol. 2010, 73, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Camire, M.E.; Kubow, S.; Donnelly, D.J. Potatoes and human health. Crit. Rev. Food Sci. Nutr. 2009, 49, 823–840. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Ocampo, G.; Cascales, J.; Medina-Fraga, A.L.; Ploschuk, E.L.; Mantese, A.I.; Crocco, C.D.; Matsusaka, D.; Sánchez, D.H.; Botto, J.F. Transcriptomic and physiological shade avoidance responses in potato (Solanum tuberosum) plants. Physiol. Plant. 2023, 175, e13991. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tang, X.; Zhu, X.; Qi, X.; Zhang, N.; Si, H. Genome-wide identification and expression analysis of the E2 gene family in potato. Mol. Biol. Rep. 2019, 46, 777–791. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Han, J.; Shangguan, L.; Yang, G.; Kayesh, E.; Zhang, Y.; Leng, X.; Fang, J. Depiction of grapevine phenology by gene expression information and a test of its workability in guiding fertilization. Plant Mol. Biol. 2014, 32, 1070–1084. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Pan, S.; Cheng, S.; Zhang, B.; Mu, D.; Ni, P.; Zhang, G.; Yang, S.; Li, R.; Wang, J.; et al. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Pieczynski, M.; Marczewski, W.; Hennig, J.; Dolata, J.; Bielewicz, D.; Piontek, P.; Wyrzykowska, A.; Krusiewicz, D.; Strzelczyk-Zyta, D.; Konopka-Postupolska, D.; et al. Down-regulation of CBP80 gene expression as a strategy to engineer a drought-tolerant potato. Plant Biotechnol. J. 2013, 11, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Si, H.J.; Xie, C.H.; Liu, J. Optimization of agrobacterium-mediated genetic transformation system of potato in vitro potato and introduction of antisense class I. patatin gene. Acta Agron. Sin. 2003, 29, 801–805. [Google Scholar]
- Al-Sady, B.; Ni, W.; Kircher, S.; Schäfer, E.; Quail, P.H. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 2006, 23, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Bo, K.; Pan, Y.; Li, Y.; Yu, D.; Li, C.; Chang, J.; Wu, S.; Wang, Z.; Zhang, X.; et al. Phytochrome-interacting factor PIF3 integrates phytochrome B and UV-B signaling pathways to regulate gibberellin- and auxin-dependent growth in cucumber hypocotyls. J. Exp. Bot. 2023, 74, 4520–4539. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.N.; Kathare, P.K.; Huq, E. Phytochromes and phytochrome interacting factors. Plant Physiol. 2018, 176, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhao, Y.; Shen, R.; Wang, B.; Xie, Y.; Ma, X.; Zheng, Z.; Wang, H. Characterization of maize phytochrome-interacting factors in light signaling and photomorphogenesis. Plant Physiol. 2019, 181, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Liu, K.; Yuan, G.; He, S.; Cong, P.; Zhang, C. Genome-wide identification and characterization of AINTEGUMENTA-LIKE (AIL) family genes in apple (Malus domestica Borkh.). Genomics 2022, 114, 110313. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Zhu, L.; Castillon, A.; Majee, M.; Downie, B.; Huq, E. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 2008, 20, 1586–1602. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wu, M.; Zhang, M.; Jiang, W.; Ren, X.; Liang, E.; Zhang, D.; Zhang, C.; Xiao, N.; Li, Y. A maize phytochrome-interacting factors protein ZmPIF1 enhances drought tolerance by inducing stomatal closure and improves grain yield in Oryza sativa. Plant Biotechnol. J. 2018, 16, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Jiang, W.; Dai, Y.; Xiao, N.; Zhang, C.; Li, H.; Lu, Y.; Wu, M.; Tao, X.; Deng, D. A maize phytochrome-interacting factor 3 improves drought and salt stress tolerance in rice. Plant Mol. Biol. 2015, 87, 413–428. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, X.; Dong, S.; Jiang, X.; Wang, L.; Yu, J.; Zhou, Y. Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol. J. 2020, 18, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Huq, E.; Al-Sady, B.; Hudson, M.; Kim, C.; Apel, K.; Quail, P.H. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 2004, 305, 1937–1941. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Oh, E. PIF4 integrates multiple environmental and hormonal signals for plant growth regulation in Arabidopsis. Mol. Cells 2016, 39, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Berriri, S.; Kumar, S.V. PIF4 coordinates thermosensory growth and immunity in Arabidopsis. Curr. Biol. 2017, 27, 243–249. [Google Scholar] [CrossRef] [PubMed]
- De Lucas, M.; Davière, J.M.; Rodríguez-Falcón, M.; Pontin, M.; Iglesias-Pedraz, J.M.; Lorrain, S.; Fankhauser, C.; Blázquez, M.A.; Titarenko, E.; Prat, S. A molecular framework for light and gibberellin control of cell elongation. Nature 2008, 451, 480–484. Available online: https://www.nature.com/articles/nature06520#citeas (accessed on 6 March 2024). [CrossRef] [PubMed]
- Halliday, K.J.; Hudson, M.; Ni, M.; Qin, M.; Quail, P.H. Poc1: An Arabidopsis mutant perturbed in phytochrome signaling because of a T DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc. Natl. Acad. Sci. USA 1999, 96, 5832–5837. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yi, H.; Choi, G.; Shin, B.; Song, P.S.; Choi, G. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 2003, 15, 2399–2407. [Google Scholar] [CrossRef] [PubMed]
- Li, P.H.; Peng, X. Study on photosynthesis effect of potato shading light. Chin. Agric. Sci. Bull. 2007, 23, 220–227. Available online: https://www.casb.org.cn/CN/Y2007/V23/I4/220 (accessed on 6 March 2024).

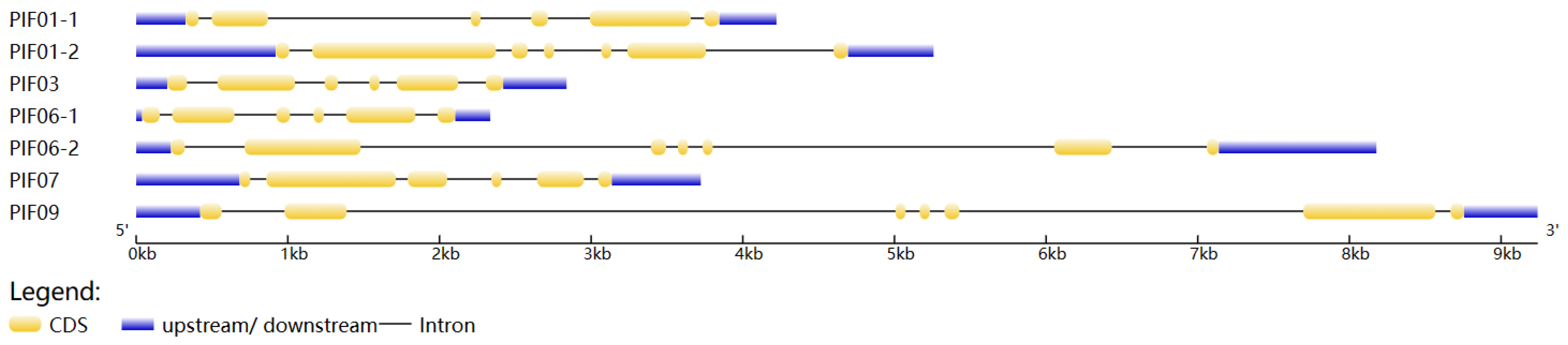
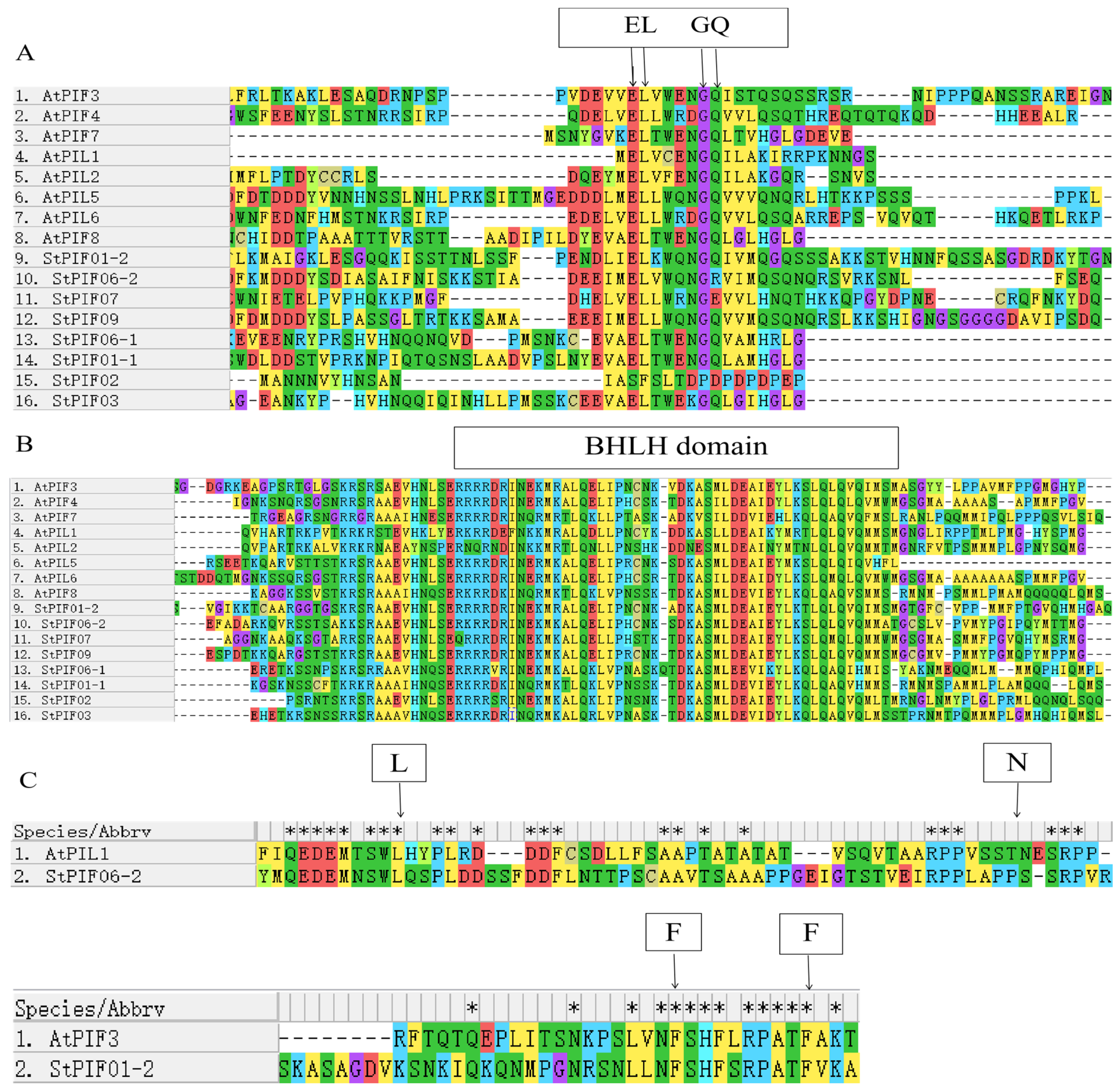
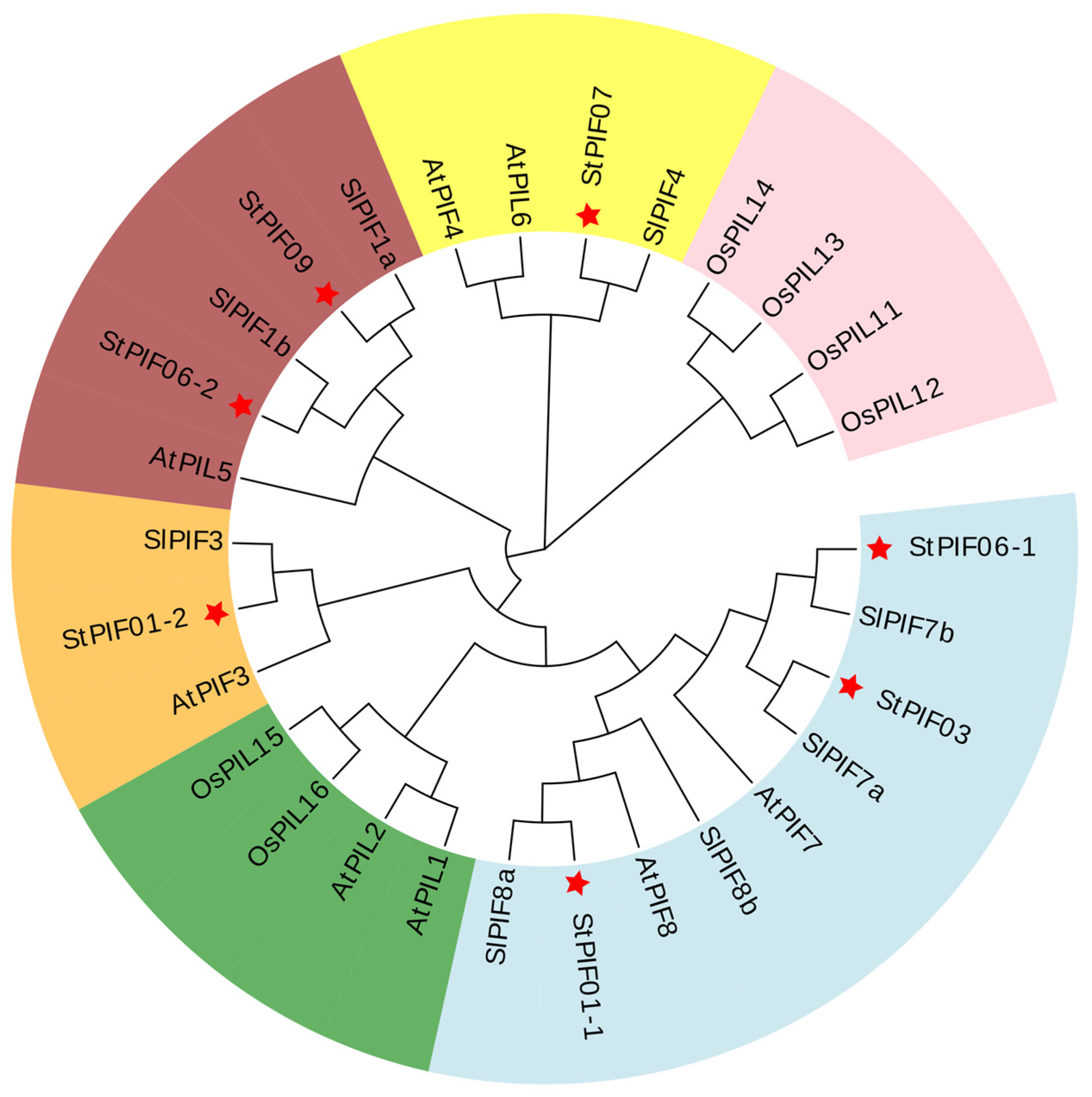
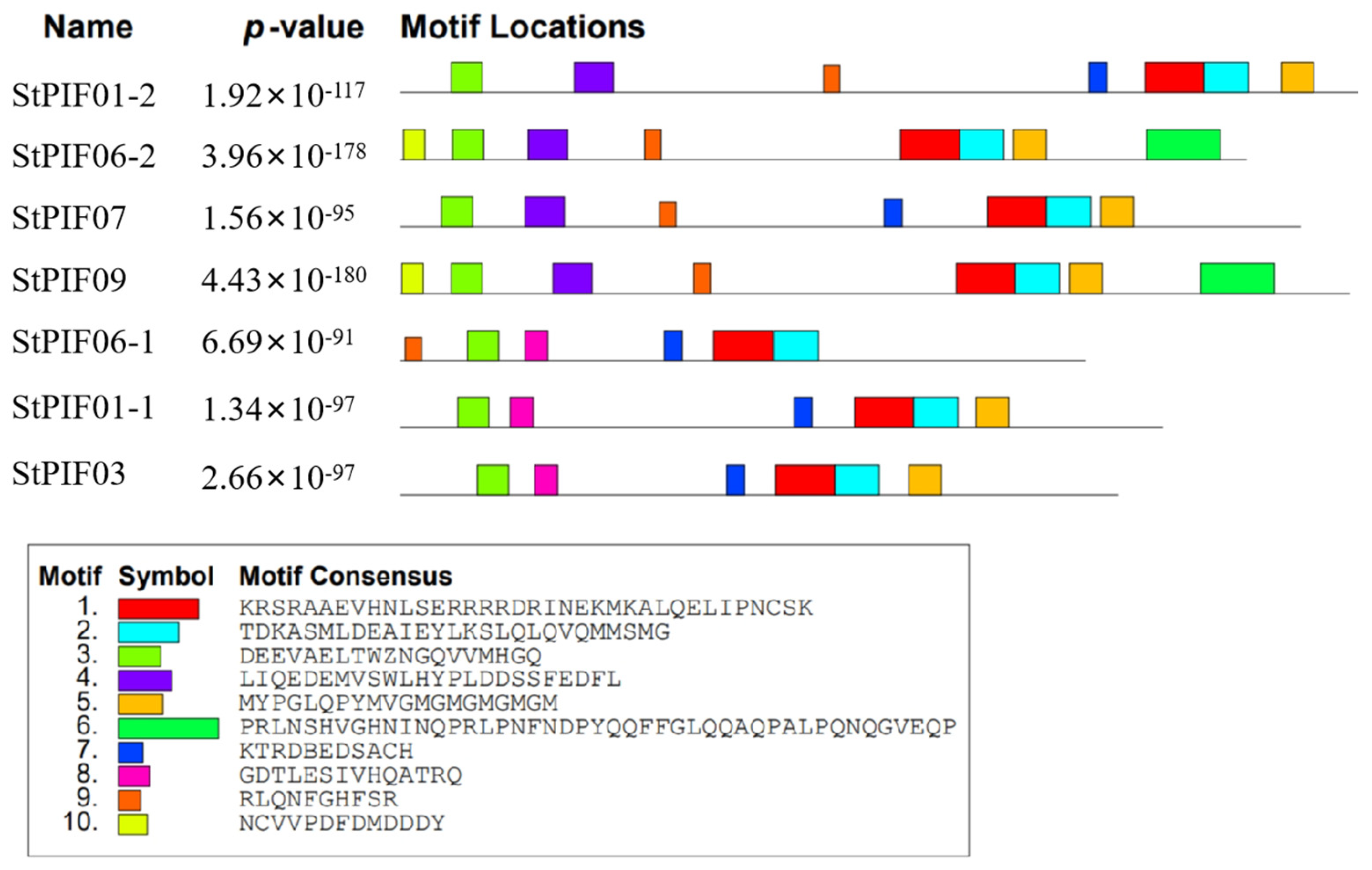

| Gene ID | Name | Subfamily | CDS Length | Protein Length | Molecular Weight | Theoretical pI | Characteristic Domains |
|---|---|---|---|---|---|---|---|
| Soltu.DM.01G041140.1 | PIF01-2 | PIF3 | 2151 | 716 | 76,772.66 | 7.26 | bHLH (460–508) |
| Soltu.DM.06G002140.1 | PIF06-2 | PIF1 | 1548 | 515 | 56,408.13 | 5.13 | bHLH (311–359) |
| Soltu.DM.07G014300.2 | PIF07 | PIF4 | 1647 | 548 | 61,114.45 | 6.93 | bHLH (372–412) |
| Soltu.DM.09G018570.7 | PIF09 | PIF4-RELATED | 1737 | 578 | 62,943.67 | 6.42 | bHLH (347–393) |
| Soltu.DM.06G025680.1 | PIF06-1 | PIF7 | 1254 | 417 | 46,935.82 | 8.71 | bHLH (198–246) |
| Soltu.DM.01G031430.2 | PIF01-1 | PIF8 | 1395 | 464 | 50,747.96 | 7.62 | bHLH (286–331) |
| Soltu.DM.02G028560.1 | PIF02 | NOT NAMED | 1131 | 376 | 41,285.79 | 5.04 | bHLH (151–197) |
| Soltu.DM.03G029660.1 | PIF03 | PIF7 | 1314 | 437 | 48,174.88 | 6.79 | bHLH (237–283) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.; Yang, J.; Zhang, N.; Gong, Y.; Liu, M.; Qiao, R.; Jiao, X.; Zhu, F.; Li, X.; Si, H. Genome-Wide Identification of Phytochrome-Interacting Factor (PIF) Gene Family in Potatoes and Functional Characterization of StPIF3 in Regulating Shade-Avoidance Syndrome. Agronomy 2024, 14, 873. https://doi.org/10.3390/agronomy14040873
Han Y, Yang J, Zhang N, Gong Y, Liu M, Qiao R, Jiao X, Zhu F, Li X, Si H. Genome-Wide Identification of Phytochrome-Interacting Factor (PIF) Gene Family in Potatoes and Functional Characterization of StPIF3 in Regulating Shade-Avoidance Syndrome. Agronomy. 2024; 14(4):873. https://doi.org/10.3390/agronomy14040873
Chicago/Turabian StyleHan, Yuwen, Jiangwei Yang, Ning Zhang, Yating Gong, Mei Liu, Run Qiao, Xinhong Jiao, Fengjiao Zhu, Xinxia Li, and Huaijun Si. 2024. "Genome-Wide Identification of Phytochrome-Interacting Factor (PIF) Gene Family in Potatoes and Functional Characterization of StPIF3 in Regulating Shade-Avoidance Syndrome" Agronomy 14, no. 4: 873. https://doi.org/10.3390/agronomy14040873
APA StyleHan, Y., Yang, J., Zhang, N., Gong, Y., Liu, M., Qiao, R., Jiao, X., Zhu, F., Li, X., & Si, H. (2024). Genome-Wide Identification of Phytochrome-Interacting Factor (PIF) Gene Family in Potatoes and Functional Characterization of StPIF3 in Regulating Shade-Avoidance Syndrome. Agronomy, 14(4), 873. https://doi.org/10.3390/agronomy14040873






