Comparative Transcriptome Analysis of Eggplant (Solanum melongena L.) Peels with Different Glossiness
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Samples and Preparation
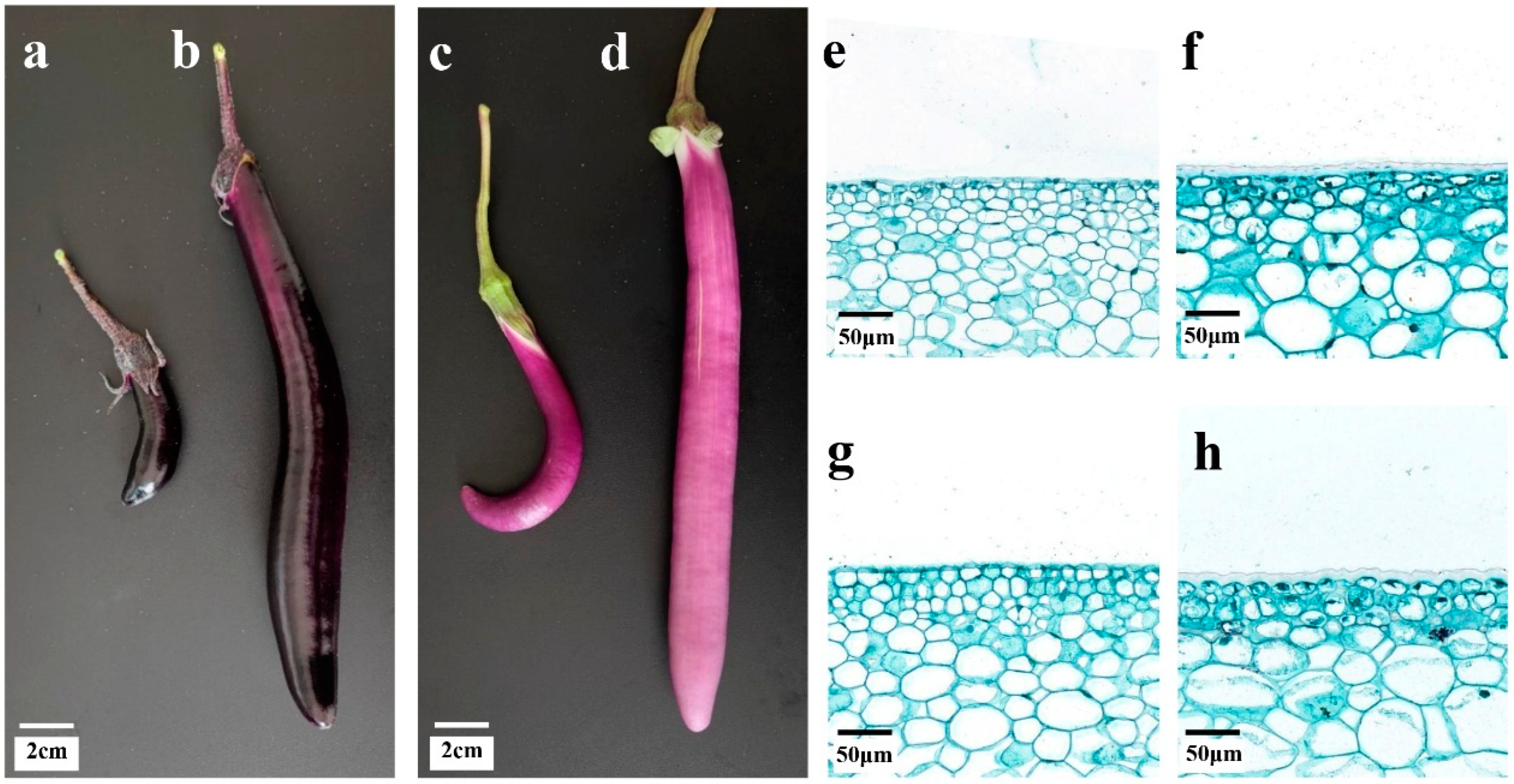
2.2. Pericarps Paraffin Sectioning
2.3. Total RNA Extraction, Library Construction, and Sequencing
2.4. Bioinformatic Analysis of the Sequencing Data and Differential Expression Analysis
2.5. Functional Enrichment and Transcription Factor Identification
2.6. Quantitative Real-Time PCR
3. Results
3.1. Morphological Characteristics of Eggplant Peel Glossiness
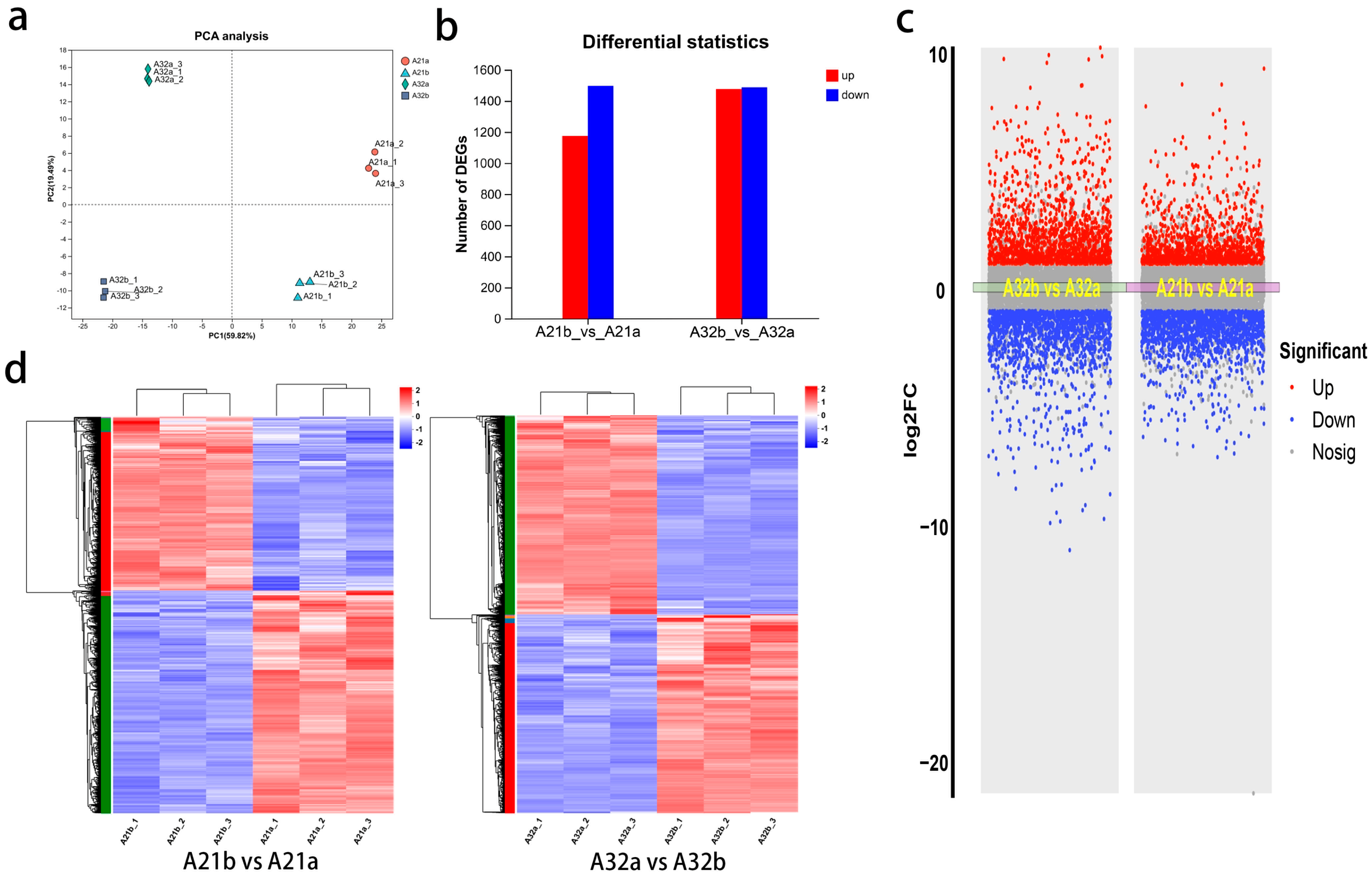
3.2. Quality Results of the Eggplant Peel Transcriptome Sequencing
3.3. Gene Expression in Eggplant Peels During Development
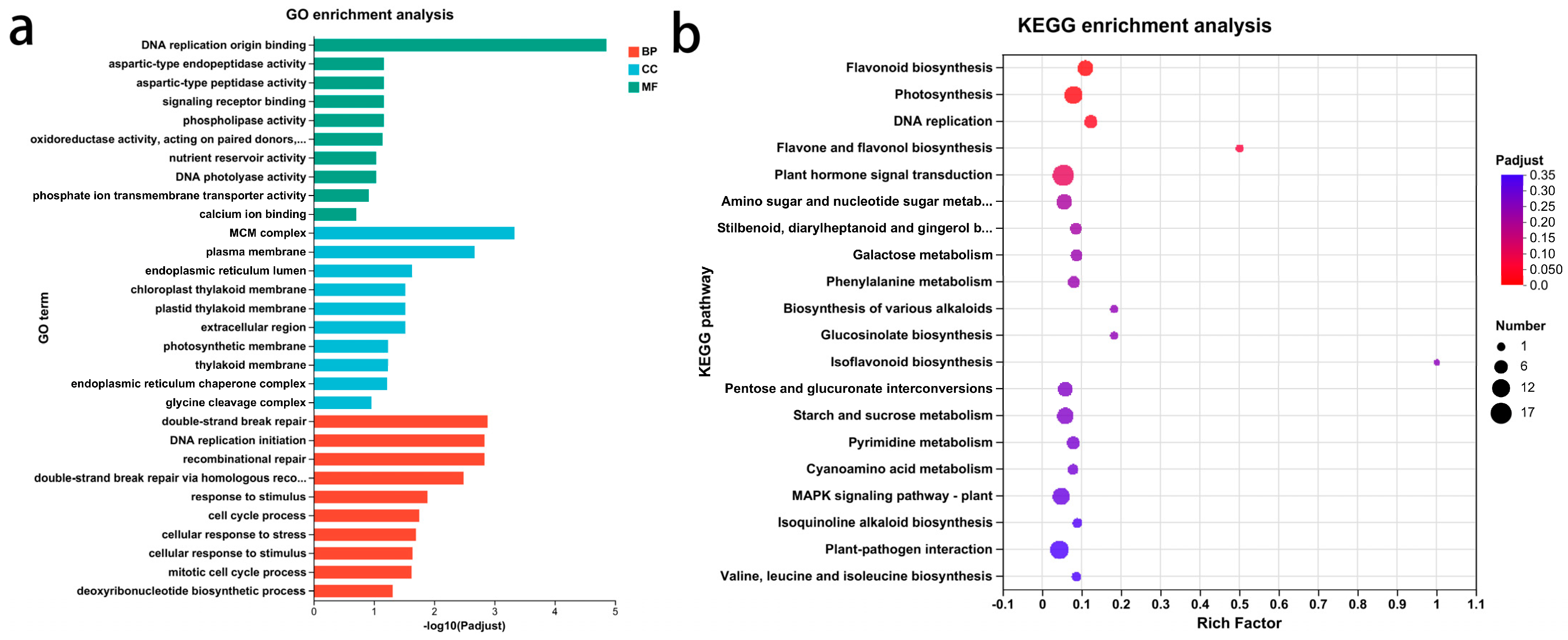
3.4. GO Functional Annotation and KEGG Pathway Analysis of DEGs Related to Glossiness
3.5. Identification of Differentially Expressed TFs
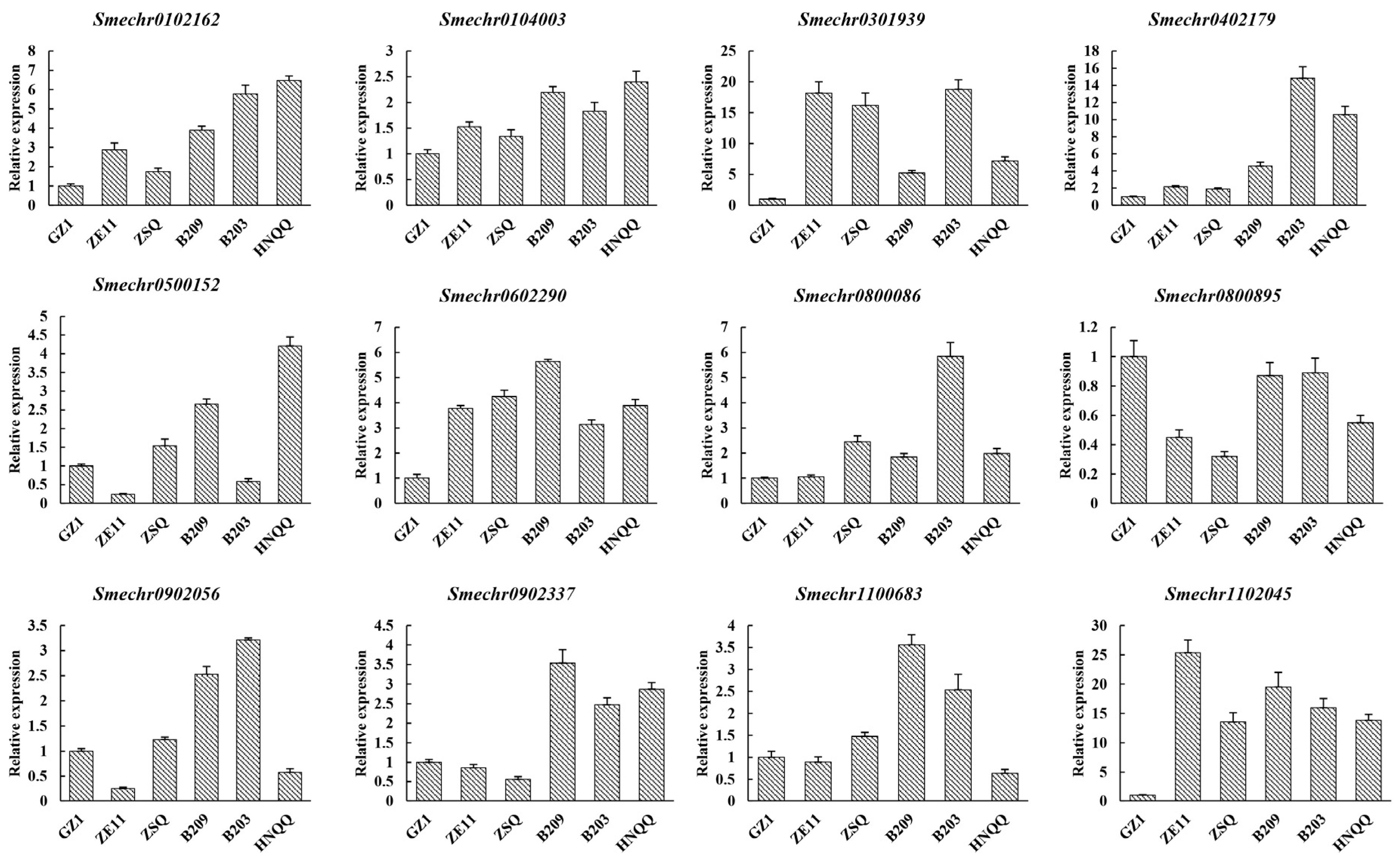
3.6. Expression of Candidate Genes Related to Peel Glossiness
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Alam, I.; Salimullah, M. Genetic engineering of eggplant (Solanum melongena L.): Progress, controversy and potential. Horticulturae 2021, 7, 78. [Google Scholar] [CrossRef]
- Jha, S.N.; Matsuoka, T. Objective Method of estimation of post-harvest freshness of eggplant fruits. IFAC Proc. Vol. 2001, 34, 67–70. [Google Scholar] [CrossRef]
- Argento, S.; Treccarichi, S.; Arena, D.; Rizzo, G.F.; Branca, F. Exploitation of a grafting technique for improving the water use efficiency of eggplant (Solanum melongena L.) grown in a cold greenhouse in mediterranean climatic conditions. Agronomy 2023, 13, 2705. [Google Scholar] [CrossRef]
- Gaccione, L.; Martina, M.; Barchi, L.; Portis, E. A compendium for novel marker-based breeding strategies in eggplant. Plants 2023, 12, 1016. [Google Scholar] [CrossRef]
- Jetter, R.; Kunst, L.; Samuels, A.L. Composition of Plant Cuticular Waxes. Ann. Plant Rev. Biol. Plant Cuticle 2007, 23, 145–181. [Google Scholar] [CrossRef]
- Wang, W.; Liu, X.; Gai, X.; Ren, J.; Liu, X.; Cai, Y.; Wang, Q.; Ren, H. Cucumis sativus L. WAX2 plays a pivotal role in wax biosynthesis, influencing pollen fertility and plant biotic and abiotic stress responses. Plant Cell Physiol. 2015, 56, 1339–1354. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; He, C.; Qin, Y.; Lin, G.; Park, W.D.; Sun, M.; Li, J.; Lu, X.; Zhang, C.; Yeh, C.T.; et al. Co-expression analysis aids in the identification of genes in the cuticular wax pathway in maize. Plant J. 2019, 97, 530–542. [Google Scholar] [CrossRef]
- Yang, S.; Liu, H.; Wei, X.; Zhao, Y.; Wang, Z.; Su, H.; Zhao, X.; Tian, B.; Zhang, X.W.; Yuan, Y. BrWAX2 plays an essential role in cuticular wax biosynthesis in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Theor. Appl. Genet. 2022, 135, 693–707. [Google Scholar] [CrossRef]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef] [PubMed]
- Rowland, O.; Zheng, H.; Hepworth, S.R.; Lam, P.; Jetter, R.; Kunst, L. CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 2006, 142, 866–877. [Google Scholar] [CrossRef]
- Tang, X.; Song, G.; Zou, J.; Ren, J.; Feng, H. BrBCAT1 mutation resulted in deficiency of epicuticular wax crystal in Chinese cabbage. Theor. Appl. Genet. 2024, 137, 123. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.C.; Zeng, Q.; Ji, Q.X.; Liu, C.F.; Liu, S.B.; Liu, Y. A comparison of the ultrastructure and composition of fruits’ cuticular wax from the wild-type ‘Newhall’ navel orange (Citrus sinensis [L.] Osbeck cv. Newhall) and its glossy mutant. Plant Cell Rep. 2012, 31, 2239–2246. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Cao, J.; Gong, S.; Hao, N.; Du, Y.; Wang, C.; Wu, T. The COPII subunit CsSEC23 mediates fruit glossiness in cucumber. Plant J. 2023, 116, 524–540. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ge, X.; An, J.; Liu, X.; Ren, H. CsCER6 and CsCER7 Influence Fruit Glossiness by Regulating Fruit Cuticular Wax Accumulation in Cucumber. Int. J. Mol. Sci. 2023, 24, 1135. [Google Scholar] [CrossRef]
- Xiong, C.; Xie, Q.; Yang, Q.; Sun, P.; Gao, S.; Li, H.; Zhang, J.; Wang, T.; Ye, Z.; Yang, C. WOOLLY, interacting with MYB transcription factor MYB31, regulates cuticular wax biosynthesis by modulating CER6 expression in tomato. Plant J. 2020, 103, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Petit, J.; Bres, C.; Just, D.; Garcia, V.; Mauxion, J.P.; Marion, D.; Bakan, B.; Joubès, J.; Domergue, F.; Rothan, C. Analyses of tomato fruit brightness mutants uncover both cutin-deficient and cutin-abundant mutants and a new hypomorphic allele of GDSL lipase. Plant Physiol 2014, 164, 888–906. [Google Scholar] [CrossRef]
- Petit, J.; Bres, C.; Mauxion, J.P.; Tai, F.W.; Martin, L.B.; Fich, E.A.; Joubès, J.; Rose, J.K.; Domergue, F.; Rothan, C. The Glycerol-3-phosphate acyltransferase GPAT6 from tomato plays a central role in fruit cutin biosynthesis. Plant Physiol 2016, 171, 894–913. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Establishment of an Identification Method for Citrus Fruit Surface Glossiness. Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2022. [Google Scholar]
- Li, L.; Zhao, M.; Wang, J.; Liu, S.; Wang, G.; Schnable, S.P. Research progress on genetic mechanisms of plant epidermal wax synthesis, transport and regulation. J. China Agric. Univ. 2023, 28, 1–19. [Google Scholar] [CrossRef]
- Park, C.S.; Go, Y.S.; Suh, M.C. Cuticular wax biosynthesis is positively regulated by WRINKLED4, an AP2/ERF-type transcription factor, in Arabidopsis stems. Plant J. 2016, 88, 257–270. [Google Scholar] [CrossRef]
- Kim, H.; Go, Y.S.; Suh, M.C. DEWAX2 transcription factor negatively regulates cuticular wax biosynthesis in Arabidopsis leaves. Plant Cell Physiol. 2018, 59, 966–977. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Li, C.; Hu, N.; Zhu, Y.; He, Z.; Sun, Y.; Wang, Z.; Wang, Y. ECERIFERUM1-6A is required for the synthesis of cuticular wax alkanes and promotes drought tolerance in wheat. Plant Physiol. 2022, 190, 1640–1657. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, C.; Wang, Y.; Wang, Y.; Ju, H.; Chen, X. Cucumber glossy fruit 1 (CsGLF1) encodes the zinc finger protein 6 that regulates fruit glossiness by enhancing cuticular wax biosynthesis. Hortic. Res. 2022, 10, 237. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Wu, H.; Wang, Y.; Zhang, Z.; Shan, L.; Zhao, X.; Wang, R.; Liu, C.; Weng, Y.; Wang, Y.; et al. The fruit glossiness locus, dull fruit (D), encodes a C(2)H(2)-type zinc finger transcription factor, CsDULL, in cucumber (Cucumis sativus L.). Hortic. Res. 2022, 9, 146. [Google Scholar] [CrossRef] [PubMed]
- Al-Abdallat, A.M.; Al-Debei, H.S.; Ayad, J.Y.; Hasan, S. Over-expression of SlSHN1 gene improves drought tolerance by increasing cuticular wax accumulation in tomato. Int. J. Mol. Sci. 2014, 15, 19499–19515. [Google Scholar] [CrossRef]
- Wu, P.; Li, S.; Yu, X.; Guo, S.; Gao, L. Identification of long-chain acyl-CoA synthetase gene family reveals that SlLACS1 is essential for cuticular wax biosynthesis in tomato. Int. J. Biol Macromol. 2024, 277, 134438. [Google Scholar] [CrossRef] [PubMed]
- Ni, F.; Yang, M.; Chen, J.; Guo, Y.; Wan, S.; Zhao, Z.; Yang, S.; Kong, L.; Chu, P.; Guan, R. BnUC1 is a key regulator of epidermal wax biosynthesis and lipid transport in Brassica napus. Int. J. Mol. Sci. 2024, 25, 9533. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Zhang, R.; Wei, B.; Nie, Z.; Xing, G.; Zhao, T.; Yang, S.; Gai, J. Key biological factors related to outcrossing-productivity of cytoplasmic-nuclear male-sterile lines in soybean [Glycine max (L.) Merr.]. Euphytica 2017, 213, 266. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Wang, J.; Wang, W.; Hu, T.; Hu, H.; Bao, C. A high-quality chromosome-level genome assembly reveals genetics for important traits in eggplant. Hortic. Res. 2020, 7, 153. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. goseq: Gene ontology testing for RNA-seq datasets. R. Bioconduct. 2012, 8, 1–25. [Google Scholar]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Liu, Z.; Liu, Y.; Ou, L.; Deng, M.-H.; Wang, J.; Song, J.; Ma, Y.; Chen, W.; Zhang, Z.; et al. Comparative transcriptome analysis between cytoplasmic male-sterile line and its maintainer during the floral bud development of pepper. Hortic. Plant J. 2020, 6, 89–98. [Google Scholar] [CrossRef]
- Hao, Y.; Luo, H.; Wang, Z.; Lu, C.; Ye, X.; Wang, H.; Miao, L. Research progress on the mechanisms of fruit glossiness in cucumber. Gene 2024, 927, 148626. [Google Scholar] [CrossRef] [PubMed]
- Hassanzadeh-Khayyat, M.; Akaberi, M.; Moalemzadeh Haghighi, H.; Sahebkar, A.; Emami, S.A. Distribution and variability of n-alkanes in waxes of conifers. J. Forestry. Res. 2019, 30, 429–433. [Google Scholar] [CrossRef]
- Ezer, R.; Manasherova, E.; Gur, A.; Schaffer, A.A.; Tadmor, Y.; Cohen, H. The dominant white color trait of the melon fruit rind is associated with epicuticular wax accumulation. Planta 2024, 260, 97. [Google Scholar] [CrossRef]
- Singh, Y.V.; Singh, K.K.; Sharma, S.K. Influence of crop nutrition on grain yield, seed quality and water productivity under two rice cultivation systems. Rice Sci. 2013, 20, 129–138. [Google Scholar] [CrossRef]
- Vidalis, N.; Kourkouvela, M.; Argyris, D.-C.; Liakopoulos, G.; Alexopoulos, A.; Petropoulos, S.A.; Karapanos, I. The impact of salinity on growth, physio-biochemical characteristics, and quality of Urospermum picroides and Reichardia picroides plants in varied cultivation regimes. Agriculture 2023, 13, 1852. [Google Scholar] [CrossRef]
- Besford, R.T.; Maw, G.A. Effect of potassium nutrition on tomato plant growth and fruit development. Plant Soil 1975, 42, 395–412. [Google Scholar] [CrossRef]
- Lu, W.; Zheng, W.; Wu, Y.; Zang, Y. Research review on features and molecular mechanism of wax formation in Brassicaceae. J. Zhejiang A&F Univ. 2021, 38, 205–213. [Google Scholar]
- Debono, A.; Yeats, T.H.; Rose, J.K.; Bird, D.; Jetter, R.; Kunst, L.; Samuels, L. Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 2009, 21, 1230–1238. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Yao, L.; Si, E.; Meng, Y.; Li, B.; Ma, X.; Yang, K.; Lai, Y.; Shang, X.; Li, C.; et al. Characterization of glossy spike mutants and identification of candidate genes regulating cuticular wax synthesis in barley (Hordeum vulgare L.). Int. J. Mol. Sci. 2022, 23, 13025. [Google Scholar] [CrossRef] [PubMed]
- Mohammadian, M.A.; Watling, J.R.; Hill, R.S. The impact of epicuticular wax on gas-exchange and photoinhibition in Leucadendron lanigerum (Proteaceae). Acta Oecologica 2007, 31, 93–101. [Google Scholar] [CrossRef]
- Li, W.; Li, J.; Wei, J.; Niu, C.; Yang, D.; Jiang, B. Response of photosynthesis, the xanthophyll cycle, and wax in Japanese yew (Taxus cuspidata L.) seedlings and saplings under high light conditions. Peer J. 2023, 11, e14757. [Google Scholar] [CrossRef]
| Sample | Raw Reads | Raw Data (Gb) | Clean Reads | Clean Reads Rates Ratio | Clean Data (Gb) | Q20 | GC Content | Mapped Reads | Mapped Rates Ratio |
|---|---|---|---|---|---|---|---|---|---|
| A32a_1 | 85,723,096 | 12.94 | 84,601,196 | 98.69% | 11.96 | 96.91% | 43.68% | 81,966,641.00 | 96.89% |
| A32a_2 | 72,334,058 | 10.92 | 71,496,558 | 98.84% | 10.09 | 96.8% | 43.6% | 69,182,538.00 | 96.76% |
| A32a_3 | 66,204,714 | 10.00 | 65,298,078 | 98.63% | 9.18 | 96.77% | 43.67% | 63,217,896.00 | 96.81% |
| A32b_1 | 66,688,094 | 10.07 | 65,590,548 | 98.35% | 9.26 | 96.74% | 43.61% | 63,501,960.00 | 96.82% |
| A32b_2 | 80,983,980 | 12.23 | 79,614,942 | 98.31% | 11.23 | 96.79% | 43.55% | 77,083,011.00 | 96.82% |
| A32b_3 | 70,561,868 | 10.65 | 69,218,874 | 98.10% | 9.76 | 96.8% | 43.62% | 67,102,207.00 | 96.94% |
| A21a_1 | 68,753,352 | 10.38 | 67,582,288 | 98.30% | 9.54 | 96.82% | 43.66% | 65,240,362.00 | 96.53% |
| A21a_2 | 68,370,232 | 10.32 | 67,442,464 | 98.64% | 9.46 | 96.8% | 43.57% | 65,221,053.00 | 96.71% |
| A21a_3 | 75,566,314 | 11.41 | 74,195,388 | 98.19% | 10.41 | 96.82% | 43.55% | 71,929,609.00 | 96.95% |
| A21b_1 | 74,872,704 | 11.31 | 73,454,914 | 98.11% | 10.35 | 96.89% | 43.74% | 71,100,144.00 | 96.79% |
| A21b_2 | 71,217,114 | 10.75 | 70,154,346 | 98.51% | 9.90 | 96.77% | 43.73% | 67,851,652.00 | 96.72% |
| A21b_3 | 64,693,436 | 9.77 | 63,827,850 | 98.66% | 8.76 | 96.6% | 43.84% | 61,653,848.00 | 96.59% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Nie, Z.; Wang, T.; Yang, S.; Zheng, J. Comparative Transcriptome Analysis of Eggplant (Solanum melongena L.) Peels with Different Glossiness. Agronomy 2024, 14, 3063. https://doi.org/10.3390/agronomy14123063
Wang H, Nie Z, Wang T, Yang S, Zheng J. Comparative Transcriptome Analysis of Eggplant (Solanum melongena L.) Peels with Different Glossiness. Agronomy. 2024; 14(12):3063. https://doi.org/10.3390/agronomy14123063
Chicago/Turabian StyleWang, Hong, Zhixing Nie, Tonglin Wang, Shuhuan Yang, and Jirong Zheng. 2024. "Comparative Transcriptome Analysis of Eggplant (Solanum melongena L.) Peels with Different Glossiness" Agronomy 14, no. 12: 3063. https://doi.org/10.3390/agronomy14123063
APA StyleWang, H., Nie, Z., Wang, T., Yang, S., & Zheng, J. (2024). Comparative Transcriptome Analysis of Eggplant (Solanum melongena L.) Peels with Different Glossiness. Agronomy, 14(12), 3063. https://doi.org/10.3390/agronomy14123063






