Abstract
Soil salinization and alkalinization are pervasive environmental issues that severely restrict plant growth and crop yield. Utilizing plant growth-promoting rhizobacteria (PGPR) is an effective strategy to enhance plant tolerance to saline–alkaline stress, though the regulatory mechanisms remain unclear. This study employed biochemical and RNA-Seq methods to uncover the critical growth-promoting effects of Trichoderma spp. on Salix linearistipularis under saline–alkaline stress. The results showed that, during saline–alkaline stress, inoculation with Trichoderma sp. M4 and M5 significantly increased the proline and soluble sugar contents in Salix linearistipularis, enhanced the activities of SOD, POD, CAT, and APX, and reduced lipid peroxidation levels, with M4 exhibiting more pronounced effects than M5. RNA-Seq analysis of revealed that 11,051 genes were upregulated after Trichoderma sp. M4 inoculation under stress conditions, with 3532 genes primarily involved in carbon metabolism, amino acid biosynthesis, and oxidative phosphorylation—processes that alleviate saline–alkaline stress. Additionally, 7519 genes were uniquely upregulated by M4 under stress, mainly enriched in secondary metabolite biosynthesis, amino acid metabolism, cyanamide metabolism, and phenylpropanoid biosynthesis. M4 mitigates saline–alkaline stress-induced damage in Salix linearistipularis seedlings by reducing oxidative damage, enhancing organic acid and amino acid metabolism, and activating phenylpropanoid biosynthesis pathways to eliminate harmful ROS. This enhances the seedlings’ tolerance to saline–alkaline stress, providing a basis for studying fungi–plant interactions under such conditions.
1. Introduction
Soil salinization and alkalinization pose major challenges to global crop production and ecological restoration [1]. The area of saline–alkaline soils continues to expand due to human activities and environmental degradation [2]. Excessive salt ions in saline–alkaline soils are toxic to plants, reducing microbial abundance and soil fertility [3]. The impact of saline–alkaline stress on plants is multifaceted, affecting the entire plant lifecycle, including leaf damage, enzyme activity, photosynthesis, and nutrient composition. This stress also impacts organ development, metabolism, root absorption, and ion utilization [4]. Thus, exploring effective strategies to mitigate the adverse effects of saline–alkaline stress on plants is imperative.
Salix linearistipularis mostly grows on the riverside and on saline and alkaline land, mainly distributed in northeastern China, Russia’s Far East, and Mongolia [5]. It is a kind of woody plant that is naturally distributed in the saline and alkaline land of the Songnen Plain of northeastern China. Salix linearistipularis has strong adaptability to saline soil [5], can be used as a greening afforestation species to improve saline land, and has great ecological value. Previous researchers have studied the biological characteristics, saline tolerance mechanism and root microorganisms of Salix linearistipularis.
Plant growth-promoting rhizobacteria (PGPR) are beneficial microorganisms that have been shown to induce plant tolerance to abiotic stresses such as drought, salinity, and nutrient deficiency [6]. Studies have demonstrated that PGPR can increase relative water content, plant height, leaf–stem ratio, biomass, photosynthetic pigments, and nutrient content, thereby improving plant tolerance to saline–alkaline stress [7]. PGPR produce various growth-regulating substances, such as siderophores and hydrogen cyanide, which enhance salt tolerance [8,9]. They also regulate plant hormone concentrations, such as indole-3-acetic acid, auxins, salicylic acid, and gibberellins, to promote growth [10]. Additionally, PGPR enhance the activities of antioxidants such as POD, SOD, APX, and CAT, reducing the entry of saline–alkaline components into plant cells [11,12,13]. Furthermore, studies have shown that PGPR improve lipid and jasmonic acid metabolism, enhancing salt tolerance [14,15]. Under saline–alkaline stress, PGPR inoculation alters plant signaling pathways, inducing systemic resistance to better cope with stress [16].
Trichoderma species, beneficial fungi that promote plant growth and immunity against plant pathogens, have garnered significant attention [17]. In rhizosphere colonization and as endophytes, Trichoderma has evolved to communicate with plants, offering hosts a wide range of benefits [18]. Research indicates that Trichoderma can alleviate the damage caused by saline–alkaline stress in maize seedlings by modulating soil environments and enzymatic activities [19]. Liu found that Trichoderma inoculation reduced ROS accumulation in cowpea tissues, effectively mitigating salt stress [20]. Zhang observed that Trichoderma inoculation improved cucumber root structure and alleviated the adverse effects of saline–alkaline stress [21]. Moreover, studies suggest that Trichoderma inoculation can regulate plant physiological functions, enhance photosynthesis, and improve salt–alkali tolerance [22]. Although previous research has reported the colonization of Trichoderma in various plant species, the mechanisms by which Trichoderma inoculation enhances Salix linearistipularis tolerance to saline–alkaline stress remain underexplored. This study further confirms, through physiological, biochemical, and molecular analyses, that strain M4 enhances the biosynthesis of lysine, glycine, and other relevant amino acids, as well as organic acids, carbon metabolism, and antioxidant enzyme activities in stressed seedlings. These findings contribute to our understanding of plant–microbe interactions under alkaline stress and propose a potential strategy for mitigating the adverse effects of alkaline stress on plant growth.
2. Materials and Methods
2.1. Experimental Materials and Design
The two endophytic Trichoderma strains used in this study were isolated from the roots of Salix linearistipularis and preserved in our laboratory. The seeds of Salix linearistipularis were collected from the experimental base of Northeast Forestry University in Anda City, Heilongjiang Province, China (latitude 46°01′–47°01′ N, longitude 124°53′–125°55′ E). The seeds were disinfected with 3% sodium hypochlorite for 10 min, washed five times with deionized water, soaked in deionized water at 25 °C for 12 h, and then placed in petri dishes for germination at 28 °C for 48 h. The germinated seeds were sown in pots and grown under natural conditions. After 30 days of growth, the seedlings were inoculated with the two Trichoderma strains using a root-drenching method [23]. Colonization was observed at 7, 14, 21, and 28 days post-inoculation, and the plants were subjected to saline–alkaline stress (0, 50, 200 mM) using a NaCl:Na2SO4:NaHCO3:Na2CO3 solution prepared at a ratio of 1:9:9:1 [24]. After 30 days of saline–alkaline stress, uniformly mature leaves above the middle of the stem were selected, with three biological replicates per treatment group, frozen in liquid nitrogen, and stored at −80 °C for further analysis.
2.2. Measurement of Physiological Indicators
The contents of malondialdehyde (MDA), soluble sugars (SS), and proline (Pro), as well as the activities of peroxidase (POD), superoxide dismutase (SOD), catalase (CAT), and ascorbate peroxidase (APX), were measured using thiobarbituric acid, anthrone colorimetry, acidic ninhydrin, guaiacol, nitro blue tetrazolium, and micro methods, respectively [25].
2.3. Construction of Transformants and Root Colonization in Salix linearistipularis
The transformants M4-GFP and M5-GFP were constructed using PEG-CaCl2 mediated protoplast transformation [23]. The transformants cake (8 mm) was inserted into sterilized PDB, at a speed of rotation of 150 r/min, and incubated in a shaker at a constant temperature of 25 °C for 7 days. The plants were then incubated at a constant temperature of 25 °C for 7 days. The number of spores was counted and diluted to 1 × 108 cfu/mL. Holes were punched around the inter-roots of the plants, and the fungal solution was evenly poured on: 21 mL of fungal solution was poured on each pot, three to four plants per pot, and untreated potted seedlings of the CK group were used as the control. After 20 days of inoculation, whole roots were washed thoroughly to remove soil and placed on slides for observation using the LSM 800 laser confocal system to evaluate the colonization status of the fungal transformants in the roots of Salix linearistipularis.
2.4. Transcriptome Sequencing
RNA from the samples was extracted using the Omega Plant RNA Extraction Kit according to the manufacturer’s instructions. Each RNA sample’s concentration was measured using a microspectrophotometer, ensuring all samples had a concentration greater than 600 ng/μL. Samples meeting the sequencing requirements were frozen and stored until sequencing.
2.5. Transcriptome Data Analysis
The quality of the raw data was evaluated using the Sanger quality score (Q). Sequencing results were filtered to remove adapters and low-quality sequences, obtaining clean reads. These clean reads were further assembled, and redundant reads were removed, to obtain unigenes. For samples with biological replicates, differential expression analysis between sample groups was performed using DESeq2 to obtain differentially expressed genes (DEGs) between two biological conditions. Post differential analysis, multiple hypothesis testing correction of the p value was conducted. DEGs were identified with a padj < 0.05 and a |log2 (Fold Change)| > 1. GO function and KEGG pathway enrichment analyses were performed on the DEGs [26]. The statistical analyses for all tables were performed using R software (version 4.0.1) and two-tailed paired t-tests; p < 0.05 was considered statistically significant. Transcriptome data were analyzed on the Metware cloud.
3. Results
3.1. Physiological Changes in Salix linearistipularis Seedlings Inoculated with Two Trichoderma Strains under Saline–Alkaline Stress
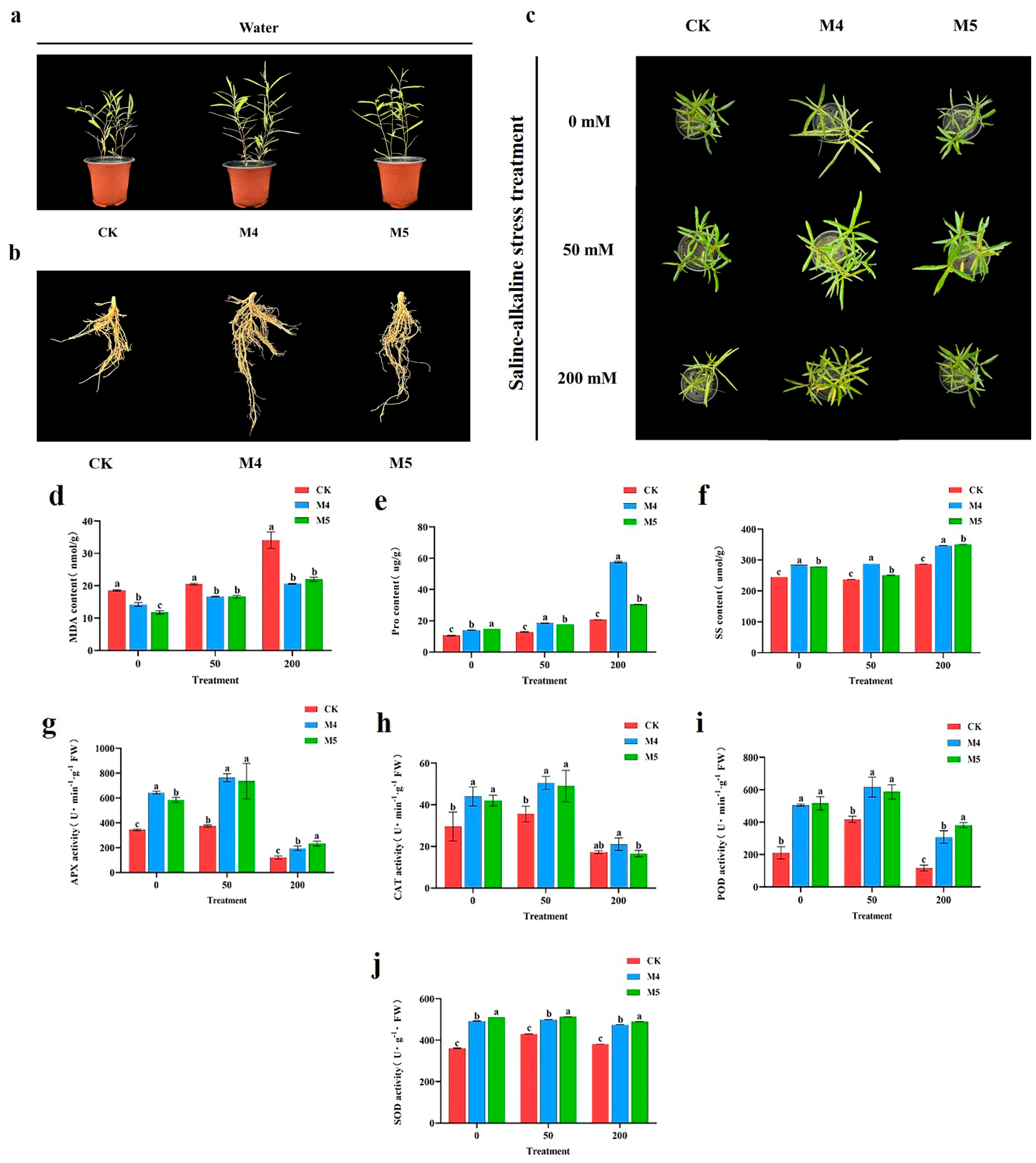
Salix linearistipularis plants, both inoculated and uninoculated with Trichoderma, were subjected to three saline–alkaline gradients (0, 50, 200 mM). Phenotypic characteristics (Figure 1a–c) showed that saline–alkaline stress inhibited the growth of Salix linearistipularis seedlings. However, inoculation with M4 and M5 promoted seedling growth, with M4-inoculated seedlings exhibiting superior height, root length, and leaf length compared to those inoculated with M5.
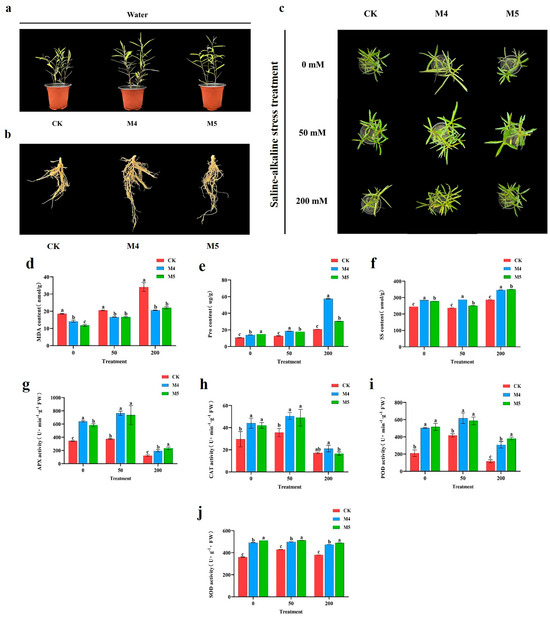
Figure 1.
Trichoderma enhances the tolerance of Salix linearistipularis to saline–alkaline stress. (a–c) Phenotypes of Salix linearistipularis inoculated or uninoculated with Trichoderma under normal or saline–alkaline stress conditions. (d) MDA content: malondialdehyde content. (e) Proline content. (f) Soluble sugar content. (g) APX activity: aseorbateperoxidase activity. (h) CAT activity: catalase activity. (i) POD activity: peroxidase activity. (j) SOD activity: superoxide dismutase activity. CK: uninoculated with Trichoderma, M4: inoculated with Trichoderma M4, M5: inoculated with Trichoderma M5. 0 mM, 50 mM, and 200 mM: saline–alkaline stress (0, 50, 200 mM) induced by NaCl:Na2SO4:NaHCO3:Na2CO3 at a ratio of 1:9:9:1. Data are from three biological repetitions (n = 3), means ± standard error (SE). Different lowercase letters indicate significant differences among different treatments (p < 0.05).
In the absence of saline–alkaline stress, significant differences were observed between the physiological indicators of M4- and M5-inoculated seedlings and the control group (p < 0.05). Under saline–alkaline stress, significant differences of the physiological indicators were observed between the treatment groups and the control (p < 0.05), but not between the two treatment groups. As saline–alkaline stress increased, the MDA content in the leaves of all groups also increased (Figure 1d).
The Pro content in the leaves increased with rising saline–alkaline stress across all three groups (Figure 1e). The soluble sugar content, however, first decreased and then increased, reaching its lowest point at 50 mM saline–alkaline stress (Figure 1f). Under the same stress gradient, the Pro and SS contents in the M4 and M5 groups were higher those in the control group, with M4 showing the highest levels (Figure 1e,f). These results suggest that the presence of Trichoderma enhances the osmotic regulation capacity of Salix linearistipularis.
The study also monitored dynamic changes in physiological indicators in Salix linearistipularis leaves infected with Trichoderma under different saline–alkaline stress gradients. Under low-concentration stress (50 mM), the activities of APX, CAT, POD, and SOD were higher compared to non-stress conditions, aiding in the removal of peroxides and free radicals. At high-concentration stress (200 mM), excessive peroxide accumulation led to a rapid decrease in the activities of these enzymes. As saline–alkaline stress increased, the activities of antioxidant enzymes initially increased and then decreased significantly. Compared to the control, APX, CAT, POD, and SOD activities in the treatment groups were higher, with M4 showing the highest activities (Figure 1g–j). These findings indicate that saline–alkaline stress significantly impacts the antioxidant system in Salix linearistipularis, causing substantial damage to the seedlings. Inoculation with Trichoderma enhances antioxidant enzyme activities, thereby improving the seedlings’ resistance to saline–alkaline stress, with M4 having a more pronounced effect than M5.
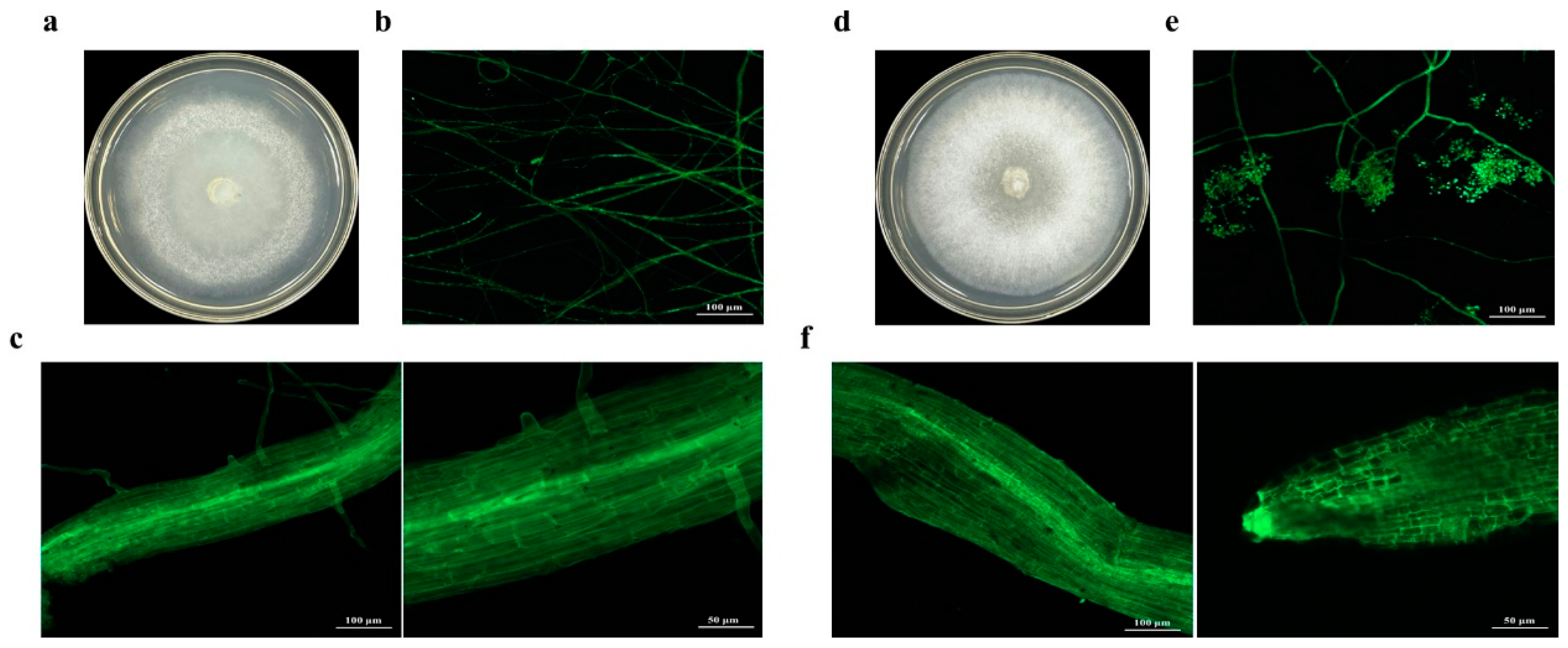
3.2. Colony Morphology of Transformants M4-GFP and M5-GFP and Their Colonization in Salix linearistipularis Roots
After six generations of subculture, transformants M4-GFP and M5-GFP were transferred to media containing 15 μg/mL and 20 μg/mL HmB, respectively (Figure 2a,d), where they grew well and emitted strong green fluorescence under a fluorescence microscope (Figure 2b,e). Using the LSM 800 laser confocal system, colonization of transformants M4-GFP and M5-GFP in the roots of Salix linearistipularis was observed. After 30 days of infection, the hyphae had further invaded and spread along the intercellular spaces, forming a network-like structure, indicating that the fungi had fully colonized the plant roots (Figure 2c,f).
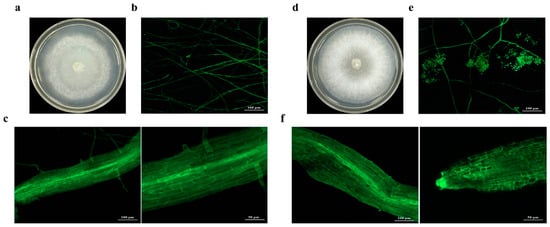
Figure 2.
Colony morphology and root colonization of Trichoderma transformants. (a,d) Colony morphology of M4-GFP and M5-GFP after 3 days on PDA medium. (b,e) Morphology of M4-GFP and M5-GFP under a fluorescence microscope. (c,f) Colonization of M4-GFP in Salix linearistipularis roots under normal conditions.
3.3. Differential Gene Expression Analysis
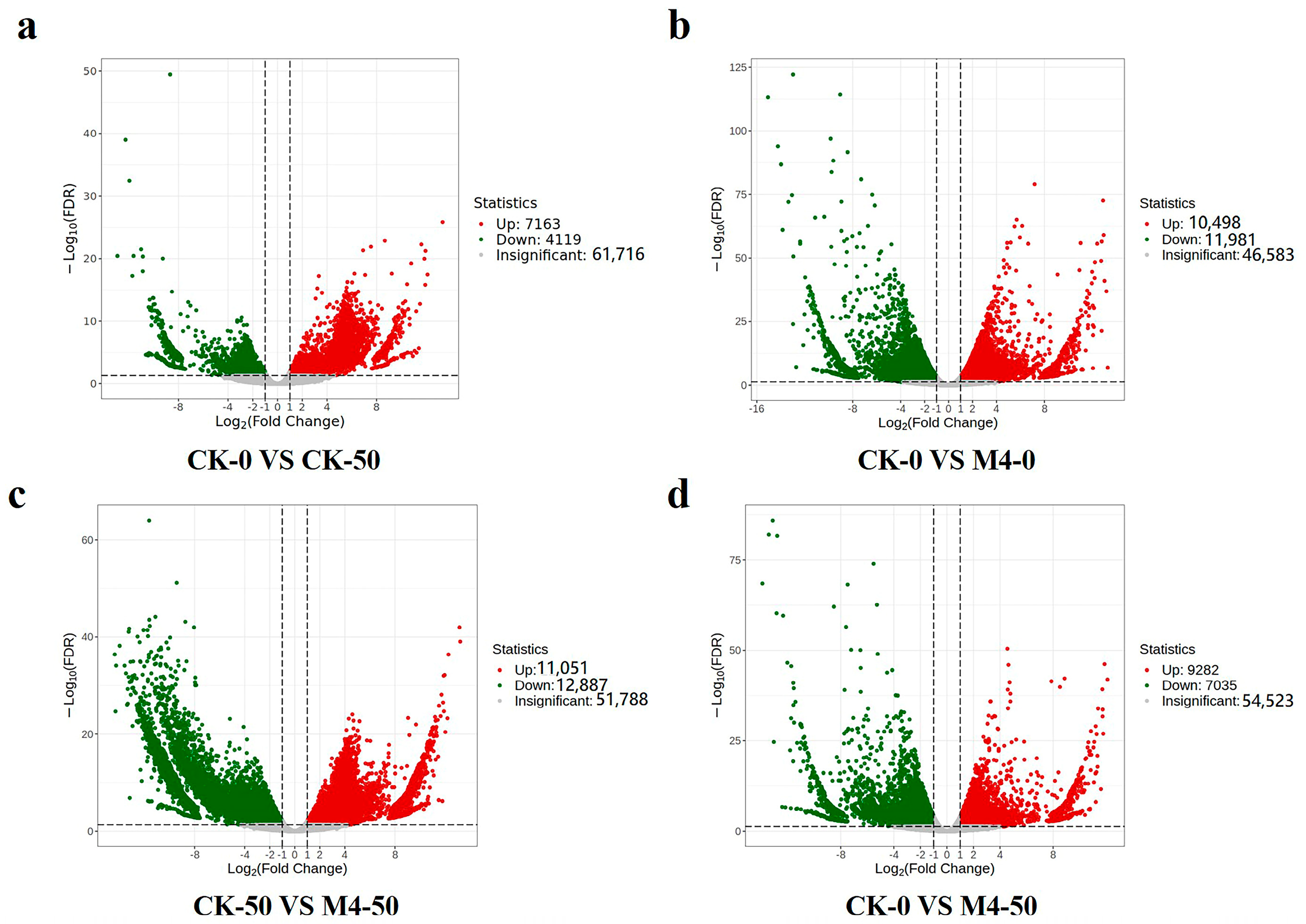
To study the regulatory mechanisms of the superior growth-promoting fungus M4 under saline–alkaline stress in Salix linearistipularis, RNA sequencing analysis was conducted on leaves of Salix linearistipularis seedlings interacting with Trichoderma M4 under 50 mM saline–alkaline stress. Differential expression analysis identified significant differentially expressed genes (DEGs) with FDR < 0.05 and log2 Fold Change > 1.
Without saline–alkaline stress, 10,498 genes were significantly upregulated and 11,981 genes were significantly downregulated in M4-treated seedlings compared to the control (Figure 3b). Under saline–alkaline stress, 7163 genes were upregulated, and 4119 genes were significantly downregulated compared to the control (Figure 3a). Inoculation with M4 under saline–alkaline stress led to significant upregulation of 11,051 genes and downregulation of 12,887 genes compared to the saline–alkaline control (Figure 3c). The expression levels of DEGs were considerably higher in M4-treated seedlings under saline–alkaline stress compared to the control (Figure 3d), indicating that M4 plays a significant regulatory role at the transcriptional level in the saline–alkaline stress response of Salix linearistipularis seedlings.
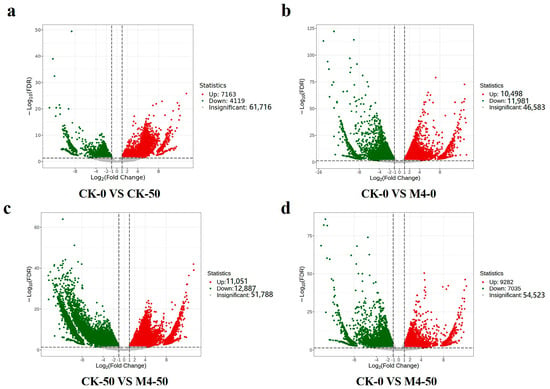
Figure 3.
The number of DEGs (differentially expressed genes) in different comparison groups. (a) Volcano plot of DEGs in CK-0 vs. CK-50. (b) Volcano plot of DEGs in CK-0 vs. M4-0. (c) Volcano plot of DEGs in CK-50 vs. M4-50. (d) Volcano plot of DEGs in CK-0 vs. M4-50.
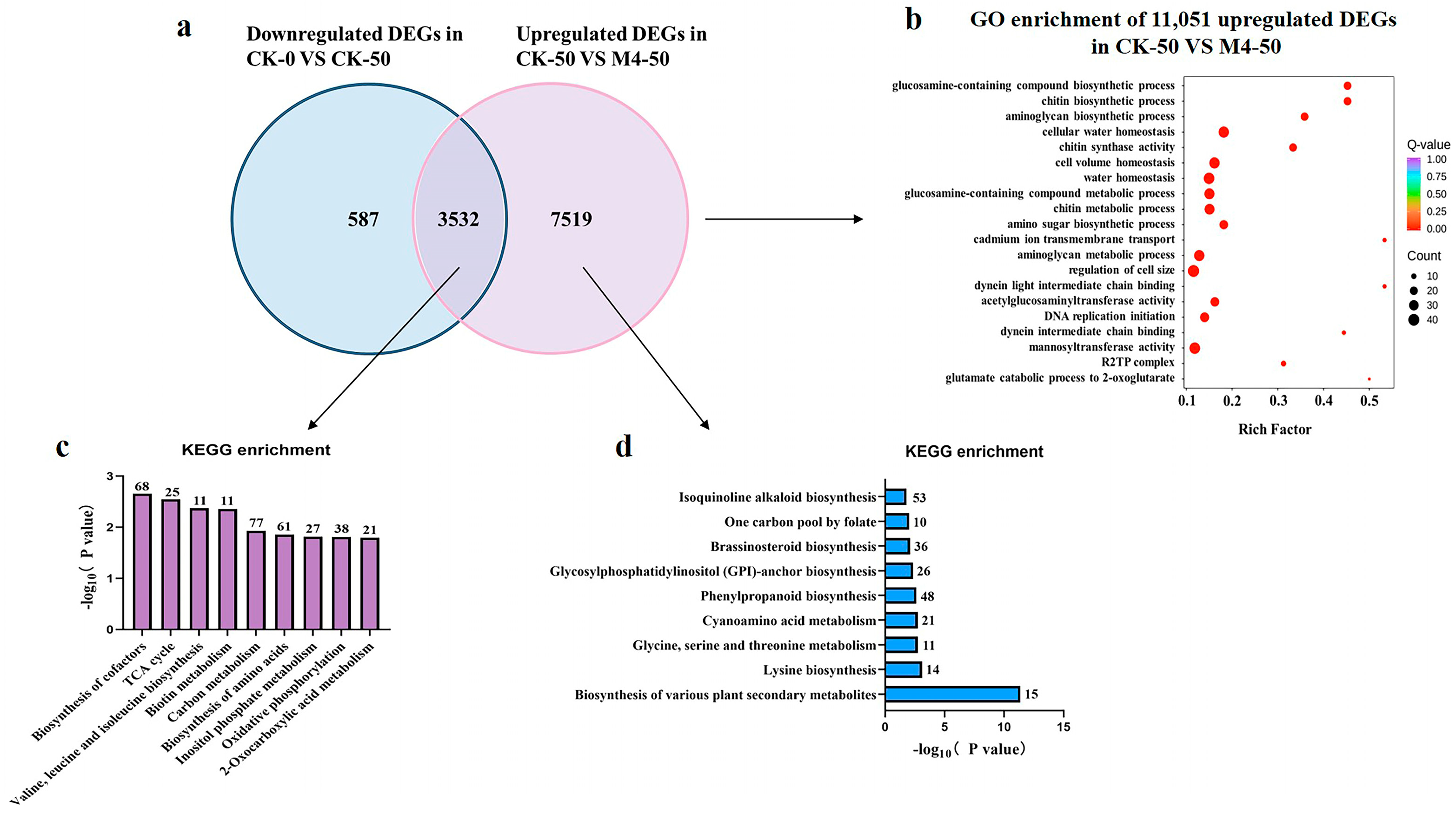
The Venn diagram shows that M4 inoculation significantly upregulated 3532 genes that were downregulated under saline–alkaline stress. Additionally, M4 uniquely induced the upregulation of 7519 genes, enhancing the tolerance of Salix linearistipularis seedlings to saline–alkaline stress (Figure 4a).
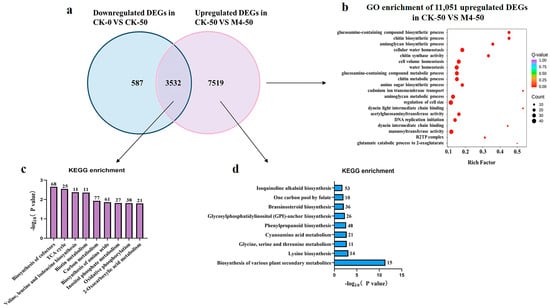
Figure 4.
RNA-Seq analysis of Salix linearistipularis seedlings inoculated with or without M4 under normal or saline–alkaline stress conditions. (b) GO analysis of 11,051 upregulated genes in CK-50 vs. M4-50. (c) KEGG enrichment analysis of genes in the overlapping portion of the Venn diagram described in (a). (d) KEGG enrichment pathways of 7519 uniquely upregulated DEGs in CK-50 vs. M4-50. KEGG: Kyoto Encyclopedia of Genes and Genomes.
To further understand the growth-promoting mechanisms of Trichoderma M4 under saline–alkaline stress, GO enrichment analysis was performed on 11,051 upregulated genes post-M4 inoculation under stress. These genes were mainly enriched in processes such as chitin biosynthetic process, glucosamine-containing compound biosynthetic process, aminoglycan biosynthetic process, cellular water homeostasis, cell volume homeostasis, and water homeostasis, which are associated with plant defense, growth, and metabolism under saline–alkaline stress. Molecular function analysis revealed an enrichment of various oxidoreductases, suggesting that M4 may enhance saline–alkaline tolerance by boosting the plant’s antioxidant system (Figure 4b).
The KEGG results showed that 3532 differentially expressed genes were primarily enriched in biosynthesis of cofactors, the TCA cycle, valine, leucine, and isoleucine biosynthesis, biotin metabolism, carbon metabolism, biosynthesis of amino acids, inositol phosphate metabolism, and the oxidative phosphorylation pathway (Figure 4c). These metabolic pathways are crucial for Salix linearistipularis’s response to saline–alkaline stress.
M4 uniquely induced the upregulation of 7519 genes under saline–alkaline stress. These DEGs were enriched in KEGG pathways including biosynthesis of other secondary metabolites, lysine biosynthesis, glycine, serine, and threonine metabolism, cyanoamino acid metabolism, and phenylpropanoid biosynthesis (Figure 4d). These results suggest that these compounds may act as multifunctional signaling molecules, regulating the expression of stress-related genes and increasing antioxidant enzyme synthesis, thereby enhancing plant stress resistance.
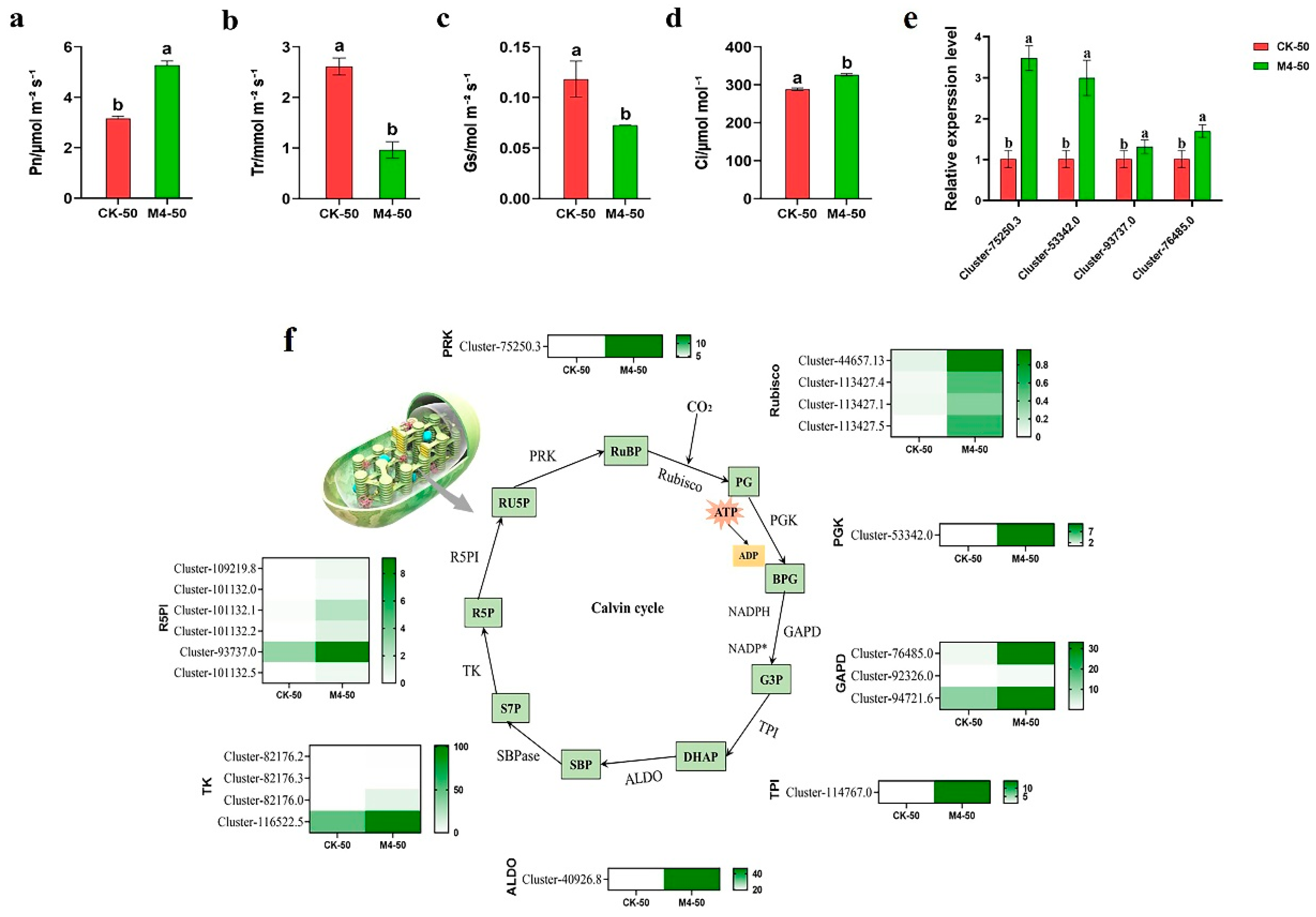
3.4. Photosynthetic Gas Exchange Parameters and Carbon Assimilation-Related Gene Expression Levels
To further investigate the effects of M4 inoculation on the tolerance of Salix linearistipularis to saline–alkaline stress, we studied photosynthesis in Salix linearistipularis leaves. Compared with CK-50, the Pn and Ci of M4-50 significantly increased by 66.70% and 13.18%, respectively (p < 0.05). Tr and Gs significantly decreased by 63.04% and 38.65%, respectively (p < 0.05) (Figure 5a–d). Transcriptome data (Figure 5f) indicated that, compared with CK-0, 21 differentially expressed genes related to the Calvin cycle (including Rubisco, PGK, GAPD, TPI, ALDO, TK, R5PI, and PRK) were upregulated in plants inoculated with M4 under 50 mM saline–alkaline stress. Consistent results were obtained from qRT-PCR analysis of genes in the Calvin cycle (Figure 5e).
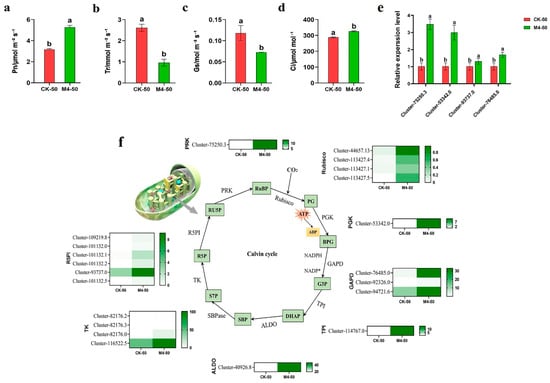
Figure 5.
Effects of M4 inoculation on photosynthetic characteristics and expression of Calvin cycle-related genes in Salix linearistipularis leaves under saline–alkaline stress. (a) Net photosynthetic rate (Photo); (b) transpiration rate (Trmmol); (c) conductance to H2O (Cond); (d) intercellular CO2 concentration (Ci); (e) qRT-PCR analysis of Calvin cycle-associated genes in mock-inoculated or M4-inoculated seedlings under saline–alkaline stress. (f) Calvin cycle-related genes transcriptional heat map. RuBP: Ribulose-1,5-bisphosphate, Rubisco: ribulose-1,5-bisphosphate carboxylase/oxygenase, PG: 2,3-phosphoglycerate, ATP: adenosine triphosphate, ADP: adenosine diphosphate; PGK: phosphogly-cerate kinase, BPG: 1,3-phosphoglycerate, GAPD: glyceraldehyde-3-phosphate dehydrogenase, NADPH: reduced nicotinamide adenine dinucleotide phosphate, NADP+: oxidation nicotinamide adenine dinucleotide phosphate, G3P: glyceraldehyde 3-phosphate, TPI: triosephosphate isomerase, DHAP: dihydroxyacetone phosphate, ALDO: fructose-bisphosphate aldolase, SBP: sedoheptulose-1,7-bisphosphate, SBPase: sedoheptulose-1,7-bisphosphatase, S7P: sedoheptulose7-phosphate, TK: transketolase, R5P: ribose 5-phosphate, R5PI: ribose-5-phosphate isomerase, RU5P: ribulose-5-phosphate, PRK: ribulose-5-bisphosphate kinase. Note: The data in Figure 5 are from three replicates (n = 3) and are shown as means ± standard error (SE). Different lowercase letters indicate significant differences (p < 0.05).
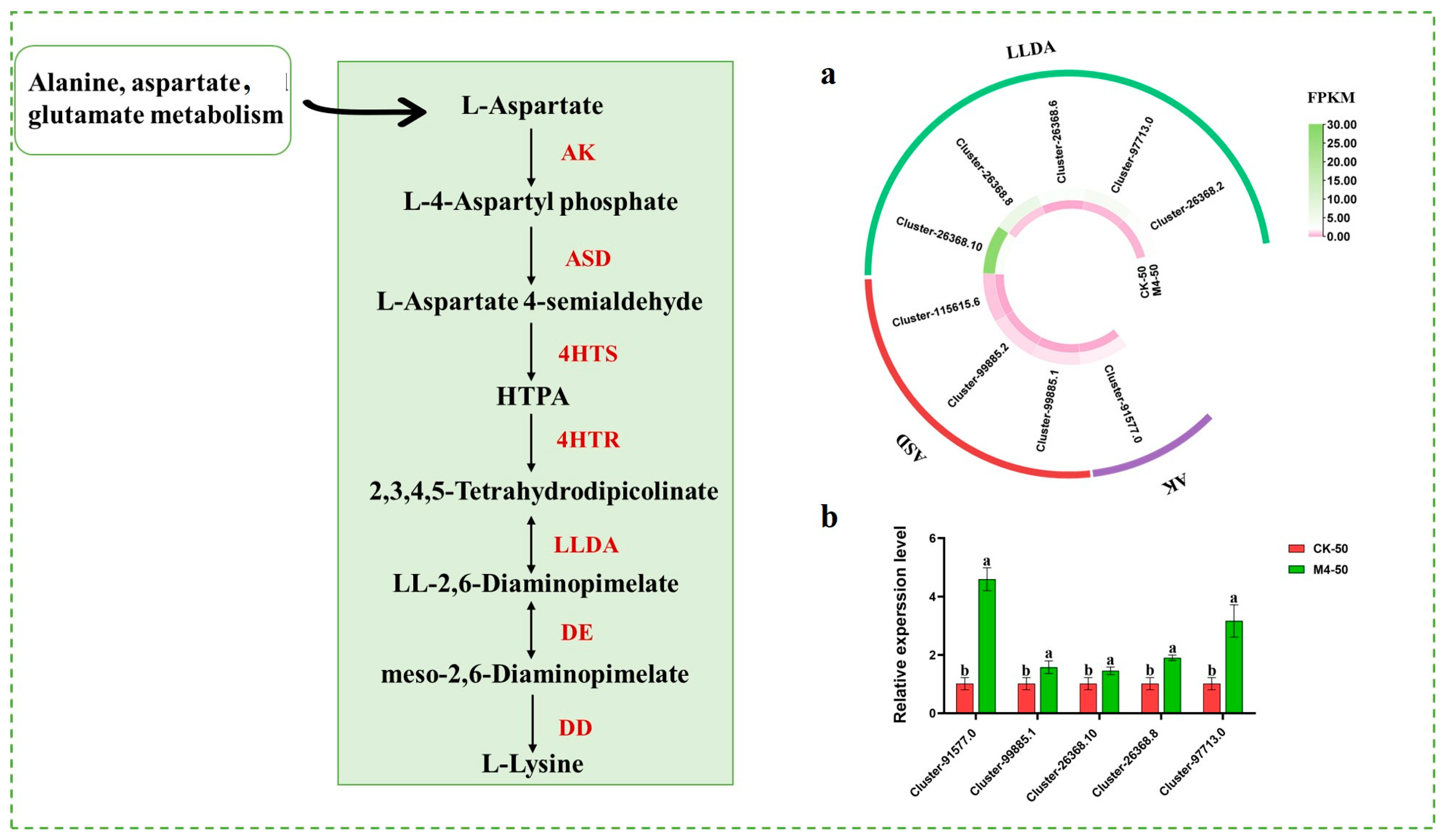
3.5. Response of Lysine Metabolic Pathway to Saline Stress
Lysine, as a multifunctional signaling molecule, can regulate the expression of stress-resistant genes in plants, promote the activation of cytochrome oxidase, and increase the synthesis of antioxidant enzymes, thereby enhancing plant stress resistance. Transcriptome data related to lysine synthesis genes (Figure 6a) showed that, under saline–alkaline stress, compared with the control group, the M4 inoculation treatment group exhibited significant upregulation trends in one AK (Cluster-91,577.0), three ASD (Cluster-99,885.1, Cluster-99,885.2, Cluster-115,615.6), and five LLDA (Cluster-26,368.10, Cluster-26,368.8, Cluster-26,368.6, Cluster-97,713.0, Cluster-26,368.2) clusters. The expression levels of lysine-related genes between inoculated and non-inoculated seedlings under saline–alkaline stress further supported this finding (Figure 6b).
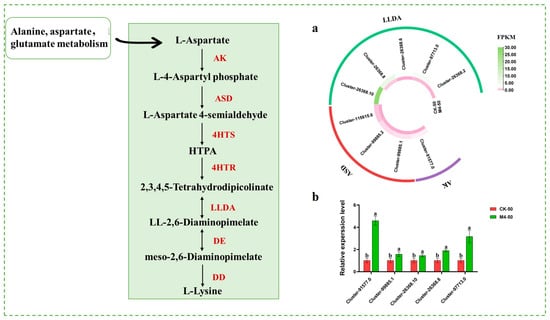
Figure 6.
Expression of genes related to lysine metabolic pathways in M4-inoculated Salix linearistipularis leaves under saline–alkaline stress. (a) Lysine metabolism-related genes transcriptional heat map; (b) qRT-PCR analysis of lysine metabolism-associated genes in mock-inoculated or M4-inoculated seedlings under saline–alkaline stress. AK: Aspartate kinase, ASD: aspartate-semialdehyde dehydrogenase, 4HTS: 4-hydroxy-tetrahydrodipicolinate synthase, 4HTR: 4-hydroxy-tetrahydrodipicolinate reductase, LLDA: LL-diaminopimelate aminotransferase, DE: diaminopimelate epimerase, DD: diaminopimelate decarboxylase. Note: The data in Figure 6 are from three replicates (n = 3) and are shown as means ± standard error (SE). Different lowercase letters indicate significant differences (p < 0.05).
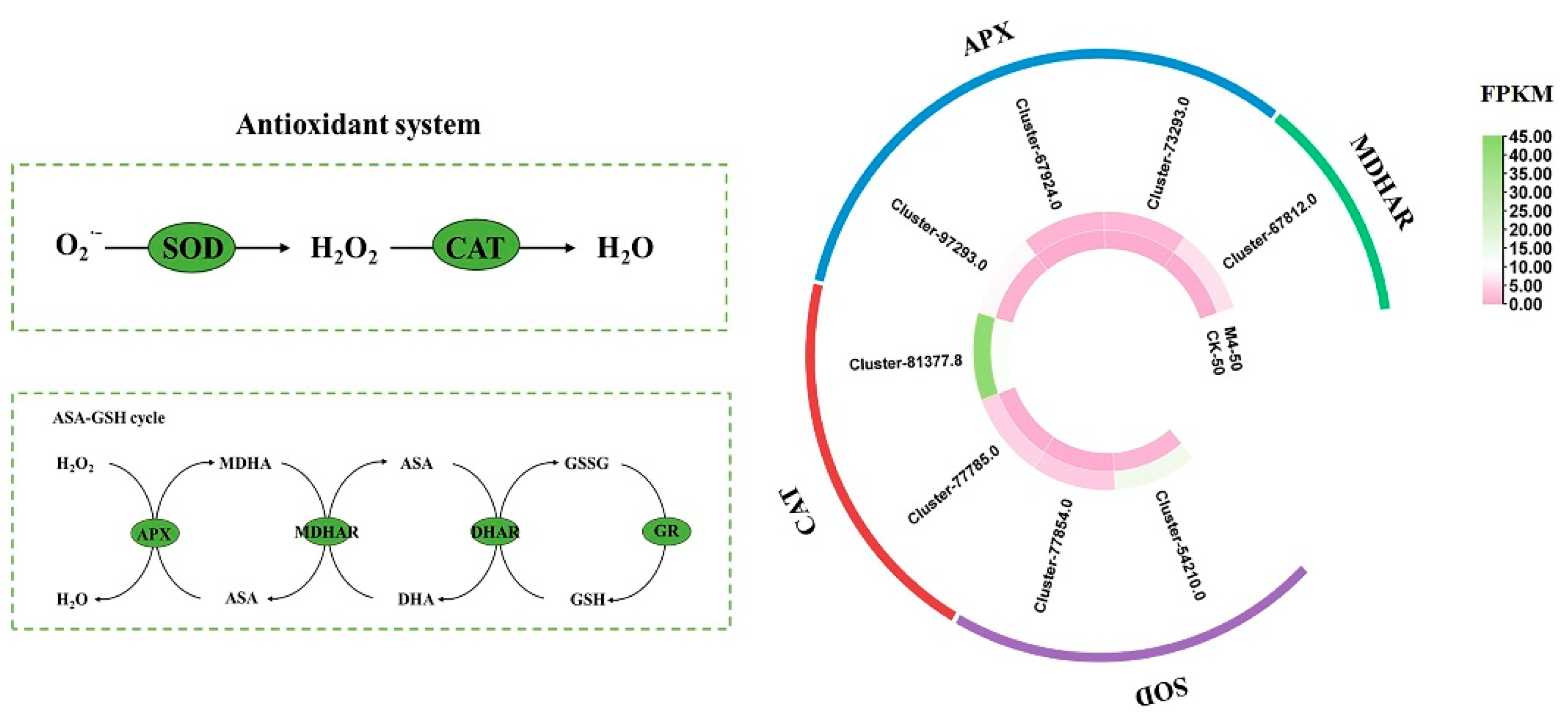
3.6. Response of the Antioxidant System to Saline–Alkaline Stress
Under saline–alkaline stress, M4 inoculation significantly reduced the MDA content in Salix linearistipularis seedlings compared to the uninoculated group (Figure 1a), while increasing the activities of SOD, POD, CAT, and APX, indicating that M4 alleviated oxidative damage induced by saline–alkaline stress (Figure 1g–j). Transcriptome analysis also showed that M4 regulated the expression of several antioxidant enzyme-related genes. Overall, M4 inoculation increased antioxidant enzyme activities, reducing oxidative damage in Salix linearistipularis seedlings under saline–alkaline stress (Figure 7).
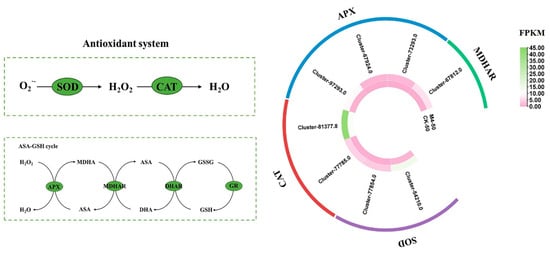
Figure 7.
Expression profiles of genes related to the antioxidant system. SOD: Superoxide dismutase, CAT: catalase, APX: ascorbate peroxidase, MDHAR: monodehydroascorbate reductase, DHAR: dehydroascorbate reductase, GR: glutathione reductase. Note: The data in Figure 7 are from three replicates (n = 3) and represent means ± standard error (SE).
4. Discussion
Analysis of physiological changes and transcriptomic data in Salix linearistipularis leaves under saline–alkaline stress confirms that inoculation with Trichoderma can enhance the tolerance of Salix linearistipularis to alkaline conditions. Trichoderma inoculation improves the physiological and biochemical responses of Salix linearistipularis to saline–alkaline stress, promoting plant growth and providing a physiological basis for enhancing the stress tolerance of symbiotic plants.
4.1. Effects of Trichoderma on Photosynthesis of Salix linearistipularis under Saline–Alkaline Stress
Photosynthesis parameters are important indicators for assessing plant growth and resistance. Under saline–alkaline stress, photosynthesis is hindered, the photosynthetic rate decreases, and plant growth and development are inhibited [27,28]. Additionally, saline–alkaline stress restricts water absorption by plant roots, and reduced or closed stomatal apertures lead to a decreased transpiration rate [29]. Under 50 mM saline–alkaline stress, the net photosynthetic rate of Salix linearistipularis leaves inoculated with M4 was significantly higher than that of CK, with a decreasing trend in transpiration rate and stomatal conductance and a slight increase in intercellular CO2 concentration. This suggests that M4 inoculation alleviated the adverse effects of saline–alkaline stress on the photosynthesis of Salix linearistipularis leaves, enhancing the photosynthetic rate. Meanwhile, Trichoderma colonization in the roots of Salix linearistipularis stimulated adventitious root formation and elongation of Salix linearistipularis roots, enhancing water absorption efficiency and improving the salt-alkaline tolerance of the Salix linearistipularis seedlings. The observed increase in intercellular CO2 concentrations may be due to the improved interaction between stomatal and non-stomatal factors influenced by M4 inoculation, as further confirmed by transcriptome data and related studies demonstrating that PGPR promotes plant growth under stress by improving photosynthesis [30].
4.2. Effects of Trichoderma Inoculation on ROS Accumulation and Osmotic Regulation in Salix linearistipularis under Saline–Alkaline Stress
Plants can produce osmotic regulators as a self-defense mechanism under stress. They can generate high levels of osmoprotectants such as soluble sugars, soluble proteins, and proline to maintain osmotic pressure and reduce cell water loss [31]. Under stress conditions, accumulated proline and soluble sugars act as low-molecular-weight antioxidants, detoxifying stress-induced toxicity and aiding in cellular osmotic regulation [32]. Transcriptome studies have also revealed that, under salt stress conditions, the presence of PGPR significantly improved water status, membrane integrity, and the expression of genes related to the accumulation of osmotic regulatory substances in rice [33]. PGPR enhances plant salt tolerance by secreting these osmotic regulatory substances [34]. Consistent with our findings, the levels of soluble sugars and proline increased gradually with the intensification of saline–alkaline stress, and plants accumulated these osmotic regulatory substances to maintain normal cell water potential, thereby ensuring normal cell growth. Compared with the control group, the SS and Pro content in the two microbial treatment groups increased, with M4 showing the most significant growth-promoting effect. This indicates that M4 enhanced the salt–alkaline tolerance of Salix linearistipularis by regulating the levels of related substances.
Malondialdehyde (MDA) is a product of lipid peroxidation and serves as a critical indicator for assessing cell membrane permeability and integrity [35]. It is commonly used to reflect the degree of cell membrane damage. Studies have shown that, under saline–alkaline stress, the molar concentration of MDA in leaves significantly increases [36,37]. This study also observed a significant upward trend in MDA content with increasing concentrations of saline–alkaline stress (Figure 1d), indicating that a large amount of reactive oxygen species (ROS) accumulated in the leaves under stress. This ROS accumulation exacerbated membrane lipid peroxidation and severely damaged the cell membrane structure. This damage is likely due to the increased Na+ content in leaves under saline–alkaline stress, disrupting ion balance and causing osmotic stress in the membrane system, thereby destabilizing the cell membrane structure. Compared with the control group, the overall MDA content in the leaves of Salix linearistipularis significantly decreased after inoculation with endophytic Trichoderma. Neshat et al. also found that the MDA content in plants significantly decreased after inoculation with plant growth-promoting rhizobacteria (PGPR) [38]. This suggests that high concentrations of saline–alkaline stress severely damage cell membranes and structure, triggering oxidative stress responses in plants. Inoculation with endophytic Trichoderma alleviates this damage to some extent, promoting plant growth and reducing cell membrane damage.
Higher plants have evolved specialized detoxification pathways to protect themselves from ROS toxicity, including pathways involving detoxifying enzymes such as catalase (CAT), superoxide dismutase (SOD), peroxidase (POD), and ascorbate peroxidase (APX), as well as the ascorbate-glutathione (ASA-GSH) cycle [31]. Previous studies have shown that plant cells can inhibit the accumulation of harmful intracellular ROS concentrations through an antioxidant defense system composed of enzymatic and non-enzymatic ROS scavengers [39]. During the early stages of stress, the activity of antioxidant enzymes in plants significantly increases [40], but under prolonged saline–alkaline stress, the activity of these enzymes tends to decrease [41]. Studies have found that, with prolonged stress, the activities of POD, CAT, SOD, and APX initially increase and then decrease [42,43]. This study confirmed this view, showing that, with increasing saline–alkaline stress concentrations, antioxidant enzyme activities initially increased and then decreased. Under low-concentration saline–alkaline stress, antioxidant enzyme activities increased to scavenge peroxides and free radicals. However, under high-concentration saline–alkaline stress, the accumulation of peroxides became too great, causing a rapid decline in enzyme activities and resulting in significant damage to Salix linearistipularis seedlings. Therefore, plants’ adaptive mechanisms involve increasing antioxidant enzyme activity under mild oxidative stress, but this response may be inhibited under severe oxidative stress [44].
Using PGPR has been proven to be an effective strategy to enhance plant salt tolerance, but its mechanisms are rarely reported. PGPR can enhance plant salt tolerance by scavenging ROS [45]. Studies have found that, under saline–alkaline stress, inoculation with plant growth-promoting rhizobacteria such as Bacillus subtilis, Enterobacter aerogenes, and Pseudomonas aeruginosa enhanced the activities of SOD, APX, and GPX in plants. These rhizobacteria mitigate the adverse effects of salinity and osmotic stress by regulating antioxidants [46,47]. Consistent with the above findings, the activities of various antioxidant enzymes increased in all treatment groups after Trichoderma inoculation. The transcriptome data from this study show that genes related to antioxidant enzymes were expressed under saline–alkaline stress with M4 inoculation. We speculate that Trichoderma enhances the ability of plants to resist adverse conditions by increasing the efficient collaboration of antioxidants and antioxidant enzymes within the plants.
4.3. Molecular Mechanisms of Trichoderma’s Growth-Promoting Effects on Salix linearistipularis under Saline–Alkaline Stress
The expression changes of differential genes in response to abiotic stress are markers of plants’ responses to stress signals. In this study, under 50 mM saline–alkaline stress, M4 inoculation led to the upregulation of 11,051 genes in Salix linearistipularis. M4 enhanced plant salt tolerance by activating complex molecular pathways, including those involved in the biosynthesis of various secondary metabolites, lysine, valine, leucine, and other amino acids, carbon metabolism, inositol phosphate metabolism, oxidative phosphorylation, cyanate metabolism, and phenylpropanoid biosynthesis. Previous research has revealed the critical role of amino acid biosynthesis-related pathways under stress conditions [48]. Among these, lysine metabolism plays a significant role in stress responses. Lysine is primarily catabolized via the saccharopine pathway, which has been shown to function in both abiotic and biotic stress responses. Furthermore, LKR/SDH expression increases under salt and osmotic stress, and downstream metabolites are enhanced. In Arabidopsis, the transcriptional memory induced by repeated salt stress depends on elevated lysine levels [49]. In plants, trimethylation of lysine 4 on histone H3 (H3K4me3) and acetylation of lysine 9 on histone H3 (H3K9ac) are closely associated with transcriptional activation [44,50]. Lysine and branched-chain amino acid degradation transfer electrons to the mitochondrial respiratory chain via ETFQO. Lysine can also be converted into N-hydroxypipecolic acid, acting as an immune signal [51,52,53,54]. Therefore, lysine metabolism is crucial for plants to withstand saline–alkaline stress. The process of oxidative phosphorylation within cells contributes to enhancing plant salt–alkaline tolerance [55]. Moreover, significant annotations in amino acid biosynthesis, carbon glucose synthesis, and oxidative phosphorylation pathways indicate the high metabolic capacity of Escherichia coli Rs-35 cells in saline environments [56]. These significantly enriched metabolic pathways are closely related to the ROS scavenging system, osmotic regulation in plants, and various amino acid-mediated salt-alkaline response pathways. Thus, it is hypothesized that, under saline–alkaline stress, M4 mitigates damage to Salix linearistipularis seedlings by modulating ROS scavenging, amino acid levels, and secondary metabolites like phenylpropanoids, thereby enhancing their ability to resist adverse conditions.
5. Conclusions
Physiological and transcriptomic analyses of the leaves of Salix linearistipularis inoculated with endophytic Trichoderma strains under different saline stress conditions demonstrated that M4 and M5 reduce MDA content and significantly increase osmotic regulators and antioxidant enzyme activities, mitigating saline–alkaline stress. The physiological and transcriptomic analyses showed that M4-inoculated plants responded faster to saline stress than M5-inoculated plants. Transcriptome analysis further revealed that M4-inoculated plants mainly responded to saline–alkaline stress by enriching pathways related to secondary metabolite biosynthesis, amino acid biosynthesis, carbon metabolism, phosphoinositol metabolism, and oxidative phosphorylation. These pathways are closely linked to ROS scavenging, osmotic regulation, and amino acid-mediated stress responses in Salix linearistipularis seedlings. At this point, it is important to highlight that, although no biochemical tests were conducted, the transcriptomic analyses provide valuable insights into the growth-promoting mechanisms associated with Trichoderma.
Author Contributions
Conceptualization, S.M.; performed the experiments, L.C., W.W., X.G., and J.S.; software, Z.H.; investigation, Z.H., L.C., and W.W.; data curation, Z.H.; writing—original draft preparation, Z.H.; writing—review and editing, S.M.; funding acquisition, S.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Special Fund for Basic Scientific Research of the Central Universities (2572023CT08) and the National Natural Science Foundation of China (U23A20151).
Informed Consent Statement
Salix linearistipularis seeds and Trichoderma were collected from the laboratory base of Northeast Forestry University, Anda City, Heilongjiang Province. The experiments were carried out in compliance with national and international guidelines. In this study, all methods were carried out in accordance with relevant guidelines.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We thank the financial support of the Fundamental Research Funds for the Central Universities for our study. The sampling work was carried out at the Anda saline–alkaline land test base of Northeast Forestry University. We are especially grateful to Li Yuhua for illuminating discussions on the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Shabala, S.; Wu, H.; Bose, J. Salt stress sensing and early signalling events in plant roots: Current knowledge and hypothesis. Plant Sci. 2015, 241, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Seki, K.; Miyazaki, T.; Ishihama, Y. The causes of soil alkalinization in the Songnen Plain of Northeast China. Paddy Water Environ. 2009, 7, 259–270. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, Q. Graphene ameliorates saline-alkaline stress-induced damage and improves growth and tolerance in alfalfa (Medicago sativa L.). Plant Physiol. Biochem. 2021, 163, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Liu, C. An Illustrated Catalogue of the Primary Colours of Saline and Alkaline Plants of Northeast China; Northeast Forestry University Press: Harbin, China, 2006. [Google Scholar]
- Tsotetsi, T.; Nephali, L.; Malebe, M.; Tugizimana, F. Bacillus for plant growth promotion and stress resilience: What have we learned? Plants 2022, 19, 2482. [Google Scholar] [CrossRef]
- Shahid, M.; Ameen, F.; Maheshwari, H.S.; Ahmed, B.; AlNadhari, S.; Khan, M.S. Colonization of Vigna radiata by a halotolerant bacterium Kosakonia sacchari improves the ionic balance, stressor metabolites, antioxidant status and yield under NaCl stress. Appl. Soil Ecol. 2021, 158, 103809. [Google Scholar] [CrossRef]
- Suchan, D.M.; Bergsveinson, J.; Manzon, L.; Pierce, A.; Kryachko, Y.; Korber, D.; Tan, Y.; Tambalo, D.D.; Khan, N.H.; Whiting, M.; et al. Transcriptomics reveal core activities of the plant growth-promoting bacterium Delftia acidovorans RAY209 during interaction with canola and soybean roots. Microb. Genom. 2020, 6, e000462. [Google Scholar] [CrossRef]
- Huang, X.F.; Zhou, D.; Guo, J.; Manter, D.K.; Reardon, K.F.; Vivanco, J.M. Bacillus spp. from rainforest soil promote plant growth under limited nitrogen conditions. J. Appl. Microbiol. 2015, 118, 672–684. [Google Scholar] [CrossRef]
- Santos, A.A.; Silveira, J.A.G.; Bonifacio, A.; Rodrigues, A.C.; Figueiredo, M.V.B. Antioxidant response of cowpea co-inoculated with plant growth-promoting bacteria under salt stress. Braz. J. Microbiol. 2018, 49, 513–521. [Google Scholar] [CrossRef]
- Abd_Allah, E.F.; Alqarawi, A.A.; Hashem, A.; Radhakrishnan, R.; Al-Huqail, A.A.; Al-Otibi, F.O.N.; Malik, J.A.; Alharbi, R.I.; Egamberdieva, D. Endophytic bacterium Bacillus subtilis (BERA 71) improves salt tolerance in chickpea plants by regulating the plant defense mechanisms. J. Plant Interact. 2018, 13, 37–44. [Google Scholar] [CrossRef]
- Khan, A.; Sirajuddin; Zhao, X.Q.; Javed, M.T.; Khan, K.S.; Bano, A.; Shen, R.F.; Masood, S. Bacillus pumilus enhances tolerance in rice (Oryza sativa L.) to combined stresses of NaCl and high boron due to limited uptake of Na+. Environ. Exp. Bot. 2016, 124, 120–129. [Google Scholar] [CrossRef]
- Arora, N.K.; Fatima, T.; Mishra, J.; Mishra, I.; Verma, S.; Verma, R.; Verma, M.; Bhattacharya, A.; Verma, P.; Mishra, P.; et al. Halo-tolerant plant growth promoting rhizobacteria for improving productivity and remediation of saline soils. J. Adv. Res. 2020, 26, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Waqas, M.; Khan, A.L.; Kamran, M.; Hamayun, M.; Kang, S.M.; Kim, Y.H.; Lee, I.J. Endophytic fungi produce gibberellins and indoleacetic acid and promotes host-plant growth during stress. Molecules 2012, 17, 10754–10773. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.-H.; Chen, X.-Y.; Yin, F.-X.; Xia, G.-M.; Yi, Y.; Zhang, Y.-B.; Liu, S.-W.; Li, F. Hybridization affects the structure and function of root microbiome by altering gene expression in roots of wheat introgression line under saline-alkali stress. Sci. Total Environ. 2022, 835, 155467. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Ma, Y.; Shadan, A. Perspective of ACC-deaminase producing bacteria in stress agriculture. J. Biotechnol. 2022, 352, 36–46. [Google Scholar] [CrossRef]
- Khan, R.A.A.; Najeeb, S.; Chen, J.; Wang, R.; Zhang, J.; Hou, J.; Liu, T. Insights into the molecular mechanism of Trichoderma stimulating plant growth and immunity against phytopathogens. Physiol. Plant. 2023, 175, e14133. [Google Scholar] [CrossRef]
- Woo, S.L.; Hermosa, R.; Lorito, M.; Monte, E. Trichoderma: A multipurpose, plant-beneficial microorganism for eco-sustainable agriculture. Nat. Rev. Microbiol. 2023, 21, 312–326. [Google Scholar] [CrossRef]
- Fu, J.; Xiao, Y.; Wang, Y.F.; Liu, Z.H.; Yang, K. Saline-alkaline stress in growing maize seedlings is alleviated by Trichoderma asperellum through regulation of the soil environment. Sci. Rep. 2021, 11, 11152. [Google Scholar] [CrossRef]
- Fu, J.; Liu, Z.; Li, Z.; Wang, Y.; Yang, K. Alleviation of the effects of saline-alkaline stress on maize seedlings by regulation of active oxygen metabolism by Trichoderma asperellum. PLoS ONE 2017, 12, e0179617. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, W.; Hu, Y.; Peng, Z.; Ren, S.; Xue, M.; Liu, Z.; Hou, J.; Xing, M.; Liu, T. A novel salt-tolerant strain Trichoderma atroviride HN082102.1 isolated from marine habitat alleviates salt stress and diminishes cucumber root rot caused by Fusarium oxysporum. BMC Microbiol. 2022, 22, 67. [Google Scholar] [CrossRef]
- Chen, J.; Guo, F.; Yang, W.; Wan, S.; Chen, W. Effect of yellow-green Trichoderma T1010 on salt tolerance physiology of peanut under salt stress. Southwest J. Agric. 2014, 27, 587–590. [Google Scholar]
- Meng, S. Dynamic Differences in Root Colonization of Fusarium oxysporum f. sp. niveum and Biocontrol Trichoderma harzianum in Watermelon and Their Response to Soil pH and Salt Concentration. Master’s Thesis, Ningxia University, Ningxia, China, 2022. [Google Scholar]
- Li, Y. Effect of Salt-Alkaline Interaction Stress on Physiological Characteristics of Salix linearistioularis. Master’s Thesis, Northeast Forestry University, Harbin, China, 2020. [Google Scholar]
- Wang, J.; Zhu, J.; Zhang, Y.; Fan, F.; Li, W.; Wang, F.; Zhong, W.; Wang, C.; Yang, J. Comparative transcriptome analysis reveals molecular response to salinity stress of salt-tolerant and sensitive genotypes of indica rice at seedling stage. Sci. Rep. 2018, 8, 2085. [Google Scholar] [CrossRef] [PubMed]
- Hou, R.; Yang, L.; Wuyun, T.; Chen, S.; Zhang, L. Genes related to osmoregulation and antioxidation play important roles in the response of Trollius chinensis seedlings to saline-alkali stress. Front. Plant Sci. 2023, 14, 1080504. [Google Scholar] [CrossRef]
- Liu, Z.; Tian, L.; Chen, M.; Zhang, L.; Lu, Q.; Wei, J.; Duan, X. Hormesis Responses of Growth and Photosynthetic Characteristics in Lonicera japonica Thunb to Cadmium Stress: Whether Electric Field Can Improve or Not? Plants 2023, 12, 933. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Che, D.; Wang, K. Effects of saline and alkaline stress on the physiological responses of four species of hydrangea roots. J. Northeast For. Univ. 2010, 38, 24–27. [Google Scholar]
- Peng, Y.; Xie, X.; Zhou, F.; Wan, H.; Zhang, C.; Zhai, R.; Zheng, Q.; Zheng, C.; Liu, Z. Growth and photosynthetic characteristics of alkali Suaeda glauca and Atriplex triangularis seedlings in response to different salinities. J. Grass Ind. 2012, 21, 64–74. [Google Scholar]
- Guo, L.; Zhang, X.; Zhao, J.; Zhang, A.; Pang, Q. Enhancement of sulfur metabolism and antioxidant machinery confers Bacillus sp. Jrh14-10–induced alkaline stress tolerance in plant. Plant Physiol. Biochem. 2023, 203, 108063. [Google Scholar] [CrossRef]
- Yu, B.; Liu, J.; Wu, D.; Liu, Y.; Cen, W.; Wang, S.; Li, R.; Luo, J. Weighted gene coexpression network analysis-based identification of key modules and hub genes associated with drought sensitivity in rice. BMC Plant Biol. 2020, 20, 478. [Google Scholar]
- Ashraf, M.; Foolad, M.R. Roles of Glycine Betaine and Proline in Improving Plant Abiotic Stress Resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Chauhan, P.S.; Lata, C.; Tiwari, S.; Chauhan, A.S.; Mishra, S.K.; Agrawal, L.; Chakrabarty, D.; Nautiyal, C.S. Transcriptional alterations reveal Bacillus amyloliquefaciens-rice cooperation under salt stress. Sci. Rep. 2019, 9, 11912. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Li, L.; Lindström, K.; Räsänen, L.A. A synergistic interaction between salt-tolerant Pseudomonas and Mesorhizobium strains improves growth and symbiotic performance of liquorice (Glycyrrhiza uralensis Fish.) under salt stress. Appl. Microbiol. Biotechnol. 2016, 100, 2829–2841. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.-L.; Ma, Y.-Y.; Liu, B.-H.; Liu, Z.-H.; Shao, H.-B. The changes of organelle ultrastructure and Ca2+ homeostasis in maize mesophyll cells during the process of drought-induced leaf senescence. Electron. J. Biotechnol. 2011, 14, 4. [Google Scholar] [CrossRef]
- He, M.; Wang, H.; Xu, P.; Liu, C.; Zhou, Y. Physiological response of Miscanthus sacchariflorus seedlings to combined salt and alkali stress. Acta Bot. Boreali-Occident. Sin. 2016, 36, 506–514. [Google Scholar]
- Liu, Q.; Wang, Z.; Zhou, X. Physiological response of Xanthium sibiricum to salt-alkali stress. J. Northeast. For. Univ. 2017, 45, 23–27. [Google Scholar]
- Neshat, M.; Abbasi, A.; Hosseinzadeh, A.; Sarikhani, M.R.; Chavan, D.D.; Rasoulnia, A. Plant growth promoting bacteria (PGPR) induce antioxidant tolerance against salinity stress through biochemical and physiological mechanisms. Physiol. Mol. Biol. Plants 2022, 28, 347–361. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, C.; Lei, J.; Dong, J.; Ren, J.; Shi, X.; Zhong, C.; Wang, X.; Zhao, X.; Yu, H. Comparative physiological and transcriptomic analyses reveal key regulatory networks and potential hub genes controlling peanut chilling tolerance. Genomics 2022, 114, 110285. [Google Scholar] [CrossRef]
- Xu, H.S.; Guo, S.M.; Zhu, L.; Xing, J.C. Growth, physiological and transcriptomic analysis of the perennial ryegrass Lolium perenne in response to saline stress. R. Soc. Open Sci. 2020, 7, 200637. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, Z.; Wang, S.; Zhao, T.; Zhang, D.; Ma, N.; Wang, Y. Saline-alkali stress tolerance is enhanced by MhPR1 in Malus halliana leaves as shown by transcriptomic analyses. Planta 2022, 256, 51. [Google Scholar] [CrossRef]
- Liu, F.; Liu, J.; Zhu, R.; Zhang, Y.; Guo, Y.; Han, G.; Tang, F. Physiological response and salt tolerance evaluation of 4 Oat species to NaCl stress. J. Grassl. Sci. 2015, 24, 183–189. [Google Scholar]
- Wu, X.; Fan, Y.; Wang, R.; Zhao, Q.; Ali, Q.; Wu, H.; Gu, Q.; Borriss, R.; Xie, Y.; Gao, X. Bacillus halotolerans KKD1 induces physiological, metabolic and molecular reprogramming in wheat under saline condition. Front. Plant Sci. 2022, 13, 978066. [Google Scholar] [CrossRef]
- Zhang, X.; Bernatavichute, Y.V.; Cokus, S.; Pellegrini, M.; Jacobsen, S.E. Genome-wide analysis of mono-, di-and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009, 10, R62. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Li, Q.; Zhou, D.; Jia, M.; Liu, Z.; Hou, Z.; Ren, Q.; Ji, S.; Sang, S.; Lu, S.; et al. B. subtilis CNBG-PGPR-1 induces methionine to regulate ethylene pathway and ROS scavenging for improving salt tolerance of tomato. Plant J. 2024, 117, 193–211. [Google Scholar] [CrossRef]
- Liu, J.; Guan, P.W.; Marker, C.N.; Smith, N.D.; Orabona, N.; Shang, S.L.; Kim, H.; Liu, Z.K. Thermodynamic properties and phase stability of the Ba-Bi system: A combined computational and experimental study. J. Alloys Compd. 2019, 771, 281–289. [Google Scholar] [CrossRef]
- Kang, S.M.; Khan, A.L.; Waqas, M.; You, Y.-H.; Kim, J.-H.; Kim, J.-G.; Hamayun, M.; Lee, I.-J. Plant growth-promoting rhizobacteria reduce adverse effects of salinity and osmotic stress by regulating phytohormones and antioxidants in Cucumis sativus. J. Plant Interact. 2014, 9, 673–682. [Google Scholar] [CrossRef]
- Li, Y.; Su, Z.; Lin, Y.; Xu, Z.; Bao, H.; Wang, F.; Liu, J.; Hu, S.; Wang, Z.; Yu, X.; et al. Utilizing transcriptomics and metabolomics to unravel key genes and metabolites of maize seedlings in response to drought stress. BMC Plant Biol. 2024, 24, 34. [Google Scholar] [CrossRef]
- Feng, X.J.; Li, J.R.; Qi, S.L.; Lin, Q.F.; Jin, J.B.; Hua, X.J. Light affects salt stress-induced transcriptional memory of P5CS1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, E8335–E8343. [Google Scholar] [CrossRef]
- Kim, J.M.; To, T.K.; Ishida, J.; Matsui, A.; Kimura, H.; Seki, M. Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 847–856. [Google Scholar] [CrossRef]
- Heinemann, B.; Künzler, P.; Eubel, H.; Braun, H.-P.; Hildebrandt, T.M. Estimating the number of protein molecules in a plant cell: Protein and amino acid homeostasis during drought. Plant Physiol. 2021, 185, 385–404. [Google Scholar] [CrossRef]
- Gipson, A.B.; Morton, K.J.; Rhee, R.J.; Simo, S.; Clayton, J.A.; Perrett, M.E.; Binkley, C.G.; Jensen, E.L.; Oakes, D.L.; Rouhier, M.F.; et al. Disruptions in valine degradation affect seed development and germination in Arabidopsis. Plant J. 2017, 90, 1029–1039. [Google Scholar] [CrossRef]
- Ding, G.; Che, P.; Ilarslan, H.; Wurtele, E.S.; Nikolau, B.J. Genetic dissection of methylcrotonyl CoA carboxylase indicates a complex role for mitochondrial leucine catabolism during seed development and germination. Plant J. 2012, 70, 562–577. [Google Scholar] [CrossRef]
- Angelovici, R.; Lipka, A.E.; Deason, N.; Gonzalez-Jorge, S.; Lin, H.; Cepela, J.; Buell, R.; Gore, M.A.; DellaPenna, D. Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. Plant Cell 2013, 25, 4827–4843. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.J.; Sun, M.H.; Kang, H.; Lu, J.; You, C.X.; Hao, Y.J. A CIPK protein kinase targets sucrose transporter MdSUT2. 2 at Ser254 for phosphorylation to enhance salt tolerance. Plant Cell Environ. 2019, 42, 918–930. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Sun, S.; Wang, R.; Ma, X.; Shen, S.; Luo, Y.; Ma, X.; Wu, T.; Li, S.; Yang, Z.; et al. Study on the mechanism of salt relief and growth promotion of Enterobacter cloacae on cotton. BMC Plant Biol. 2023, 23, 656. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).