Genome-Wide Association Studies for Key Agronomic and Quality Traits in Potato (Solanum tuberosum L.)
Abstract
1. Introduction
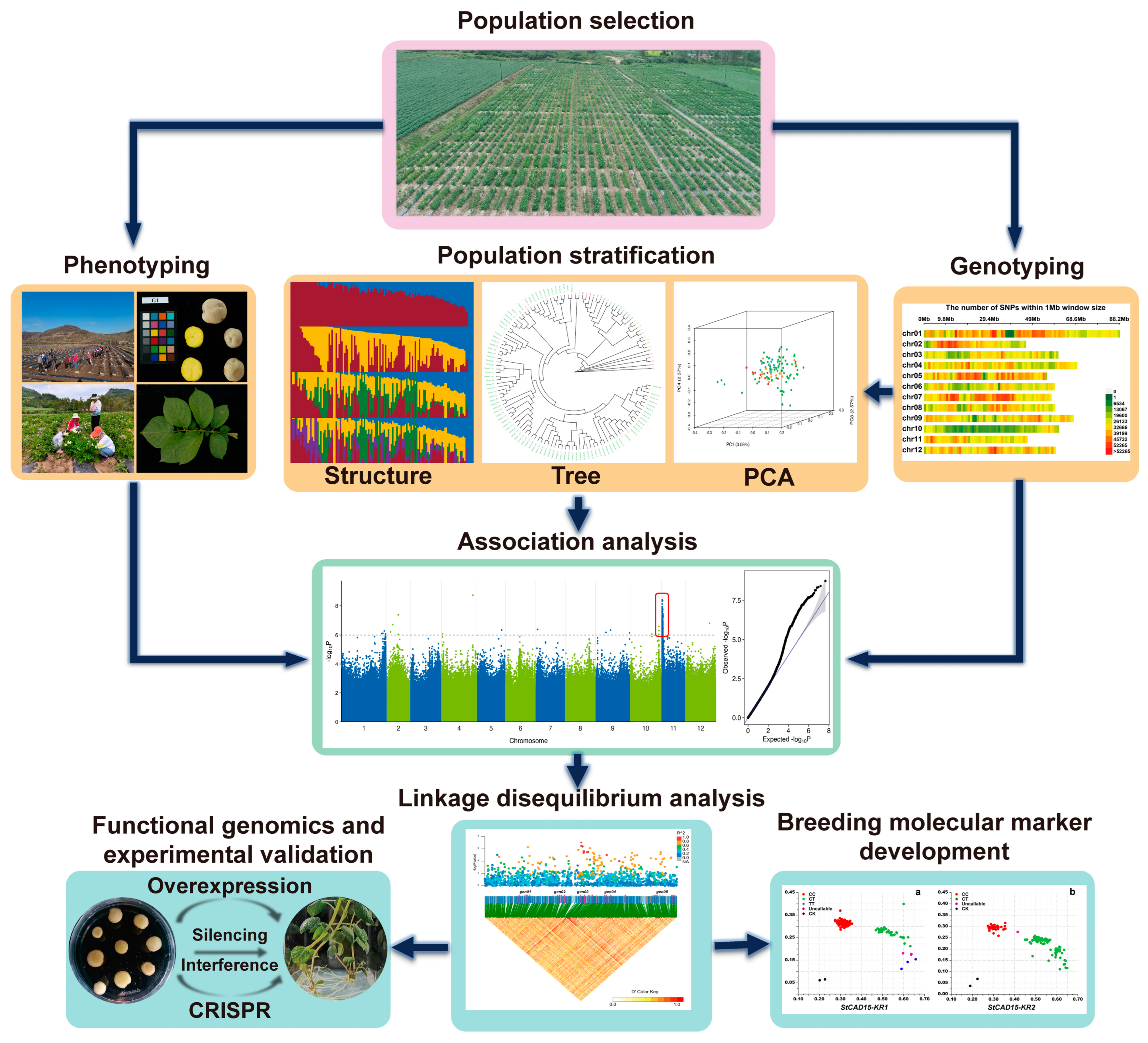
2. Overview of Steps for Conducting GWASs
2.1. Population Selection, Phenotyping, and Genotyping
2.2. Population Stratification and Linkage Disequilibrium Analysis
2.3. GWAS Analysis Models
3. GWASs for Key Agronomic Traits in Potato
3.1. Plant Morphology
3.2. Tuber Phenotype
3.3. Root and Stolon Phenotype
3.4. Yield Components
3.5. Maturity
3.6. Dormancy and Germination
3.7. Disease and Pest Resistance
3.8. Drought and Bruising Resistance
4. GWASs for Key Quality Traits in Potato
4.1. Starch
4.2. Sugar
4.3. Glycoalkaloids
4.4. Mineral Elements
4.5. Organic Acids
5. Summary and Prospects
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- FAOSTAT. Available online: https://www.fao.org/faostat (accessed on 14 July 2023).
- Zaheer, K.; Akhtar, M.H. Potato production, usage, and nutrition—A review. Crit. Rev. Food Sci. Nutr. 2014, 56, 711–721. [Google Scholar] [CrossRef] [PubMed]
- George, T.S.; Taylor, M.A.; Dodd, I.C.; White, P.J. Climate change and consequences for potato production: A review of tolerance to emerging abiotic stress. Potato Res. 2018, 60, 239–268. [Google Scholar] [CrossRef]
- Furrer, A.N.; Chegeni, M.; Ferruzzi, M.G. Impact of potato processing on nutrients, phytochemicals, and human health. Crit. Rev. Food Sci. Nutr. 2018, 58, 146–168. [Google Scholar] [CrossRef] [PubMed]
- Dahal, K.; Li, X.Q.; Tai, H.; Creelman, A.; Bizimungu, B. Improving potato stress tolerance and tuber yield under a climate change scenario-a current overview. Front. Plant Sci. 2019, 10, 563. [Google Scholar] [CrossRef] [PubMed]
- Morris, W.L.; Taylor, M.A. Improving flavor to increase consumption. Am. J. Potato Res. 2019, 96, 195–200. [Google Scholar] [CrossRef]
- Nasir, M.W.; Toth, Z. Effect of drought stress on potato production: A review. Agronomy 2022, 12, 635. [Google Scholar] [CrossRef]
- Naeem, M.; Demirel, U.; Yousaf, M.F.; Caliskan, S.; Caliskan, M.E. Overview on domestication, breeding, genetic gain and improvement of tuber quality traits of potato using fast forwarding technique (GWAS): A review. Plant Breed. 2021, 140, 519–542. [Google Scholar] [CrossRef]
- Aksoy, E.; Demirel, U.; Bakhsh, A.; Zia, M.A.B.; Naeem, M.; Saeed, F.; Çalışkan, S.; Çalışkan, M.E. Recent Advances in Potato (Solanum tuberosum L.) Breeding. In Advances in Plant Breeding Strategies: Vegetable Crops: Volume 8: Bulbs, Roots and Tubers; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Cham, Switzerland, 2021; Volume 8, pp. 409–487. [Google Scholar]
- Pandey, J.; Scheuring, D.C.; Koym, J.W.; Coombs, J.; Novy, R.G.; Thompson, A.L.; Holm, D.G.; Douches, D.S.; Miller, J.C.; Vales, M.I. Genetic diversity and population structure of advanced clones selected over forty years by a potato breeding program in the USA. Sci. Rep. 2021, 11, 8344. [Google Scholar] [CrossRef]
- Machida-Hirano, R. Diversity of potato genetic resources. Breed. Sci. 2015, 65, 26–40. [Google Scholar] [CrossRef]
- Bradshaw, J.E.; Bryan, G.J.; Ramsay, G. Genetic resources (including wild and cultivated Solanum species) and progress in their utilisation in potato breeding. Potato Res. 2006, 49, 49–65. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhao, B.; Zhang, Q.; Yu, B.; Cheng, L.; Jin, R.; Wang, Y.; Zhang, J.; Wang, D.; Zhang, F. Screening for chip-processing potato line from introgression of wild species’ germplasms with post-harvest storage and chip qualities. Am. J. Potato Res. 2013, 90, 425–439. [Google Scholar] [CrossRef]
- Watanabe, K. Potato genetics, genomics, and applications. Breed. Sci. 2015, 65, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, J.E. Review and analysis of limitations in ways to improve conventional potato breeding. Potato Res. 2017, 60, 171–193. [Google Scholar] [CrossRef]
- Mir, R.R.; Reynolds, M.; Pinto, F.; Khan, M.A.; Bhat, M.A. High-throughput phenotyping for crop improvement in the genomics era. Plant Sci. 2019, 282, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Manrique-Carpintero, N.C.; Coombs, J.J.; Pham, G.M.; Laimbeer, F.P.E.; Braz, G.T.; Jiang, J.; Veilleux, R.E.; Buell, C.R.; Douches, D.S. Genome reduction in tetraploid potato reveals genetic load, haplotype variation, and loci associated with agronomic traits. Front. Plant Sci. 2018, 9, 944. [Google Scholar] [CrossRef]
- Hagely, K.B.; Jo, H.; Kim, J.-H.; Hudson, K.A.; Bilyeu, K. Molecular-assisted breeding for improved carbohydrate profiles in soybean seed. Theor. Appl. Genet. 2020, 133, 1189–1200. [Google Scholar] [CrossRef]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef]
- Slater, A.T.; Cogan, N.O.I.; Forster, J.W. Cost analysis of the application of marker-assisted selection in potato breeding. Mol. Breed. 2013, 32, 299–310. [Google Scholar] [CrossRef]
- Ramakrishnan, A.P.; Ritland, C.E.; Blas Sevillano, R.H.; Riseman, A. Review of potato molecular markers to enhance trait selection. Am. J. Potato Res. 2015, 92, 455–472. [Google Scholar] [CrossRef]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and prospects of association mapping in plants. Plant Genome 2008, 1, 5–20. [Google Scholar] [CrossRef]
- Han, B.; Huang, X. Sequencing-based genome-wide association study in rice. Curr. Opin. Plant Biol. 2013, 16, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Scheben, A.; Batley, J.; Edwards, D. Genotyping-by-sequencing approaches to characterize crop genomes: Choosing the right tool for the right application. Plant Biotechnol. J. 2017, 15, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Endelman, J.B.; Jansky, S.H. Genetic mapping with an inbred line-derived F2 population in potato. Theor. Appl. Genet. 2016, 129, 935–943. [Google Scholar] [CrossRef] [PubMed]
- Marfil, C.F.; Camadro, E.L.; Masuelli, R.W. Phenotypic instability and epigenetic variability in a diploid potato of hybrid origin, Solanum ruiz-lealii. BMC Plant Biol. 2009, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Hardigan, M.A.; Laimbeer, F.P.E.; Newton, L.; Crisovan, E.; Hamilton, J.P.; Vaillancourt, B.; Wiegert-Rininger, K.; Wood, J.C.; Douches, D.S.; Farre, E.M.; et al. Genome diversity of tuber-bearing Solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato. Proc. Natl. Acad. Sci. USA 2017, 114, E9999–E10008. [Google Scholar] [CrossRef]
- Xu, Y.; Li, P.; Yang, Z.; Xu, C. Genetic mapping of quantitative trait loci in crops. Crop J. 2017, 5, 175–184. [Google Scholar] [CrossRef]
- Rodriguez, M.; Scintu, A.; Posadinu, C.M.; Xu, Y.; Nguyen, C.V.; Sun, H.; Bitocchi, E.; Bellucci, E.; Papa, R.; Fei, Z.; et al. GWAS Based on RNA-Seq SNPs and High-Throughput Phenotyping Combined with Climatic Data Highlights the Reservoir of Valuable Genetic Diversity in Regional Tomato Landraces. Genes 2020, 11, 1387. [Google Scholar] [CrossRef]
- Arafa, R.A.; Rakha, M.T.; Soliman, N.E.K.; Moussa, O.M.; Kamel, S.M.; Shirasawa, K. Rapid identification of candidate genes for resistance to tomato late blight disease using next-generation sequencing technologies. PLoS ONE 2017, 12, e0189951. [Google Scholar] [CrossRef]
- Tripodi, P.; Soler, S.; Campanelli, G.; Diez, M.J.; Esposito, S.; Sestili, S.; Figas, M.R.; Leteo, F.; Casanova, C.; Platani, C.; et al. Genome wide association mapping for agronomic, fruit quality, and root architectural traits in tomato under organic farming conditions. BMC Plant Biol. 2021, 21, 481. [Google Scholar] [CrossRef]
- Fu, G.; Yu, S.; Wu, K.; Yang, M.; Altaf, M.A.; Wu, Z.; Deng, Q.; Lu, X.; Fu, H.; Wang, Z.; et al. Genome-wide association study and candidate gene identification for agronomic traits in 182 upward-growing fruits of C. frutescens and C. annuum. Sci. Rep. 2024, 14, 14691. [Google Scholar] [CrossRef]
- Han, K.; Lee, H.Y.; Ro, N.Y.; Hur, O.S.; Lee, J.H.; Kwon, J.K.; Kang, B.C. QTL mapping and GWAS reveal candidate genes controlling capsaicinoid content in Capsicum. Plant Biotechnol. J. 2018, 16, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Ro, N.; Haile, M.; Hur, O.; Geum, B.; Rhee, J.; Hwang, A.; Kim, B.; Lee, J.; Hahn, B.S.; Lee, J.; et al. Genome-Wide Association Study of Resistance to Phytophthora capsici in the Pepper (Capsicum spp.) Collection. Front. Plant Sci. 2022, 13, 902464. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, A.; Nimmakayala, P.; Davenport, B.; Natarajan, P.; Tonapi, K.; Kadiyala, S.S.; Lopez-Ortiz, C.; Ibarra-Munoz, L.; Chakrabarti, M.; Benedito, V.; et al. Genetic tapestry of Capsicum fruit colors: A comparative analysis of four cultivated species. Theor. Appl. Genet. 2024, 137, 130. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Qian, Z.; Zhang, J.; Yang, J.; Wu, M.; Barchi, L.; Zhao, H.; Sun, H.; Cui, Y.; Wen, C. Impact of fruit shape selection on genetic structure and diversity uncovered from genome-wide perfect SNPs genotyping in eggplant. Mol. Breed. 2019, 39, 140. [Google Scholar] [CrossRef]
- Cericola, F.; Portis, E.; Lanteri, S.; Toppino, L.; Barchi, L.; Acciarri, N.; Pulcini, L.; Sala, T.; Rotino, G.L. Linkage disequilibrium and genome-wide association analysis for anthocyanin pigmentation and fruit color in eggplant. BMC Genom. 2014, 15, 896. [Google Scholar] [CrossRef]
- Ro, N.; Haile, M.; Ko, H.C.; Cho, G.T.; Lee, J.; Kim, B.; Lee, S.; Kim, S.H. Genome-Wide Association Study of Phenolic Content and Antioxidant Properties in Eggplant Germplasm. Genes 2023, 14, 1315. [Google Scholar] [CrossRef]
- Prodhomme, C.; Esselink, D.; Borm, T.; Visser, R.G.F.; van Eck, H.J.; Vossen, J.H. Comparative Subsequence Sets Analysis (CoSSA) is a robust approach to identify haplotype specific SNPs; mapping and pedigree analysis of a potato wart disease resistance gene Sen3. Plant Methods 2019, 15, 60. [Google Scholar] [CrossRef]
- Jo, K.R.; Choi, J.-G.; Kwon, D.-H.; Park, Y.-E.; Kim, S.-J. Revealing genetic variations associated with chip-processing properties in potato (Solanum tuberosum L.). Agronomy 2023, 13, 642. [Google Scholar] [CrossRef]
- Fang, C.; Luo, J. Metabolic GWAS-based dissection of genetic bases underlying the diversity of plant metabolism. Plant J. 2019, 97, 91–100. [Google Scholar] [CrossRef]
- Brouckaert, M.; Peng, M.; Hofer, R.; El Houari, I.; Darrah, C.; Storme, V.; Saeys, Y.; Vanholme, R.; Goeminne, G.; Timokhin, V.I.; et al. QT-GWAS: A novel method for unveiling biosynthetic loci affecting qualitative metabolic traits. Mol. Plant 2023, 16, 1212–1227. [Google Scholar] [CrossRef]
- Jia, J.; Zhao, G.; Li, D.; Wang, K.; Kong, C.; Deng, P.; Yan, X.; Zhang, X.; Lu, Z.; Xu, S.; et al. Genome resources for the elite bread wheat cultivar Aikang 58 and mining of elite homeologous haplotypes for accelerating wheat improvement. Mol. Plant 2023, 16, 1893–1910. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Xu, J.; Chen, S.; Chen, J.; Liang, Y.; Zhang, C.; Li, Q.; Lai, J.; Li, L. Large-scale translatome profiling annotates the functional genome and reveals the key role of genic 3’ untranslated regions in translatomic variation in plants. Plant Commun. 2021, 2, 100181. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, R. Genomic-led potato breeding for increasing genetic gains: Achievements and outlook. Crop Breed. Genet. Genom. 2020, 2, e200010. [Google Scholar] [CrossRef]
- Alseekh, S.; Kostova, D.; Bulut, M.; Fernie, A.R. Genome-wide association studies: Assessing trait characteristics in model and crop plants. Cell. Mol. Life Sci. 2021, 78, 5743–5754. [Google Scholar] [CrossRef]
- Hirschhorn, J.N.; Daly, M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005, 6, 95–108. [Google Scholar] [CrossRef]
- Lindqvist-Kreuze, H.; De Boeck, B.; Unger, P.; Gemenet, D.; Li, X.; Pan, Z.; Sui, Q.; Qin, J.; Woldegjorgis, G.; Negash, K.; et al. Global multi-environment resistance QTL for foliar late blight resistance in tetraploid potato with tropical adaptation. G3 2021, 11, jkab251. [Google Scholar] [CrossRef]
- Hamazaki, K.; Kajiya-Kanegae, H.; Yamasaki, M.; Ebana, K.; Yabe, S.; Nakagawa, H.; Iwata, H. Choosing the optimal population for a genome-wide association study: A simulation of whole-genome sequences from rice. Plant Genome 2020, 13, e20005. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef]
- Vos, P.G.; Uitdewilligen, J.G.; Voorrips, R.E.; Visser, R.G.; van Eck, H.J. Development and analysis of a 20K SNP array for potato (Solanum tuberosum): An insight into the breeding history. Theor. Appl. Genet. 2015, 128, 2387–2401. [Google Scholar] [CrossRef]
- Patel, A.P.; Peloso, G.M.; Pirruccello, J.P.; Johansen, C.T.; Dube, J.B.; Larach, D.B.; Ban, M.R.; Dallinge-Thie, G.M.; Gupta, N.; Boehnke, M.; et al. Targeted exonic sequencing of GWAS loci in the high extremes of the plasma lipids distribution. Atherosclerosis 2016, 250, 63–68. [Google Scholar] [CrossRef]
- Fan, H.; Ives, A.R.; Surget-Groba, Y. Reconstructing phylogeny from reduced-representation genome sequencing data without assembly or alignment. Mol. Ecol. Resour. 2018, 18, 1482–1491. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Ning, W.; Wei, D.; Jiang, M.; Zhu, K.; Wang, X.; Edwards, D.; Odeny, D.A.; Cheng, S. Plant genome resequencing and population genomics: Current status and future prospects. Mol. Plant 2023, 16, 1252–1268. [Google Scholar] [CrossRef] [PubMed]
- Geibel, J.; Reimer, C.; Weigend, S.; Weigend, A.; Pook, T.; Simianer, H. How array design creates SNP ascertainment bias. PLoS ONE 2021, 16, e0245178. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop breeding chips and genotyping platforms: Progress, challenges, and perspectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef]
- Pavan, S.; Delvento, C.; Ricciardi, L.; Lotti, C.; Ciani, E.; D’Agostino, N. Recommendations for choosing the genotyping method and best practices for quality control in crop genome-wide association studies. Front. Genet. 2020, 11, 447. [Google Scholar] [CrossRef]
- Yuan, Y.; Bayer, P.E.; Batley, J.; Edwards, D. Current status of structural variation studies in plants. Plant Biotechnol. J. 2021, 19, 2153–2163. [Google Scholar] [CrossRef]
- Xu, X.; Bai, G. Whole-genome resequencing: Changing the paradigms of SNP detection, molecular mapping and gene discovery. Mol. Breed. 2015, 35, 33. [Google Scholar] [CrossRef]
- Gabur, I.; Chawla, H.S.; Snowdon, R.J.; Parkin, I.A.P. Connecting genome structural variation with complex traits in crop plants. Theor. Appl. Genet. 2019, 132, 733–750. [Google Scholar] [CrossRef]
- Consortium, T.P.G.S. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef]
- Aversano, R.; Contaldi, F.; Ercolano, M.R.; Grosso, V.; Iorizzo, M.; Tatino, F.; Xumerle, L.; Dal Molin, A.; Avanzato, C.; Ferrarini, A.; et al. The Solanum commersonii genome sequence provides insights into adaptation to stress conditions and genome evolution of wild potato relatives. Plant Cell 2015, 27, 954–968. [Google Scholar] [CrossRef]
- Leisner, C.P.; Hamilton, J.P.; Crisovan, E.; Manrique-Carpintero, N.C.; Marand, A.P.; Newton, L.; Pham, G.M.; Jiang, J.; Douches, D.S.; Jansky, S.H.; et al. Genome sequence of M6, a diploid inbred clone of the high-glycoalkaloid-producing tuber-bearing potato species Solanum chacoense, reveals residual heterozygosity. Plant J. 2018, 94, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Tang, D.; Huang, W.; Yang, Z.; Zhang, Y.; Hamilton, J.P.; Visser, R.G.F.; Bachem, C.W.B.; Robin Buell, C.; Zhang, Z.; et al. Haplotype-resolved genome analyses of a heterozygous diploid potato. Nat. Genet. 2020, 52, 1018–1023. [Google Scholar] [CrossRef]
- Wang, F.; Xia, Z.; Zou, M.; Zhao, L.; Jiang, S.; Zhou, Y.; Zhang, C.; Ma, Y.; Bao, Y.; Sun, H.; et al. The autotetraploid potato genome provides insights into highly heterozygous species. Plant Biotechnol. J. 2022, 20, 1996–2005. [Google Scholar] [CrossRef]
- Lang, D.; Zhang, S.; Ren, P.; Liang, F.; Sun, Z.; Meng, G.; Tan, Y.; Li, X.; Lai, Q.; Han, L.; et al. Comparison of the two up-to-date sequencing technologies for genome assembly: HiFi reads of Pacific Biosciences Sequel II system and ultralong reads of Oxford Nanopore. Gigascience 2020, 9, giaa123. [Google Scholar] [CrossRef]
- Sun, H.; Jiao, W.B.; Krause, K.; Campoy, J.A.; Goel, M.; Folz-Donahue, K.; Kukat, C.; Huettel, B.; Schneeberger, K. Chromosome-scale and haplotype-resolved genome assembly of a tetraploid potato cultivar. Nat. Genet. 2022, 54, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Li, C.; Li, G.; Wang, P.; Peng, Z.; Cheng, L.; Li, H.; Zhang, Z.; Li, Y.; Huang, W.; et al. Genome architecture and tetrasomic inheritance of autotetraploid potato. Mol. Plant 2022, 15, 1211–1226. [Google Scholar] [CrossRef] [PubMed]
- Hoopes, G.; Meng, X.; Hamilton, J.P.; Achakkagari, S.R.; de Alves Freitas Guesdes, F.; Bolger, M.E.; Coombs, J.J.; Esselink, D.; Kaiser, N.R.; Kodde, L.; et al. Phased, chromosome-scale genome assemblies of tetraploid potato reveal a complex genome, transcriptome, and predicted proteome landscape underpinning genetic diversity. Mol. Plant 2022, 15, 520–536. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. GigaScience 2018, 8, giy154. [Google Scholar] [CrossRef] [PubMed]
- Tsepilov, Y.A.; Ried, J.S.; Strauch, K.; Grallert, H.; van Duijn, C.M.; Axenovich, T.I.; Aulchenko, Y.S. Development and application of genomic control methods for genome-wide association studies using non-additive models. PLoS ONE 2013, 8, e81431. [Google Scholar] [CrossRef]
- He, J.; Meng, S.; Zhao, T.; Xing, G.; Yang, S.; Li, Y.; Guan, R.; Lu, J.; Wang, Y.; Xia, Q.; et al. An innovative procedure of genome-wide association analysis fits studies on germplasm population and plant breeding. Theor. Appl. Genet. 2017, 130, 2327–2343. [Google Scholar] [CrossRef]
- Zhu, C.; Yu, J. Nonmetric multidimensional scaling corrects for population structure in association mapping with different sample types. Genetics 2009, 182, 875–888. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ralph, P. Local PCA shows how the effect of population structure differs along the genome. Genetics 2019, 211, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, D.; Liu, H.; Christopher, A. Robust methods for population stratification in genome wide association studies. BMC Bioinform. 2013, 14, 132. [Google Scholar] [CrossRef] [PubMed]
- Akond, Z.; Ahsan, M.A.; Alam, M.; Mollah, M.N.H. Robustification of GWAS to explore effective SNPs addressing the challenges of hidden population stratification and polygenic effects. Sci. Rep. 2021, 11, 13060. [Google Scholar] [CrossRef]
- Tibbs Cortes, L.; Zhang, Z.; Yu, J. Status and prospects of genome-wide association studies in plants. Plant Genome 2021, 14, e20077. [Google Scholar] [CrossRef]
- Ahrens, C.W.; Rymer, P.D.; Stow, A.; Bragg, J.; Dillon, S.; Umbers, K.D.L.; Dudaniec, R.Y. The search for loci under selection: Trends, biases and progress. Mol. Ecol. 2018, 27, 1342–1356. [Google Scholar] [CrossRef]
- Waples, R.K.; Larson, W.A.; Waples, R.S. Estimating contemporary effective population size in non-model species using linkage disequilibrium across thousands of loci. Heredity 2016, 117, 233–240. [Google Scholar] [CrossRef]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Primers 2021, 1, 59. [Google Scholar] [CrossRef]
- Berhe, M.; Dossa, K.; You, J.; Mboup, P.A.; Diallo, I.N.; Diouf, D.; Zhang, X.; Wang, L. Genome-wide association study and its applications in the non-model crop Sesamum indicum. BMC Plant Biol. 2021, 21, 283. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Wen, Y.J.; Zhang, H.; Ni, Y.L.; Huang, B.; Zhang, J.; Feng, J.Y.; Wang, S.B.; Dunwell, J.M.; Zhang, Y.M.; Wu, R. Methodological implementation of mixed linear models in multi-locus genome-wide association studies. Brief. Bioinf. 2018, 19, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.M.; Zaitlen, N.A.; Wade, C.M.; Kirby, A.; Heckerman, D.; Daly, M.J.; Eskin, E. Efficient control of population structure in model organism association mapping. Genetics 2008, 178, 1709–1723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Lippert, C.; Listgarten, J.; Liu, Y.; Kadie, C.M.; Davidson, R.I.; Heckerman, D. FaST linear mixed models for genome-wide association studies. Nat. Methods 2011, 8, 833–835. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Jia, Z.; Dunwell, J.M. Editorial: The applications of new multi-locus gwas methodologies in the genetic dissection of complex traits. Front. Plant Sci. 2019, 10, 100. [Google Scholar] [CrossRef]
- Buzdugan, L.; Kalisch, M.; Navarro, A.; Schunk, D.; Fehr, E.; Buhlmann, P. Assessing statistical significance in multivariable genome wide association analysis. Bioinformatics 2016, 32, 1990–2000. [Google Scholar] [CrossRef]
- Wang, S.B.; Feng, J.Y.; Ren, W.L.; Huang, B.; Zhou, L.; Wen, Y.J.; Zhang, J.; Dunwell, J.M.; Xu, S.; Zhang, Y.M. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 2016, 6, 19444. [Google Scholar] [CrossRef]
- Segura, V.; Vilhjalmsson, B.J.; Platt, A.; Korte, A.; Seren, U.; Long, Q.; Nordborg, M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 2012, 44, 825–830. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Southam, L.; Panoutsopoulou, K.; Rayner, N.W.; Chapman, K.; Durrant, C.; Ferreira, T.; Arden, N.; Carr, A.; Deloukas, P.; Doherty, M.; et al. The effect of genome-wide association scan quality control on imputation outcome for common variants. Eur. J. Hum. Genet. 2011, 19, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Tam, V.; Patel, N.; Turcotte, M.; Bosse, Y.; Pare, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Alamin, M.; Sultana, M.H.; Lou, X.; Jin, W.; Xu, H. Dissecting complex traits using omics data: A review on the linear mixed models and their application in gwas. Plants 2022, 11, 3277. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, A.; Li, J.; Hu, W.; Yu, J.; Khan, S.U.; Khan, M.H.U.; Xie, G.; Wang, J.; Wang, L. Uncovering genomic regions controlling plant architectural traits in hexaploid wheat using different GWAS models. Sci. Rep. 2021, 11, 6767. [Google Scholar] [CrossRef]
- Enyew, M.; Feyissa, T.; Carlsson, A.S.; Tesfaye, K.; Hammenhag, C.; Seyoum, A.; Geleta, M. Genome-wide analyses using multi-locus models revealed marker-trait associations for major agronomic traits in Sorghum bicolor. Front. Plant Sci. 2022, 13, 999692. [Google Scholar] [CrossRef]
- Malik, P.; Kumar, J.; Sharma, S.; Meher, P.K.; Balyan, H.S.; Gupta, P.K.; Sharma, S. GWAS for main effects and epistatic interactions for grain morphology traits in wheat. Physiol. Mol. Biol. Plants 2022, 28, 651–668. [Google Scholar] [CrossRef]
- Kim, Y.U.; Lee, B.W. Differential mechanisms of potato yield loss induced by high day and night temperatures during tuber initiation and bulking: Photosynthesis and tuber growth. Front. Plant Sci. 2019, 10, 300. [Google Scholar] [CrossRef]
- Haverkort, A.J.; Struik, P.C. Yield levels of potato crops: Recent achievements and future prospects. Field Crop. Res. 2015, 182, 76–85. [Google Scholar] [CrossRef]
- Solomon, F.; Asrat, A.; Daniel, T.; Zenebe, G.M.; Eshetu, A. Evaluation of potato (Solanum tuberosum L.) varieties for yield and yield components. J. Hortic. For. 2019, 11, 48–53. [Google Scholar] [CrossRef]
- Thiele, G.; Dufour, D.; Vernier, P.; Mwanga, R.O.M.; Parker, M.L.; Schulte Geldermann, E.; Teeken, B.; Wossen, T.; Gotor, E.; Kikulwe, E.; et al. A review of varietal change in roots, tubers and bananas: Consumer preferences and other drivers of adoption and implications for breeding. Int. J. Food Sci. Technol. 2021, 56, 1076–1092. [Google Scholar] [CrossRef]
- Tiwari, J.K.; Buckseth, T.; Zinta, R.; Bhatia, N.; Dalamu, D.; Naik, S.; Poonia, A.K.; Kardile, H.B.; Challam, C.; Singh, R.K.; et al. Germplasm, breeding, and genomics in potato improvement of biotic and abiotic stresses tolerance. Front. Plant Sci. 2022, 13, 805671. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zou, M.; Jiang, S.; Dong, X.; Deng, K.; Na, T.; Wang, J.; Xia, Z.; Wang, F. Insights into the genetic determination of the autotetraploid potato plant height. Genes 2023, 14, 507. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; XIE, R.; Guo, J.; Guo, B.; Yi, L.; Hou, J. Candidate genes related to numbers of main stems and tubers per plant of potato mined using GWAS. J. Northwest A F Univ. (Nat. Sci. Ed.) 2023, 51, 22–36. [Google Scholar] [CrossRef]
- Berdugo-Cely, J.; Valbuena, R.I.; Sanchez-Betancourt, E.; Barrero, L.S.; Yockteng, R. Genetic diversity and association mapping in the Colombian Central Collection of Solanum tuberosum L. Andigenum group using SNPs markers. PLoS ONE 2017, 12, e0173039. [Google Scholar] [CrossRef] [PubMed]
- Massa, A.N.; Manrique-Carpintero, N.C.; Coombs, J.; Haynes, K.G.; Bethke, P.C.; Brandt, T.L.; Gupta, S.K.; Yencho, G.C.; Novy, R.G.; Douches, D.S. Linkage analysis and QTL mapping in a tetraploid russet mapping population of potato. BMC Genet. 2018, 19, 87. [Google Scholar] [CrossRef]
- Zia, M.A.B.; Demirel, U.; Nadeem, M.A.; Caliskan, M.E. Genome-wide association study identifies various loci underlying agronomic and morphological traits in diversified potato panel. Physiol. Mol. Biol. Plants 2020, 26, 1003–1020. [Google Scholar] [CrossRef]
- Ospina Nieto, C.A.; Lammerts van Bueren, E.T.; Allefs, S.; Vos, P.G.; van der Linden, G.; Maliepaard, C.A.; Struik, P.C. Association mapping of physiological and morphological traits related to crop development under contrasting nitrogen inputs in a diverse set of potato cultivars. Plants 2021, 10, 1727. [Google Scholar] [CrossRef]
- Rak, K.; Bethke, P.C.; Palta, J.P. QTL mapping of potato chip color and tuber traits within an autotetraploid family. Mol. Breed. 2017, 37, 15. [Google Scholar] [CrossRef]
- Zia, M.A.B.; DemİRel, U.; Nadeem, M.A.; Ali, F.; Dawood, A.; Ijaz, M.; ÇAliŞKan, M.E. Genome-wide association studies (GWAS) revealed a genetic basis associated with floral traits in potato germplasm. Turk. J. Agric. For. 2022, 46, 90–103. [Google Scholar] [CrossRef]
- Kaiser, N.R.; Billings, G.; Coombs, J.; Buell, C.R.; Enciso-Rodríguez, F.; Douches, D.S. Self-fertility and resistance to the Colorado potato beetle (Leptinotarsa decemlineata) in a diploid Solanum chacoense recombinant inbred line population. Crop Sci. 2021, 61, 3392–3414. [Google Scholar] [CrossRef]
- Manrique-Carpintero, N.C.; Coombs, J.J.; Cui, Y.; Veilleux, R.E.; Buell, C.R.; Douches, D. Genetic map and qtl analysis of agronomic traits in a diploid potato population using single nucleotide polymorphism markers. Crop Sci. 2015, 55, 2566–2579. [Google Scholar] [CrossRef]
- Massa, A.N.; Manrique-Carpintero, N.C.; Coombs, J.J.; Zarka, D.G.; Boone, A.E.; Kirk, W.W.; Hackett, C.A.; Bryan, G.J.; Douches, D.S. Genetic linkage mapping of economically important traits in cultivated tetraploid potato (Solanum tuberosum L.). G3 2015, 5, 2357–2364. [Google Scholar] [CrossRef] [PubMed]
- Braun, S.R.; Endelman, J.B.; Haynes, K.G.; Jansky, S.H. Quantitative trait loci for resistance to common scab and cold-induced sweetening in diploid potato. Plant Genome 2017, 10, plantgenome2016.10.0110. [Google Scholar] [CrossRef] [PubMed]
- Mengist, M.F.; Alves, S.; Griffin, D.; Creedon, J.; McLaughlin, M.J.; Jones, P.W.; Milbourne, D. Genetic mapping of quantitative trait loci for tuber-cadmium and zinc concentration in potato reveals associations with maturity and both overlapping and independent components of genetic control. Theor. Appl. Genet. 2018, 131, 929–945. [Google Scholar] [CrossRef] [PubMed]
- Caraza-Harter, M.V.; Endelman, J.B. The genetic architectures of vine and skin maturity in tetraploid potato. Theor. Appl. Genet. 2022, 135, 2943–2951. [Google Scholar] [CrossRef]
- Xue, W.; Haynes, K.G.; Clarke, C.R.; Qu, X. Genetic dissection of early blight resistance in tetraploid potato. Front. Plant Sci. 2022, 13, 851538. [Google Scholar] [CrossRef]
- Pandey, J.; Scheuring, D.C.; Koym, J.W.; Endelman, J.B.; Vales, M.I. Genomic selection and genome-wide association studies in tetraploid chipping potatoes. Plant Genome 2023, 16, e20297. [Google Scholar] [CrossRef]
- Campbell, R.; Ducreux, L.; Cowan, G.; Young, V.; Chinoko, G.; Chitedze, G.; Kwendani, S.; Chiipanthenga, M.; Bita, C.E.; Mwenye, O.; et al. Allelic variants of a potato HEAT SHOCK COGNATE 70 gene confer improved tuber yield under a wide range of environmental conditions. Food Energy Secur. 2023, 12, e377. [Google Scholar] [CrossRef]
- Yousaf, M.F.; Demirel, U.; Naeem, M.; Naawe, E.K.; Caliskan, M.E. SNP markers associated with some root, stolon, and tuber traits in tetraploid Potatoes (Solanum tuberosum L.) grown under diverse growing systems. Potato Res. 2023, 1, 1–19. [Google Scholar] [CrossRef]
- Aliche, E.B.; Oortwijn, M.; Theeuwen, T.P.J.M.; Bachem, C.W.B.; van Eck, H.J.; Visser, R.G.F.; van der Linden, C.G. Genetic mapping of tuber size distribution and marketable tuber yield under drought stress in potatoes. Euphytica 2019, 215, 186. [Google Scholar] [CrossRef]
- Prashar, A.; Hornyik, C.; Young, V.; McLean, K.; Sharma, S.K.; Dale, M.F.; Bryan, G.J. Construction of a dense SNP map of a highly heterozygous diploid potato population and QTL analysis of tuber shape and eye depth. Theor. Appl. Genet. 2014, 127, 2159–2171. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Massa, A.N.; Douches, D.; Coombs, J.; Akdemir, D.; Yencho, G.C.; Whitworth, J.L.; Novy, R.G. Linkage and QTL mapping for tuber shape and specific gravity in a tetraploid mapping population of potato representing the russet market class. BMC Plant Biol. 2021, 21, 507. [Google Scholar] [CrossRef] [PubMed]
- Pandey, J.; Scheuring, D.C.; Koym, J.W.; Vales, M.I. Genomic regions associated with tuber traits in tetraploid potatoes and identification of superior clones for breeding purposes. Front. Plant Sci. 2022, 13, 952263. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zou, M.; Deng, K.; Xia, C.; Jiang, S.; Zhang, C.; Ma, Y.; Dong, X.; He, M.; Na, T.; et al. Insights into the genetic determination of tuber shape and eye depth in potato natural population based on autotetraploid potato genome. Front. Plant Sci. 2023, 14, 1080666. [Google Scholar] [CrossRef] [PubMed]
- Bisognin, D.A.; Manrique-Carpintero, N.C.; Douches, D.S. QTL analysis of tuber dormancy and sprouting in potato. Am. J. Potato Res. 2018, 95, 374–382. [Google Scholar] [CrossRef]
- Yousaf, M.F.; Demirel, U.; Naeem, M.; Caliskan, M.E. Association mapping reveals novel genomic regions controlling some root and stolon traits in tetraploid potato (Solanum tuberosum L.). 3 Biotech 2021, 11, 174. [Google Scholar] [CrossRef]
- Getahun, B.B.; Tiruneh, M.A.; Aliche, E.; Malossetti, M.; Visser, R.G.F.; van der Linden, C.G. Genotype-by-environment interaction for quantitative trait loci affecting nitrogen use efficiency and associated traits in potato. Potato Res. 2022, 65, 777–807. [Google Scholar] [CrossRef]
- Odilbekov, F.; Selga, C.; Ortiz, R.; Chawade, A.; Liljeroth, E. QTL mapping for resistance to early blight in a tetraploid potato population. Agronomy 2020, 10, 728. [Google Scholar] [CrossRef]
- Lindqvist-Kreuze, H.; Gastelo, M.; Perez, W.; Forbes, G.A.; de Koeyer, D.; Bonierbale, M. Phenotypic stability and genome-wide association study of late blight resistance in potato genotypes adapted to the tropical highlands. Phytopathology 2014, 104, 624–633. [Google Scholar] [CrossRef]
- Mosquera, T.; Alvarez, M.F.; Jimenez-Gomez, J.M.; Muktar, M.S.; Paulo, M.J.; Steinemann, S.; Li, J.; Draffehn, A.; Hofmann, A.; Lubeck, J.; et al. Targeted and untargeted approaches unravel novel candidate genes and diagnostic snps for quantitative resistance of the potato (Solanum tuberosum L.) to Phytophthora infestans causing the late blight disease. PLoS ONE 2016, 11, e0156254. [Google Scholar] [CrossRef]
- Santa, J.D.; Berdugo-Cely, J.; Cely-Pardo, L.; Soto-Suarez, M.; Mosquera, T.; Galeano, M.C. QTL analysis reveals quantitative resistant loci for Phytophthora infestans and Tecia solanivora in tetraploid potato (Solanum tuberosum L.). PLoS ONE 2018, 13, e0199716. [Google Scholar] [CrossRef] [PubMed]
- Juyo Rojas, D.K.; Soto Sedano, J.C.; Ballvora, A.; Leon, J.; Mosquera Vasquez, T. Novel organ-specific genetic factors for quantitative resistance to late blight in potato. PLoS ONE 2019, 14, e0213818. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zou, M.; Zhao, L.; Xia, Z.; Wang, J. Genome-wide association mapping of late blight tolerance trait in potato (Solanum tuberosum L.). Front. Genet. 2021, 12, 714575. [Google Scholar] [CrossRef] [PubMed]
- Sood, S.; Bhardwaj, V.; Bairwa, A.; Dalamu; Sharma, S.; Sharma, A.K.; Kumar, A.; Lal, M.; Kumar, V. Genome-wide association mapping and genomic prediction for late blight and potato cyst nematode resistance in potato (Solanum tuberosum L.). Front. Plant Sci. 2023, 14, 1211472. [Google Scholar] [CrossRef] [PubMed]
- Habe, I.; Miyatake, K.; Nunome, T.; Yamasaki, M.; Hayashi, T. QTL analysis of resistance to bacterial wilt caused by Ralstonia solanacearum in potato. Breed. Sci. 2019, 69, 592–600. [Google Scholar] [CrossRef]
- Yuan, J.; Bizimungu, B.; De Koeyer, D.; Rosyara, U.; Wen, Z.; Lagüe, M. Genome-wide association study of resistance to potato common scab. Potato Res. 2019, 63, 253–266. [Google Scholar] [CrossRef]
- Kaiser, N.R.; Coombs, J.J.; Felcher, K.J.; Hammerschmidt, R.; Zuehlke, M.L.; Buell, C.R.; Douches, D.S. Genome-wide association analysis of common scab resistance and expression profiling of tubers in response to thaxtomin a treatment underscore the complexity of common scab resistance in tetraploid potato. Am. J. Potato Res. 2020, 97, 513–522. [Google Scholar] [CrossRef]
- Koizumi, E.; Igarashi, T.; Tsuyama, M.; Ogawa, K.; Asano, K.; Kobayashi, A.; Sanetomo, R.; Hosaka, K. Association of genome-wide snp markers with resistance to common scab of potato. Am. J. Potato Res. 2021, 98, 149–156. [Google Scholar] [CrossRef]
- Zorrilla, C.; Navarro, F.; Vega-Semorile, S.; Palta, J. QTL for pitted scab, hollow heart, and tuber calcium identified in a tetraploid population of potato derived from an Atlantic × Superior cross. Crop Sci. 2021, 61, 1630–1651. [Google Scholar] [CrossRef]
- Obidiegwu, J.E.; Sanetomo, R.; Flath, K.; Tacke, E.; Hofferbert, H.R.; Hofmann, A.; Walkemeier, B.; Gebhardt, C. Genomic architecture of potato resistance to Synchytrium endobioticum disentangled using SSR markers and the 8.3k SolCAP SNP genotyping array. BMC Genet. 2015, 16, 38. [Google Scholar] [CrossRef]
- Bartkiewicz, A.; Chilla, F.; Terefe-Ayana, D.; Lubeck, J.; Strahwald, J.; Tacke, E.; Hofferbert, H.R.; Flath, K.; Linde, M.; Debener, T. Improved genetic resolution for linkage mapping of resistance to potato wart in monoparental dihaploids with potential diagnostic value in tetraploid potato varieties. Theor. Appl. Genet. 2018, 131, 2555–2566. [Google Scholar] [CrossRef] [PubMed]
- Prodhomme, C.; van Arkel, G.; Plich, J.; Tammes, J.E.; Rijk, J.; van Eck, H.J.; Visser, R.G.F.; Vossen, J.H. A Hitchhiker’s guide to the potato wart disease resistance galaxy. Theor. Appl. Genet. 2020, 133, 3419–3439. [Google Scholar] [CrossRef] [PubMed]
- Prodhomme, C.; Vos, P.G.; Paulo, M.J.; Tammes, J.E.; Visser, R.G.F.; Vossen, J.H.; van Eck, H.J. Distribution of P1(D1) wart disease resistance in potato germplasm and GWAS identification of haplotype-specific SNP markers. Theor. Appl. Genet. 2020, 133, 1859–1871. [Google Scholar] [CrossRef] [PubMed]
- Angelin-Bonnet, O.; Thomson, S.; Vignes, M.; Biggs, P.J.; Monaghan, K.; Bloomer, R.; Wright, K.; Baldwin, S. Investigating the genetic components of tuber bruising in a breeding population of tetraploid potatoes. BMC Plant Biol. 2023, 23, 238. [Google Scholar] [CrossRef] [PubMed]
- Diaz, P.; Sarmiento, F.; Mathew, B.; Ballvora, A.; Mosquera Vasquez, T. Genomic regions associated with physiological, biochemical and yield-related responses under water deficit in diploid potato at the tuber initiation stage revealed by GWAS. PLoS ONE 2021, 16, e0259690. [Google Scholar] [CrossRef]
- Srivastava, A.; Bhardwaj, V.; Singh, B.P.; Khurana, S.M.P. Potato Diversity and Its Genetic Enhancement. In Gene Pool Diversity and Crop Improvement; Rajpal, V., Rao, S., Raina, S., Eds.; Springer: Cham, Switzerland, 2016; Volume 10, pp. 187–226. [Google Scholar]
- Fleisher, D.H.; Timlin, D.J.; Yang, Y.; Reddy, V.R. Potato stem density effects on canopy development and production. Potato Res. 2011, 54, 137–155. [Google Scholar] [CrossRef]
- Gao, S.; Chu, C. Gibberellin Metabolism and Signaling: Targets for Improving Agronomic Performance of Crops. Plant Cell Physiol. 2020, 61, 1902–1911. [Google Scholar] [CrossRef]
- Han, Y.; Yang, J.; Zhang, N.; Gong, Y.; Liu, M.; Qiao, R.; Jiao, X.; Zhu, F.; Li, X.; Si, H. Genome-Wide Identification of Phytochrome-Interacting Factor (PIF) Gene Family in Potatoes and Functional Characterization of StPIF3 in Regulating Shade-Avoidance Syndrome. Agronomy 2024, 14, 873. [Google Scholar] [CrossRef]
- Deng, J.; Deng, X.; Yao, H.; Ji, S.; Dong, L. Gibberellins Play an Essential Role in the Bud Growth of Petunia hybrida. Curr. Issues Mol. Biol. 2024, 46, 9906–9915. [Google Scholar] [CrossRef]
- Nasir, M.W.; Toth, Z. Response of different potato genotypes to drought stress. Agriculture 2021, 11, 763. [Google Scholar] [CrossRef]
- Kavi Kishor, P.B.; Hima Kumari, P.; Sunita, M.S.; Sreenivasulu, N. Role of proline in cell wall synthesis and plant development and its implications in plant ontogeny. Front. Plant Sci. 2015, 6, 544. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Umar, S.; Khan, N.A.; Khan, M.I.R.J.E.; Botany, E. A new perspective of phytohormones in salinity tolerance: Regulation of proline metabolism. Environ. Exp. Bot. 2014, 100, 34–42. [Google Scholar] [CrossRef]
- Ovchinnikova, A.; Krylova, E.; Gavrilenko, T.; Smekalova, T.; Zhuk, M.; Knapp, S.; Spooner, D.M. Taxonomy of cultivated potatoes (Solanum section Petota: Solanaceae). Bot. J. Linn. Soc. 2011, 165, 107–155. [Google Scholar] [CrossRef]
- Clot, C.R.; Polzer, C.; Prodhomme, C.; Schuit, C.; Engelen, C.J.M.; Hutten, R.C.B.; van Eck, H.J. The origin and widespread occurrence of Sli-based self-compatibility in potato. Theor. Appl. Genet. 2020, 133, 2713–2728. [Google Scholar] [CrossRef] [PubMed]
- Germain, H.; Gray-Mitsumune, M.; Lafleur, E.; Matton, D.P. ScORK17, a transmembrane receptor-like kinase predominantly expressed in ovules is involved in seed development. Planta 2008, 228, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Lu, X.; Gou, X. Receptor-like protein kinases in plant reproduction: Current understanding and future perspectives. Plant Commun. 2022, 3, 100273. [Google Scholar] [CrossRef]
- van Eck, H.J. Genetics of morphological and tuber traits. In Genetics of Morphological and Tuber Traits; Vreugdenhil, D., Bradshaw, J., Gebhardt, C., Govers, F., MacKerron, D.K.L., Taylor, M.A., Ross, H.A., Eds.; Elsevier Science BV: Mauterndorf, Austria, 2007; pp. 91–115. [Google Scholar]
- Chen, N.; Zhu, W.; Xu, J.; Duan, S.; Bian, C.; Hu, J.; Wang, W.; Li, G.; Jin, L. Molecular marker development and primary physical map construction for the tuber shape Ro gene locus in diploid potato (Solanum tuberosum L.). Mol. Breed. 2018, 39, 6. [Google Scholar] [CrossRef]
- Zhang, F.; Qu, L.; Gu, Y.; Xu, Z.-H.; Xue, H.-W. Resequencing and genome-wide association studies of autotetraploid potato. Mol. Hortic. 2022, 2, 6. [Google Scholar] [CrossRef]
- Wang, A.; Hou, Q.; Si, L.; Huang, X.; Luo, J.; Lu, D.; Zhu, J.; Shangguan, Y.; Miao, J.; Xie, Y.; et al. The PLATZ Transcription Factor GL6 Affects Grain Length and Number in Rice. Plant Physiol. 2019, 180, 2077–2090. [Google Scholar] [CrossRef]
- Liwka, J.S.; Wasilewicz-Flis, I.; Jakuczun, H.; Gebhardt, C. Tagging quantitative trait loci for dormancy, tuber shape, regularity of tuber shape, eye depth and flesh colour in diploid potato originated from six Solanum species. Plant Breed. 2008, 127, 49–55. [Google Scholar]
- Parra-Galindo, M.-A.; Piñeros-Niño, C.; Soto-Sedano, J.C.; Mosquera-Vasquez, T. Chromosomes I and X harbor consistent genetic factors associated with the anthocyanin variation in potato. Agronomy 2019, 9, 366. [Google Scholar] [CrossRef]
- Parra-Galindo, M.A.; Soto-Sedano, J.C.; Mosquera-Vasquez, T.; Roda, F. Pathway-based analysis of anthocyanin diversity in diploid potato. PLoS ONE 2021, 16, e0250861. [Google Scholar] [CrossRef] [PubMed]
- Iwama, K. Physiology of the potato: New insights into root system and repercussions for crop management. Potato Res. 2008, 51, 333–353. [Google Scholar] [CrossRef]
- Du, J.; Kirui, A.; Huang, S.; Wang, L.; Barnes, W.J.; Kiemle, S.N.; Zheng, Y.; Rui, Y.; Ruan, M.; Qi, S.; et al. Mutations in the Pectin Methyltransferase QUASIMODO2 Influence Cellulose Biosynthesis and Wall Integrity in Arabidopsis. Plant Cell 2020, 32, 3576–3597. [Google Scholar] [CrossRef] [PubMed]
- Bidhendi, A.J.; Geitmann, A. Relating the mechanics of the primary plant cell wall to morphogenesis. J. Exp. Bot. 2016, 67, 449–461. [Google Scholar] [CrossRef]
- Zhao, S.; Han, X.; Liu, B.; Wang, S.; Guan, W.; Wu, Z.; Theodorakis, P.E. Shelf-life prediction model of fresh-cut potato at different storage temperatures. J. Food Eng. 2022, 317, 110867. [Google Scholar] [CrossRef]
- Kloosterman, B.; Abelenda, J.A.; Gomez Mdel, M.; Oortwijn, M.; de Boer, J.M.; Kowitwanich, K.; Horvath, B.M.; van Eck, H.J.; Smaczniak, C.; Prat, S.; et al. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 2013, 495, 246–250. [Google Scholar] [CrossRef]
- Sattar, M.A.; Sultana, N.; Hossain, M.M.; Rashid, M.H.; Islam, A.A. Genetic variability, correlation and path analysis in potato (Solanum tuberosum L.). Bangladesh J. Plant Breed. Genet. 2007, 20, 33–38. [Google Scholar] [CrossRef]
- Khan, M.S.; van Eck, H.J.; Struik, P.C.J.P.R. Model-based evaluation of maturity type of potato using a diverse set of standard cultivars and a segregating diploid population. Potato Res. 2013, 56, 127–146. [Google Scholar] [CrossRef]
- Danan, S.; Veyrieras, J.-B.; Lefebvre, V. Construction of a potato consensus map and QTL meta-analysis offer new insights into the genetic architecture of late blight resistance and plant maturity traits. BMC Plant Biol. 2011, 11, 16. [Google Scholar] [CrossRef]
- Gong, H.-L.; Dusengemungu, L.; Igiraneza, C.; Rukundo, P. Molecular regulation of potato tuber dormancy and sprouting: A mini-review. Plant Biotechnol. Rep. 2021, 15, 417–434. [Google Scholar] [CrossRef]
- Si, H.; Zhang, C.; Zhang, N.; Wen, Y.; Wang, D. Control of potato tuber dormancy and sprouting by expression of sense and antisense genes of pyrophosphatase in potato. Acta Physiol. Plant. 2016, 38, 69. [Google Scholar] [CrossRef]
- Paluchowska, P.; Sliwka, J.; Yin, Z. Late blight resistance genes in potato breeding. Planta 2022, 255, 127. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.; Zheng, L.; Yang, P.; Wang, J.; Tian, M.; Pan, Y.; Zhao, D.; Yang, Z.; Zhu, J. Detection of Alternaria solani with high accuracy and sensitivity during the latent period of potato early blight. Front. Microbiol. 2022, 13, 1016996. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.R.; Kramer, C.G.; Kotha, R.R.; Wanner, L.A.; Luthria, D.L.; Kramer, M. Cultivar resistance to common scab disease of potato is dependent on the pathogen species. Phytopathology 2019, 109, 1544–1554. [Google Scholar] [CrossRef]
- van de Vossenberg, B.; Prodhomme, C.; Vossen, J.H.; van der Lee, T.A.J. Synchytrium endobioticum, the potato wart disease pathogen. Mol. Plant Pathol. 2022, 23, 461–474. [Google Scholar] [CrossRef]
- Kromann, P.; Miethbauer, T.; Ortiz, O.; Forbes, G.A. Review of Potato Biotic Constraints and Experiences with Integrated Pest Management Interventions. In Integrated Pest Management; Pimentel, D., Peshin, R., Eds.; Springer: Dordrecht, The Netherlands, 2014; Volume 3, pp. 245–268. [Google Scholar]
- Wright, P.J.; Triggs, C.M.; Anderson, J.A.D. Effects of specific gravity and cultivar on susceptibility of potato (Solanum tuberosum) tubers to blackspot bruising and bacterial soft rot. N. Z. J. Crop Hortic. Sci. 2005, 33, 353–361. [Google Scholar] [CrossRef]
- Coetzer, C.; Corsini, D.; Love, S.; Pavek, J.; Tumer, N.J.J.o.A.; Chemistry, F. Control of enzymatic browning in potato (Solanum tuberosum L.) by sense and antisense RNA from tomato polyphenol oxidase. J. Agric. Food Chem. 2001, 49, 652–657. [Google Scholar] [CrossRef]
- Rodriguez Galdon, B.; Rios Mesa, D.; Rodriguez Rodriguez, E.M.; Diaz Romero, C. Influence of the cultivar on the organic acid and sugar composition of potatoes. J. Sci. Food Agric. 2010, 90, 2301–2309. [Google Scholar] [CrossRef]
- Schonhals, E.M.; Ding, J.; Ritter, E.; Paulo, M.J.; Cara, N.; Tacke, E.; Hofferbert, H.R.; Lubeck, J.; Strahwald, J.; Gebhardt, C. Physical mapping of QTL for tuber yield, starch content and starch yield in tetraploid potato (Solanum tuberosum L.) by means of genome wide genotyping by sequencing and the 8.3 K SolCAP SNP array. BMC Genom. 2017, 18, 642. [Google Scholar] [CrossRef]
- Khlestkin, V.K.; Erst, T.V.; Rozanova, I.V.; Efimov, V.M.; Khlestkina, E.K. Genetic loci determining potato starch yield and granule morphology revealed by genome-wide association study (GWAS). PeerJ 2020, 8, e10286. [Google Scholar] [CrossRef] [PubMed]
- Khlestkin, V.K.; Rozanova, I.V.; Efimov, V.M.; Khlestkina, E.K. Starch phosphorylation associated SNPs found by genome-wide association studies in the potato (Solanum tuberosum L.). BMC Genet. 2019, 20, 29. [Google Scholar] [CrossRef] [PubMed]
- Levina, A.V.; Hoekenga, O.; Gordin, M.; Broeckling, C.; De Jong, W.S. Genetic analysis of potato tuber metabolite composition: Genome-wide association studies applied to a nontargeted metabolome. Crop Sci. 2020, 61, 591–603. [Google Scholar] [CrossRef]
- Vos, P.G.; Paulo, M.J.; Bourke, P.M.; Maliepaard, C.A.; van Eeuwijk, F.A.; Visser, R.G.F.; van Eck, H.J. GWAS in tetraploid potato: Identification and validation of SNP markers associated with glycoalkaloid content. Mol. Breed. 2022, 42, 76. [Google Scholar] [CrossRef] [PubMed]
- Klaassen, M.T.; Willemsen, J.H.; Vos, P.G.; Visser, R.G.F.; van Eck, H.J.; Maliepaard, C.; Trindade, L.M. Genome-wide association analysis in tetraploid potato reveals four QTLs for protein content. Mol. Breed. 2019, 39, 151. [Google Scholar] [CrossRef]
- Berdugo-Cely, J.A.; Ceron-Lasso, M.D.S.; Yockteng, R. Phenotypic and molecular analyses in diploid and tetraploid genotypes of Solanum tuberosum L. reveal promising genotypes and candidate genes associated with phenolic compounds, ascorbic acid contents, and antioxidant activity. Front. Plant Sci. 2022, 13, 1007104. [Google Scholar] [CrossRef]
- Toubiana, D.; Cabrera, R.; Salas, E.; Maccera, C.; Franco Dos Santos, G.; Cevallos, D.; Lindqvist-Kreuze, H.; Lopez, J.M.; Maruenda, H. Morphological and metabolic profiling of a tropical-adapted potato association panel subjected to water recovery treatment reveals new insights into plant vigor. Plant J. 2020, 103, 2193–2210. [Google Scholar] [CrossRef]
- Yang, H.; Liao, Q.; Ma, L.; Luo, W.; Xiong, X.; Luo, Y.; Yang, X.; Du, C.; He, Y.; Li, X.; et al. Features and genetic basis of chlorogenic acid formation in diploid potatoes. Food Chem. Mol. Sci. 2021, 3, 100039. [Google Scholar] [CrossRef]
- Bali, S.; Robinson, B.R.; Sathuvalli, V.; Bamberg, J.; Goyer, A. Single Nucleotide Polymorphism (SNP) markers associated with high folate content in wild potato species. PLoS ONE 2018, 13, e0193415. [Google Scholar] [CrossRef]
- Khlestkin, V.; Erst, T.; Igoshin, A.; Rozanova, I.; Khlestkina, E. Meta-analysis of genetic factors for potato starch phosphorylation. Agronomy 2022, 12, 1343. [Google Scholar] [CrossRef]
- Byrne, S.; Meade, F.; Mesiti, F.; Griffin, D.; Kennedy, C.; Milbourne, D. Genome-wide association and genomic prediction for fry color in potato. Agronomy 2020, 10, 90. [Google Scholar] [CrossRef]
- Levina, A.V.; Hoekenga, O.A.; Gordin, M.; Broeckling, C.; De Jong, W.S. Applying network and genetic analysis to the potato metabolome. Front. Plant Sci. 2023, 14, 1108351. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Ma, Z.; Chen, H.; Gao, H. Toward an understanding of potato starch structure, function, biosynthesis, and applications. Food Front. 2023, 4, 980–1000. [Google Scholar] [CrossRef]
- Dupuis, J.H.; Liu, Q. Potato starch: A review of physicochemical, functional and nutritional properties. Am. J. Potato Res. 2019, 96, 127–138. [Google Scholar] [CrossRef]
- Blennow, A.; Hansen, M.; Schulz, A.; Jorgensen, K.; Donald, A.M.; Sanderson, J. The molecular deposition of transgenically modified starch in the starch granule as imaged by functional microscopy. J. Struct. Biol. 2003, 143, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.; Song, B.; Liu, X.; Xie, C.; Li, M.; Lin, Y.; Zhang, H.; Liu, J. Promoter regions of potato vacuolar invertase gene in response to sugars and hormones. Plant Physiol. Biochem. 2013, 69, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Benkeblia, N. Potato Glycoalkaloids: Occurrence, biological activities and extraction for biovalorisation–a review. Int. J. Food Sci. Technol. 2020, 55, 2305–2313. [Google Scholar] [CrossRef]
- Mader, J.; Rawel, H.; Kroh, L.W. Composition of phenolic compounds and glycoalkaloids alpha-solanine and alpha-chaconine during commercial potato processing. J. Agric. Food Chem. 2009, 57, 6292–6297. [Google Scholar] [CrossRef]
- Tack, F.M.G. Trace Elements in Potato. Potato Res. 2015, 57, 311–325. [Google Scholar] [CrossRef]
- Palta, J.P. Improving potato tuber quality and production by targeted calcium nutrition: The discovery of tuber roots leading to a new concept in potato nutrition. Potato Res. 2010, 53, 267–275. [Google Scholar] [CrossRef]
- Kratzke, M.G.; Jiwan, P.P. Calcium accumulation in potato tubers: Role of the basal roots. HortScience 1986, 21, 1022–1024. [Google Scholar] [CrossRef]
- Haynes, K.G.; Yencho, G.C.; Clough, M.E.; Henninger, M.R.; Sterrett, S.B. Genetic variation for potato tuber micronutrient content and implications for biofortification of potatoes to reduce micronutrient malnutrition. Am. J. Potato Res. 2012, 89, 192–198. [Google Scholar] [CrossRef]
- Sanderson, D.V.; Voutchkov, M.; Benkeblia, N. Bioaccumulation of cadmium in potato tuber grown on naturally high levels cadmium soils in Jamaica. Sci. Total Environ. 2019, 649, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Joly, N.; Souidi, K.; Depraetere, D.; Wils, D.; Martin, P. Potato by-products as a source of natural chlorogenic acids and phenolic compounds: Extraction, characterization, and antioxidant capacity. Molecules 2020, 26, 177. [Google Scholar] [CrossRef]
- Guo, J.; Guo, J.; Li, L.; Bai, X.; Huo, X.; Shi, W.; Gao, L.; Dai, K.; Jing, R.; Hao, C. Combined linkage analysis and association mapping identifies genomic regions associated with yield-related and drought-tolerance traits in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2023, 136, 250. [Google Scholar] [CrossRef]
| Traits | Population Origin | Population Size | Planting Environments | Number of Loci | Chromosome Location of Key Loci | References |
|---|---|---|---|---|---|---|
| Plant height | Tetraploid | 370 | Two years, one location | 92 | 1, 5 | Zhao et al., 2023 [104] |
| Number of main stems | Tetraploid | 251 | Two years, four locations | 7 | 2, 4, 5 | Han et al., 2023 [105] |
| Stem color | Tetraploid | 466 | Eight years, three locations | 2 | 4, 7 | Berdugo-Cely et al., 2017 [106] |
| Growth habit | Tetraploid | 162 | Two years, three locations | 2 | 5 | Massa et al., 2018 [107] |
| Leaf shape | Tetraploid | 237 | Two years, one location | 9 | 1 | Zia et al., 2020 [108] |
| Number of leaflets | Tetraploid | 237 | Two years, one location | 3 | 3, 4, 8 | Zia et al., 2020 [108] |
| Canopy cover | Tetraploid | 189 | Two years, two locations | 8 | 1, 2, 3 | Ospina Nieto et al., 2021 [109] |
| Flower color | Tetraploid | 466 | Eight years, three locations | 13 | 3, 7 | Berdugo-Cely et al., 2017 [106] |
| Tetraploid | 110 | Four years, two locations | 1 | 10 | Rak et al., 2017 [110] | |
| Tetraploid | 237 | Two years, one location | 1 | 4 | Zia et al., 2020 [108] | |
| Stamen and pistil length | Tetraploid | 237 | Two years, one location | 5 | 2, 5 | Zia et al., 2022 [111] |
| Self-fertility | Diploid | 164 | Two years, one location | 1 | 3 | Kaiser et al., 2021 [112] |
| Maturity | Diploid | 98 | Two years, one location | 1 | 5 | Manrique-Carpintero et al., 2015 [113] |
| Tetraploid | 156 | Two years, one location | 2 | 5 | Massa et al., 2015 [114] | |
| Diploid | 110 | Two years, one location | 1 | 5 | Braun et al., 2017 [115] | |
| Tetraploid | 162 | Two years, three locations | 1 | 5 | Massa et al., 2018 [107] | |
| Tetraploid | 188 | Two years, one location | 2 | 4, 5 | Mengist et al., 2018 [116] | |
| Tetraploid | 237 | Two years, one location | 3 | 5, 9 | Zia et al., 2020 [108] | |
| Tetraploid | 586 | Two years, one location | 3 | 4, 5, 6 | Caraza-Harter and Endelman, 2022 [117] | |
| Tetraploid | 241 | Two years, one location | 3 | 5 | Xue et al., 2022 [118] | |
| Tetraploid | 384 | Four years, one location | 6 | 5 | Pandey et al., 2023 [119] | |
| Yield | Diploid | 98 | Two years, one location | 3 | 2, 5, 12 | Manrique-Carpintero et al., 2015 [113] |
| Tetraploid | 110 | Four years, two locations | 6 | 2, 5 | Rak et al., 2017 [110] | |
| Tetraploid | 290 | Two years, two locations | 192 | 4 | Campbell et al., 2023 [120] | |
| Tetraploid | 192 | Three years, two locations | 6 | 5, 8, 9 | Yousaf et al., 2023 [121] | |
| Average tuber weight | Diploid | 98 | Two years, one location | 6 | 4, 5 | Manrique-Carpintero et al., 2015 [113] |
| Diploid | 110 | Two years, one location | 2 | 1 | Braun et al., 2017 [115] | |
| Tetraploid | 110 | Four years, two locations | 6 | 5 | Rak et al., 2017 [110] | |
| Tetraploid | 103 | Three years, two locations | 3 | 5 | Aliche et al., 2019 [122] | |
| Tetraploid | 192 | Three years, two locations | 20 | 4 | Yousaf et al., 2023 [121] | |
| Tuber number per plant | Diploid | 98 | Two years, one location | 2 | 5 | Manrique-Carpintero et al., 2015 [113] |
| Tetraploid | 110 | Four years, two locations | 3 | 4, 5, 10 | Rak et al., 2017 [110] | |
| Tetraploid | 103 | Three years, two locations | 1 | 3 | Aliche et al., 2019 [122] | |
| Tetraploid | 251 | Two years, four locations | 13 | 1 | Han et al., 2023 [105] | |
| Tetraploid | 192 | Three years, two locations | 3 | 6, 7 | Yousaf et al., 2023 [121] | |
| Tuber shape | Diploid | 186 | Three years, one location | 7 | 2, 10 | Prashar et al., 2014 [123] |
| Tetraploid | 110 | Four years, two locations | 8 | 5, 6 | Rak et al., 2017 [110] | |
| Tetraploid | 466 | Eight years, three locations | 1 | 7 | Berdugo-Cely et al., 2017 [106] | |
| Tetraploid | 162 | Two years, three locations | 1 | 5 | Massa et al., 2018 [107] | |
| Tetraploid | 237 | Two years, one location | 3 | 4, 10 | Zia et al., 2020 [108] | |
| Tetraploid | 205 | Two years, two locations | 13 | 4, 10 | Park et al., 2021 [124] | |
| Tetraploid | 214 | Three years, two locations | 11 | 10 | Pandey et al., 2022 [125] | |
| Tetraploid | 370 | Two years, one location | 146 | 1, 2 | Zhao et al., 2023 [126] | |
| Tetraploid | 192 | Three years, two locations | 21 | 10 | Yousaf et al., 2023 [121] | |
| Tuber flesh color | Tetraploid | 214 | Three years, two locations | 5 | 3 | Pandey et al., 2022 [125] |
| Tuber skin color | Tetraploid | 466 | Eight years, three locations | 6 | 7 | Berdugo-Cely et al., 2017 [106] |
| Tetraploid | 237 | Two years, one location | 17 | 1, 8, 12 | Zia et al., 2020 [108] | |
| Tetraploid | 214 | Three years, two locations | 16 | 3, 11 | Pandey et al., 2022 [125] | |
| Eye depth | Diploid | 186 | Three years, one location | 4 | 10 | Prashar et al., 2014 [123] |
| Tetraploid | 214 | Three years, two locations | 3 | 3, 5, 10 | Pandey et al., 2022 [125] | |
| Tetraploid | 370 | Two years, one location | 53 | 5, 6 | Zhao et al., 2023 [126] | |
| Tuber skin russeting texture degree | Tetraploid | 214 | Three years, two locations | 12 | 1, 4, 5, 12 | Pandey et al., 2022 [125] |
| Tuber skin maturity | Tetraploid | 586 | Two years, one location | 3 | 4, 5, 9 | Caraza-Harter and Endelman, 2022 [117] |
| Dormancy and germination | Diploid | 129 | One year, one location | 14 | 2, 3, 7, 11 | Bisognin et al., 2018 [127] |
| Root diameter | Tetraploid | 192 | Two years, one location | 9 | 9, 11, 12 | Yousaf et al., 2023 [121] |
| Stolon diameter | Tetraploid | 192 | Three years, two locations | 8 | 5, 6, 11 | Yousaf et al., 2023 [121] |
| Stolon length | Tetraploid | 192 | Two years, one location | 41 | 4, 6, 9 | Yousaf et al., 2021 [128] |
| Stolon weight | Tetraploid | 192 | Three years, two locations | 12 | 3, 4 | Yousaf et al., 2023 [121] |
| Plant vigor | Diploid | 98 | Two years, one location | 3 | 3, 5, 10 | Manrique-Carpintero et al., 2015 [113] |
| Nitrogen use efficiency | Tetraploid | 88 | Two years, two locations | 77 | 3, 5, 6 | Getahun et al., 2022 [129] |
| Early blight resistance | Tetraploid | 162 | Two years, three locations | 1 | 5 | Massa et al., 2018 [107] |
| Tetraploid | 80 | Three years, one location | 33 | 1, 5, 11 | Odilbekov et al., 2020 [130] | |
| Tetraploid | 241 | Two years, one location | 9 | 5 | Xue et al., 2022 [118] | |
| Late blight resistance | Tetraploid | 103 | Six years, two locations | 3 | 7, 9 | Lindqvist-Kreuze et al., 2014 [131] |
| Tetraploid | 156 | Two years, one location | 2 | 9 | Massa et al., 2015 [114] | |
| Tetraploid | 184 | Two years, two locations | 17 | 5 | Mosquera et al., 2016 [132] | |
| Tetraploid | 94 | Two years, one location | 6 | 1, 4, 5, 8 | Santa et al., 2018 [133] | |
| Diploid | 150 | Three years, one location | 16 | 3, 12 | Juyo Rojas et al., 2019 [134] | |
| Tetraploid | 284 | Two years, one location | 964 | 1, 4, 9 | Wang et al., 2021 [135] | |
| Diploid/tetraploid | 380 | Four years, three locations | 30 | 3, 9 | Lindqvist-Kreuze et al., 2021 [48] | |
| Tetraploid | 222 | Three years, one location | 7 | 11 | Sood et al., 2023 [136] | |
| Bacterial wilt resistance | Diploid | 94 | One year, one location | 5 | 1, 10, 11 | Habe et al., 2019 [137] |
| Verticillium wilt resistance | Tetraploid | 162 | Two years, three locations | 1 | 5 | Massa et al., 2018 [107] |
| Common scab resistance | Diploid | 110 | Two years, one location | 2 | 11 | Braun et al., 2017 [115] |
| Tetraploid | 143 | Three years, one location | 3 | 2, 4, 12 | Yuan et al., 2019 [138] | |
| Tetraploid | 198 | Two years, one location | 3 | 1, 2 | Kaiser et al., 2020 [139] | |
| Tetraploid | 165 | Twelve years, two locations | 6 | 1 | Koizumi et al., 2021 [140] | |
| Tetraploid | 151 | Four years, one location | 7 | 1, 3, 6, 10 | Zorrilla et al., 2021 [141] | |
| Wart resistance | Tetraploid | 133 | One year, one location | 67 | 12 | Obidiegwu et al., 2015 [142] |
| Tetraploid | 215 | One year, one location | 186 | 11 | Bartkiewicz et al., 2018 [143] | |
| Tetraploid | 330 | Public database | 70 | 11 | Prodhomme et al., 2020 [144] | |
| Tetraploid | 569 | Public database | 333 | 10, 11, 12 | Prodhomme et al., 2020 [145] | |
| Tuber end rot resistance | Diploid | 98 | Two years, one location | 4 | 1, 3, 5, 6 | Manrique-Carpintero et al., 2015 [113] |
| Cyst nematode resistance | Tetraploid | 222 | Three years, one location | 20 | 3, 10 | Sood et al., 2023 [136] |
| Potato tuber moth resistance | Tetraploid | 94 | Two years, one location | 14 | 2, 7, 12 | Santa et al., 2018 [133] |
| Colorado potato beetle resistance | Diploid | 136 | Two years, one location | 11 | 2 | Kaiser et al., 2021 [112] |
| Tuber bruising resistance | Tetraploid | 158 | One year, one location | 5 | 1, 7, 8 | Angelin-Bonnet et al., 2023 [146] |
| Drought tolerance | Diploid | 104 | One year, one location | 38 | 3, 11, 12 | Diaz et al., 2021 [147] |
| Hollow heart tolerance | Tetraploid | 151 | Four years, one location | 5 | 3, 5, 6, 12 | Zorrilla et al., 2021 [141] |
| Traits | Population Origin | Population Size | Planting Environments | Number of Loci | Chromosome Location of Key Loci | References |
|---|---|---|---|---|---|---|
| Specific gravity | Diploid | 98 | Two years, one location | 2 | 5, 9 | Manrique-Carpintero et al., 2015 [113] |
| Tetraploid | 205 | Two years, two locations | 11 | 1, 5 | Park et al., 2021 [124] | |
| Starch content | Tetraploid | 264/184 | Three years, two locations | 117 | 1, 11, 12 | Schonhals et al., 2017 [185] |
| Tetraploid | 90 | One year, one location | 15 | 4, 5, 11 | Khlestkin et al., 2020 [186] | |
| Starch granule morphology | Tetraploid | 90 | One year, one location | 38 | 1, 2 | Khlestkin et al., 2020 [186] |
| Tetraploid | 90 | One year, one location | 14 | 1 | Khlestkin et al., 2019 [187] | |
| Glucose content | Tetraploid | 162 | Two years, three locations | 4 | 4, 5 | Massa et al., 2018 [107] |
| Anthocyanin content | Diploid | 96 | One year, one location | 7 | 1, 10 | Parra-Galindo et al., 2019 [165] |
| Diploid | 96 | One year, one location | 22 | 1, 10 | Parra-Galindo et al., 2021 [166] | |
| Glycoalkaloid content | Diploid | 136 | Two years, one location | 23 | 2, 6, 7 | Kaiser et al., 2021 [112] |
| Tetraploid | 185 | One year, one location | 21 | 2, 7, 11 | Levina et al., 2020 [188] | |
| Tetraploid | 275 | Two years, two locations | 603 | 1, 11 | Vos et al., 2022 [189] | |
| Protein content | Tetraploid | 277 | Three years, four locations | 12 | 5 | Klaassen et al., 2019 [190] |
| Phenol content | Diploid/tetraploid | 404 | One year, one location | 23 | 9 | Berdugo-Cely et al., 2022 [191] |
| Ascorbic acid content | Diploid/tetraploid | 404 | One year, one location | 11 | 2, 3, 4, 6 | Berdugo-Cely et al., 2022 [191] |
| Fumarate content | Tetraploid | 258 | One year, one location | 11 | 1, 4, 5 | Toubiana et al., 2020 [192] |
| Chlorogenic acid content | Diploid | 271 | Two years, one location | 18 | 4, 8, 10 | Yang et al., 2021 [193] |
| Citric acid content | Tetraploid | 162 | Two years, three locations | 1 | 5 | Massa et al., 2018 [107] |
| Folate content | Diploid | 94 | One year, one location | 42 | 4, 6 | Bali et al., 2018 [194] |
| Phosphorus content | Tetraploid | 90 | Three years, one location | 4 | 5 | Khlestkin et al., 2022 [195] |
| Calcium content | Tetraploid | 151 | Four years, one location | 6 | 1, 3, 7, 8 | Zorrilla et al., 2021 [141] |
| Cadmium content | Tetraploid | 188 | Two years, one location | 4 | 3, 5, 6, 7 | Mengist et al., 2018 [116] |
| Zinc content | Tetraploid | 188 | Two years, one location | 5 | 1, 3, 5 | Mengist et al., 2018 [116] |
| Antioxidant activity | Diploid/tetraploid | 404 | One year, one location | 50 | 3, 4, 6, 9 | Berdugo-Cely et al., 2022 [191] |
| Cold-induced sweetening | Diploid | 110 | Two years, one location | 2 | 4, 6 | Braun et al., 2017 [115] |
| Chip color | Tetraploid | 110 | Four years, two locations | 17 | 2, 3, 9 | Rak et al., 2017 [110] |
| Tetraploid | 274 | Three years, one location | 30 | 4, 10 | Byrne et al., 2020 [196] | |
| Tetraploid | 393 | Two years, one location | 2 | 4, 10 | Jo et al., 2023 [40] | |
| Tetraploid | 184 | Public database | 22 | 3, 7 | Levina et al., 2023 [197] | |
| Tetraploid | 384 | Four years, one location | 12 | 1, 3, 7 | Pandey et al., 2023 [119] | |
| Bud-end and stem-end fry color | Tetraploid | 162 | Two years, three locations | 7 | 4 | Massa et al., 2018 [107] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, J.; Cheng, L.; Wang, Y.; Zhang, F. Genome-Wide Association Studies for Key Agronomic and Quality Traits in Potato (Solanum tuberosum L.). Agronomy 2024, 14, 2214. https://doi.org/10.3390/agronomy14102214
Yuan J, Cheng L, Wang Y, Zhang F. Genome-Wide Association Studies for Key Agronomic and Quality Traits in Potato (Solanum tuberosum L.). Agronomy. 2024; 14(10):2214. https://doi.org/10.3390/agronomy14102214
Chicago/Turabian StyleYuan, Jianlong, Lixiang Cheng, Yuping Wang, and Feng Zhang. 2024. "Genome-Wide Association Studies for Key Agronomic and Quality Traits in Potato (Solanum tuberosum L.)" Agronomy 14, no. 10: 2214. https://doi.org/10.3390/agronomy14102214
APA StyleYuan, J., Cheng, L., Wang, Y., & Zhang, F. (2024). Genome-Wide Association Studies for Key Agronomic and Quality Traits in Potato (Solanum tuberosum L.). Agronomy, 14(10), 2214. https://doi.org/10.3390/agronomy14102214






