Identification of SNPs Associated with Grain Quality Traits in Spring Barley Collection Grown in Southeastern Kazakhstan
Abstract
1. Introduction
2. Materials and Methods
2.1. Barley Germplasm Collection and Genotyping
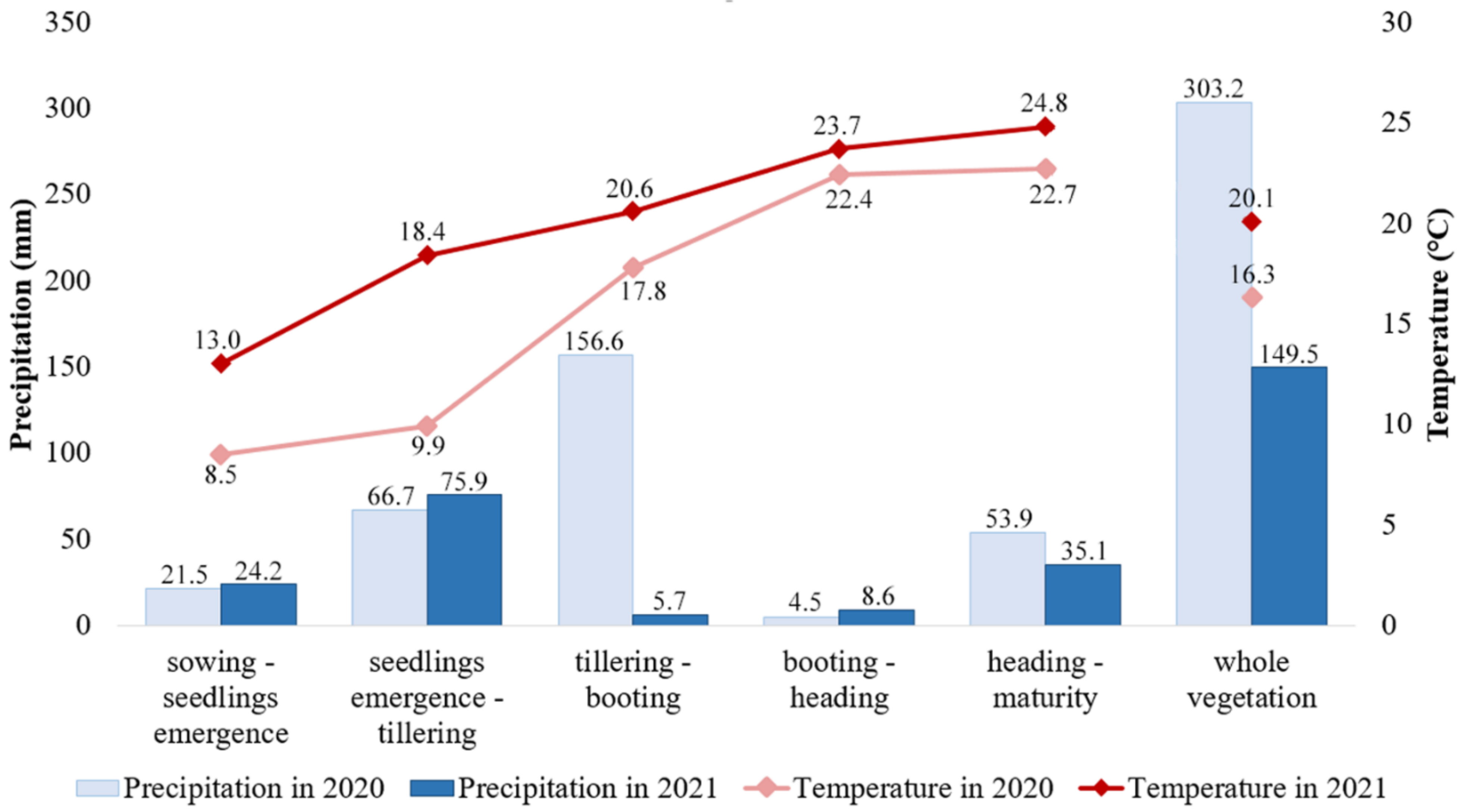
2.2. Field Experiment, Analysis of Grain Quality Traits, and Statistics
2.3. Genetic Structure of the Population and the GWAS
3. Results
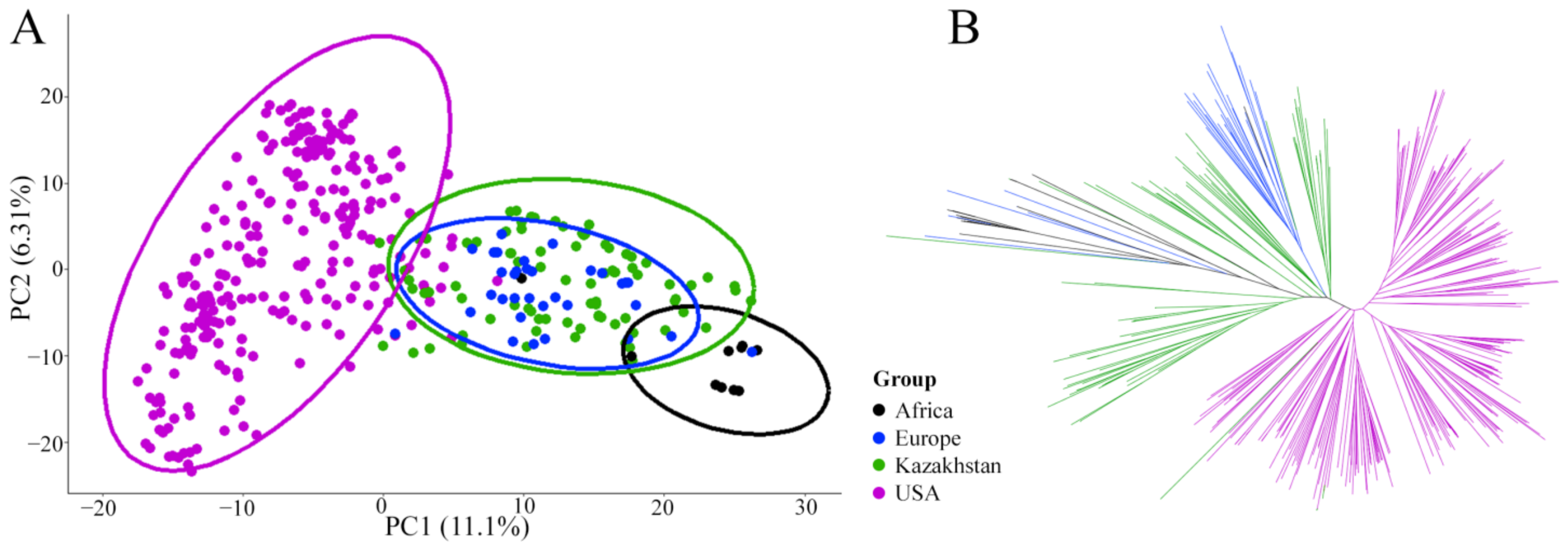
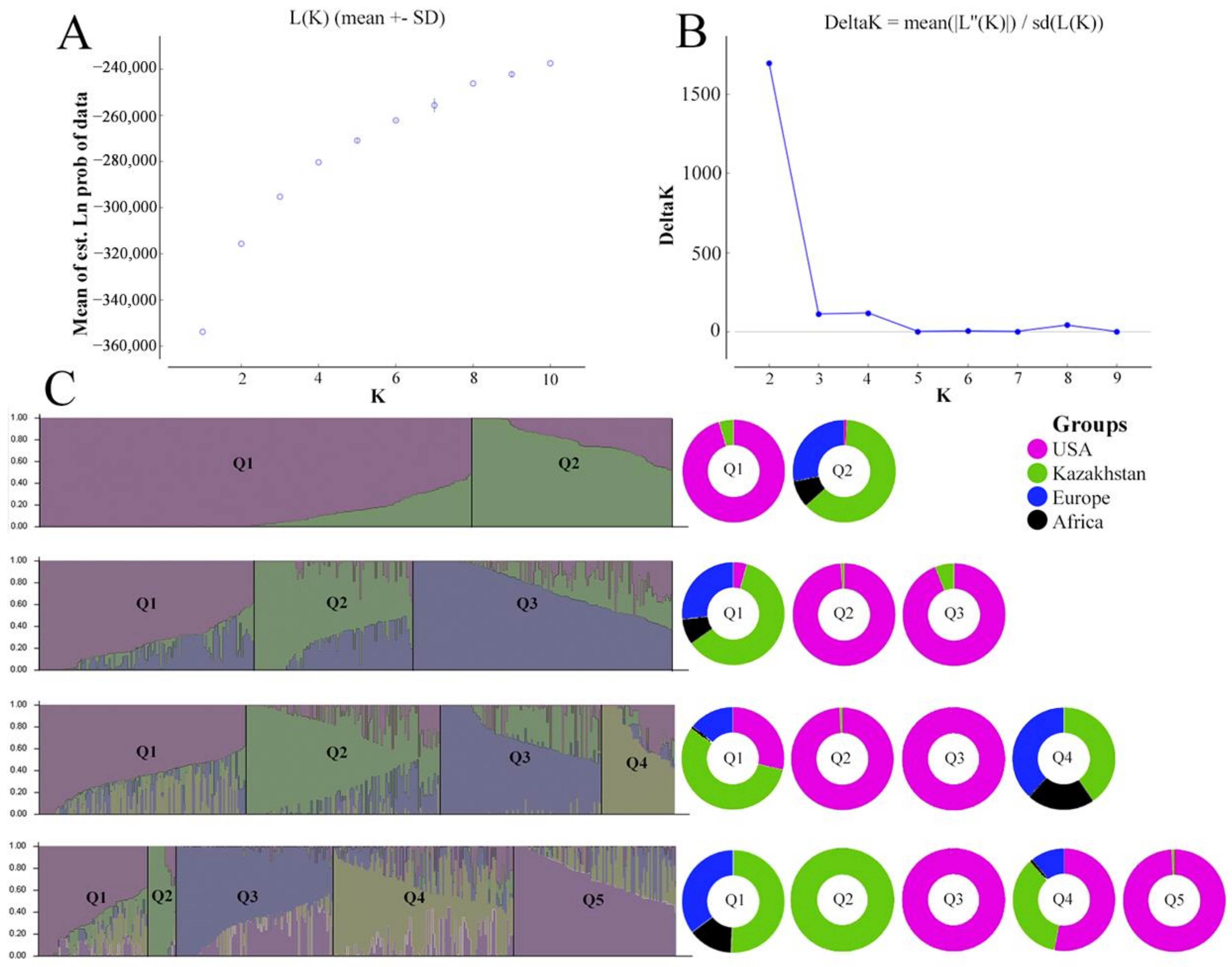
3.1. Genetic Structure of the Barley Population
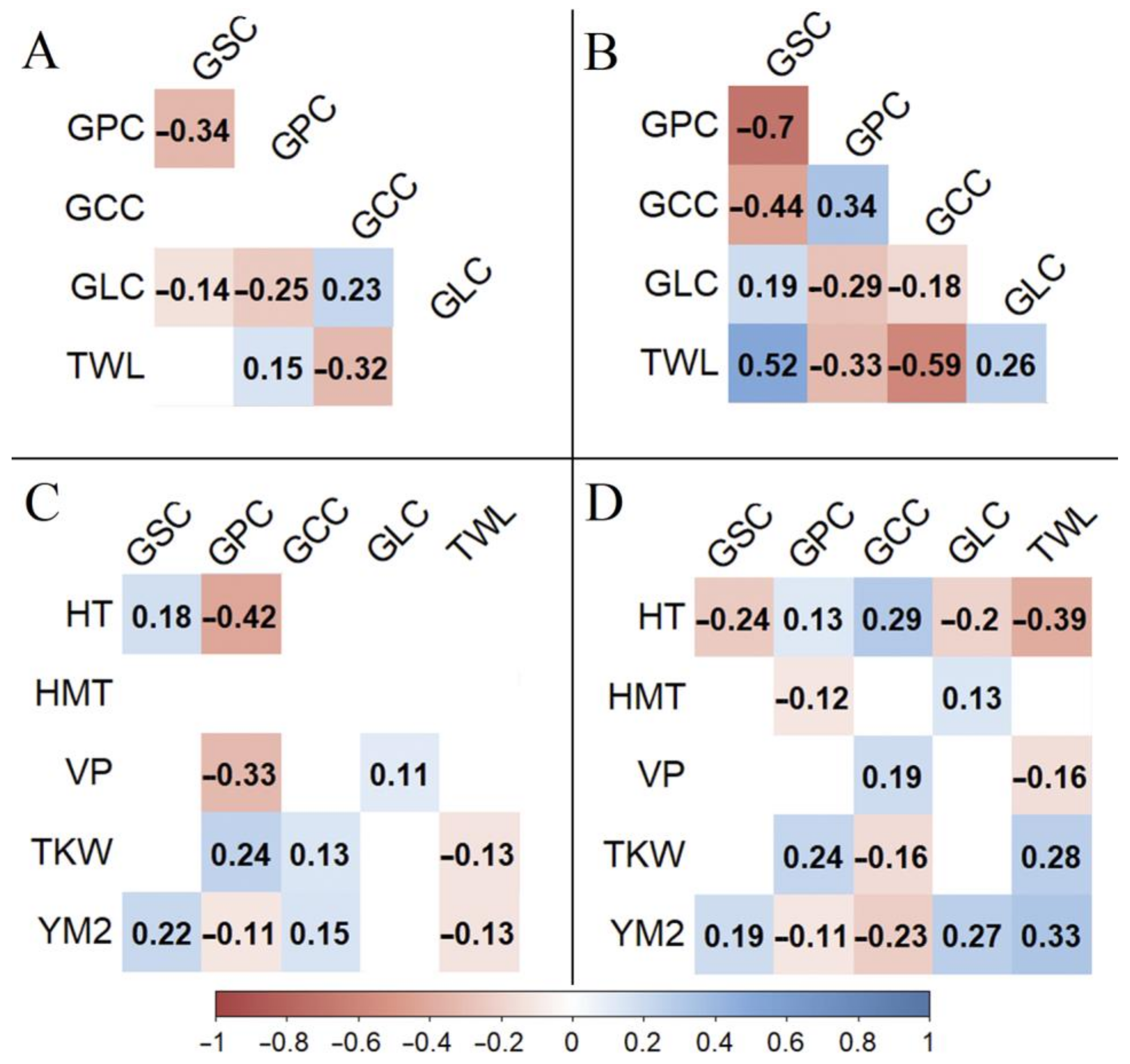
3.2. Grain Quality Traits
3.3. Association Analysis and Novel QTLs
4. Discussion
4.1. Genetic Structure of the Studied Barley Collection
4.2. Grain Quality Trait Variation in the Studied Barley Collection
4.3. QTLs Associated with Grain Quality Traits in the Studied Barley Collection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Statista Database. Available online: https://www.statista.com/ (accessed on 12 January 2023).
- Agency for Strategic Planning and Reforms of the Republic of Kazakhstan, Bureau of National Statistics. Available online: https://stat.gov.kz/ (accessed on 12 January 2023).
- Langridge, P. Economic and Academic Importance of Barley. In The Barley Genome. Compendium of Plant Genomes; Stein, N., Muehlbauer, G.J., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–10. [Google Scholar]
- Henry, R.J. The carbohydrates of barley grains—A review. J. Inst. Brew. 1988, 94, 71–78. [Google Scholar] [CrossRef]
- Hussain, A.; Ali, S.; Hussain, A.; Hussain, Z.; Manzoor, M.F.; Hussain, A.; Hussain, M. Compositional profile of barley landlines grown in different regions of Gilgit-Baltistan. Food Sci. Nutr. 2021, 9, 2605–2611. [Google Scholar] [CrossRef] [PubMed]
- Gebeyaw, M. Impact of malt barley varieties on malt quality: A review. Agric. Rev. 2021, 42, 116–119. [Google Scholar] [CrossRef]
- Bleidere, M.; Gaile, Z. Grain quality traits important in feed barley. Proc. Latv. Acad. Sci. Sect. B Nat. Exact Appl. Sci. 2012, 66, 1–9. [Google Scholar] [CrossRef]
- Smith, A.M. The biosynthesis of starch granules. Biomacromolecules 2001, 2, 335–341. [Google Scholar] [CrossRef]
- Collins, H.M.; Betts, N.S.; Dockter, C.; Berkowitz, O.; Braumann, I.; Cuesta-Seijo, J.A.; Skadhauge, B.; Whelan, J.; Bulone, V.; Fincher, G.B. Genes that mediate starch metabolism in developing and germinated barley grain. Front. Plant Sci. 2021, 12, 641325. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.C.; Zhang, G.P.; Zhou, M.X. Protein and hordein content in barley seeds as affected by nitrogen level and their relationship to beta-amylase activity. J. Cereal Sci. 2006, 43, 102–107. [Google Scholar] [CrossRef]
- Jaeger, A.; Zannini, E.; Sahin, A.W.; Arendt, E.K. Barley protein properties, extraction and applications, with a focus on brewers’ spent grain protein. Foods 2021, 10, 1389. [Google Scholar] [CrossRef]
- Kaczmarczyk, A.; Bowra, S.; Elek, Z.; Vincze, E. Quantitative RT-PCR based platform for rapid quantification of the transcripts of highly homologous multigene families and their members during grain development. BMC Plant Biol. 2012, 12, 184. [Google Scholar] [CrossRef]
- Vinje, M.A.; Walling, J.G.; Henson, C.A.; Duke, S.H. Comparative gene expression analysis of the β-amylase and hordein gene families in the developing barley grain. Gene 2019, 693, 127–136. [Google Scholar] [CrossRef]
- Jamar, C.; Loffet, F.; Frettinger, P.; Ramsay, L.; Fauconnier, M.L.; Du Jardin, P. NAM-1 gene polymorphism and grain protein content in Hordeum. J. Plant Physiol. 2010, 167, 497–501. [Google Scholar] [CrossRef]
- Cai, S.; Yu, G.; Chen, X.; Huang, Y.; Jiang, X.; Zhang, G.; Jin, X. Grain protein content variation and its association analysis in barley. BMC Plant Biol. 2013, 13, 35. [Google Scholar] [CrossRef]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef] [PubMed]
- Hagenblad, J.; Vanhala, T.; Madhavan, S.; Leino, M.W. Protein content and HvNAM alleles in Nordic barley (Hordeum vulgare) during a century of breeding. Hereditas 2022, 159, 12. [Google Scholar] [CrossRef] [PubMed]
- Fedak, G.; Roche, I.D.L. Lipid and fatty acid composition of barley kernels. Can. J. Plant Sci. 1977, 57, 257–260. [Google Scholar] [CrossRef]
- Kannangara, R.; Branigan, C.; Liu, Y.; Penfield, T.; Rao, V.; Mouille, G.; Hofte, H.; Pauly, M.; Riechmann, J.L.; Broun, P. The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 2007, 19, 1278–1294. [Google Scholar] [CrossRef]
- Taketa, S.; Amano, S.; Tsujino, Y.; Sato, T.; Saisho, D.; Kakeda, K.; Nomura, M.; Suzuki, T.; Matsumoto, T.; Sato, K.; et al. Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 4062–4067. [Google Scholar] [CrossRef]
- Little, A.; Schwerdt, J.G.; Shirley, N.J.; Khor, S.F.; Neumann, K.; O’Donovan, L.A.; Lahnstein, J.; Collins, H.M.; Henderson, M.; Fincher, G.B.; et al. Revised phylogeny of the cellulose synthase gene superfamily: Insights into cell wall evolution. Plant Physiol. 2018, 177, 1124–1141. [Google Scholar] [CrossRef]
- Xu, Y.; Li, P.; Yang, Z.; Xu, C. Genetic mapping of quantitative trait loci in crops. Crop J. 2017, 5, 175–184. [Google Scholar] [CrossRef]
- Emebiri, L.C.; Moody, D.B.; Panozzo, J.F.; Chalmers, K.J.; Kretschmer, J.M.; Ablett, G.A. Identification of QTLs associated with variations in grain protein concentration in two-row barley. Aust. J. Agric. Res. 2003, 54, 1211–1221. [Google Scholar] [CrossRef]
- Li, J.Z.; Huang, X.Q.; Heinrichs, F.; Ganal, M.W.; Röder, M.S. Analysis of QTLs for yield components, agronomic traits, and disease resistance in an advanced backcross population of spring barley. Genome 2006, 49, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Haleem, H.; Bowman, J.; Giroux, M.; Kanazin, V.; Talbert, H.; Surber, L.; Blake, T. Quantitative trait loci of acid detergent fiber and grain chemical composition in hulled× hull-less barley population. Euphytica 2010, 172, 405–418. [Google Scholar] [CrossRef]
- Fan, C.; Zhai, H.; Wang, H.; Yue, Y.; Zhang, M.; Li, J.; Wen, S.; Guo, G.; Zeng, Y.; Ni, Z.; et al. Identification of QTLs controlling grain protein concentration using a high-density SNP and SSR linkage map in barley (Hordeum vulgare L.). BMC Plant Biol. 2017, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Borem, A.; Mather, D.E.; Rasmusson, D.C.; Fulcher, R.G.; Hayes, P.M. Mapping quantitative trait loci for starch granule traits in barley. J. Cereal Sci. 1999, 29, 153–160. [Google Scholar] [CrossRef]
- Han, F.; Ullrich, S.E.; Romagosa, I.; Clancy, J.A.; Froseth, J.A.; Wesenberg, D.M. Quantitative genetic analysis of acid detergent fibre content in barley grain. J. Cereal Sci. 2003, 38, 167–172. [Google Scholar] [CrossRef]
- Marquez-Cedillo, L.A.; Hayes, P.M.; Jones, B.L.; Kleinhofs, A.; Legge, W.G.; Rossnagel, B.G.; Sato, K.; Ullrich, S.E.; Wesenberg, D.M. QTL analysis of malting quality in barley based on the doubled-haploid progeny of two North American varieties representing different germplasm groups. Theor. Appl. Genet. 2000, 101, 173–184. [Google Scholar] [CrossRef]
- Marquez-Cedillo, L.A.; Hayes, P.M.; Kleinhofs, A.; Legge, W.G.; Rossnagel, B.G.; Sato, K.; Ullrich, S.E.; Wesenberg, D.M. QTL analysis of agronomic traits in barley based on the doubled haploid progeny of two elite North American varieties representing different germplasm groups. Theor. Appl. Genet. 2001, 103, 625–637. [Google Scholar] [CrossRef]
- Peñalba, J.V.; Wolf, J.B. From molecules to populations: Appreciating and estimating recombination rate variation. Nat. Rev. Genet. 2020, 21, 476–492. [Google Scholar] [CrossRef]
- Stumpf, M.P.; McVean, G.A. Estimating recombination rates from population-genetic data. Nat. Rev. Genet. 2003, 4, 959–968. [Google Scholar] [CrossRef]
- Berger, G.L.; Liu, S.; Hall, M.D.; Brooks, W.S.; Chao, S.; Muehlbauer, G.J.; Baik, B.-K.; Steffenson, B.; Griffey, C.A. Marker-trait associations in Virginia Tech winter barley identified using genome-wide mapping. Theor. Appl. Genet. 2013, 126, 693–710. [Google Scholar] [CrossRef]
- Pauli, D.; Muehlbauer, G.J.; Smith, K.P.; Cooper, B.; Hole, D.; Obert, D.E.; Ullrich, S.E.; Blake, T.K. Association mapping of agronomic QTLs in US spring barley breeding germplasm. Plant Genome 2014, 7, plantgenome2013-11. [Google Scholar] [CrossRef]
- Genievskaya, Y.; Almerekova, S.; Sariev, B.; Chudinov, V.; Tokhetova, L.; Sereda, G.; Ortaev, A.; Tsygankov, V.; Blake, T.; Chao, S.; et al. Marker-trait associations in two-rowed spring barley accessions from Kazakhstan and the USA. PLoS ONE 2018, 13, e0205421. [Google Scholar] [CrossRef] [PubMed]
- Almerekova, S.; Sariev, B.; Abugalieva, A.; Chudinov, V.; Sereda, G.; Tokhetova, L.; Ortaev, A.; Tsygankov, V.; Blake, T.; Chao, S.; et al. Association mapping for agronomic traits in six-rowed spring barley from the USA harvested in Kazakhstan. PLoS ONE 2019, 14, e0221064. [Google Scholar] [CrossRef]
- Gutiérrez, L.; Germán, S.; Pereyra, S.; Hayes, P.M.; Pérez, C.A.; Capettini, F.; Locatelli, A.; Berberian, N.M.; Falconi, E.E.; Estrada, R.; et al. Multi-environment multi-QTL association mapping identifies disease resistance QTL in barley germplasm from Latin America. Theor. Appl. Genet. 2015, 128, 501–516. [Google Scholar] [CrossRef] [PubMed]
- Gyawali, S.; Chao, S.; Vaish, S.S.; Singh, S.P.; Rehman, S.; Vishwakarma, S.R.; Verma, R.P.S. Genome wide association studies (GWAS) of spot blotch resistance at the seedling and the adult plant stages in a collection of spring barley. Mol. Breed. 2018, 38, 62. [Google Scholar] [CrossRef]
- Amezrou, R.; Verma, R.P.S.; Chao, S.; Brueggeman, R.S.; Belqadi, L.; Arbaoui, M.; Rehman, S.; Gyawali, S. Genome-wide association studies of net form of net blotch resistance at seedling and adult plant stages in spring barley collection. Mol. Breed. 2018, 38, 58. [Google Scholar] [CrossRef]
- Fan, Y.; Zhou, G.; Shabala, S.; Chen, Z.H.; Cai, S.; Li, C.; Zhou, M. Genome-wide association study reveals a new QTL for salinity tolerance in barley (Hordeum vulgare L.). Front. Plant Sci. 2016, 7, 946. [Google Scholar] [CrossRef]
- Thabet, S.G.; Moursi, Y.S.; Sallam, A.; Karam, M.A.; Alqudah, A.M. Genetic associations uncover candidate SNP markers and genes associated with salt tolerance during seedling developmental phase in barley. Environ. Exp. Bot. 2021, 188, 104499. [Google Scholar] [CrossRef]
- Pasam, R.K.; Sharma, R.; Malosetti, M.; van Eeuwijk, F.A.; Haseneyer, G.; Kilian, B.; Graner, A. Genome-wide association studies for agronomical traits in a world wide spring barley collection. BMC Plant Biol. 2012, 12, 16. [Google Scholar] [CrossRef]
- Li, M.; Geng, L.; Xie, S.; Wu, D.; Ye, L.; Zhang, G. Genome-wide association study on total starch, amylose and amylopectin in barley grain reveals novel putative alleles. Int. J. Mol. Sci. 2021, 22, 553. [Google Scholar] [CrossRef]
- Hassan, A.S.; Houston, K.; Lahnstein, J.; Shirley, N.; Schwerdt, J.G.; Gidley, M.J.; Waugh, R.; Little, A.; Burton, R.A. A genome wide association study of arabinoxylan content in 2-row spring barley grain. PLoS ONE 2017, 12, e0182537. [Google Scholar] [CrossRef] [PubMed]
- Youssef, H.M.; Eggert, K.; Koppolu, R.; Alqudah, A.M.; Poursarebani, N.; Fazeli, A.; Sakuma, S.; Tagiri, A.; Rutten, T.; Govind, G.; et al. VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat. Genet. 2017, 49, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Alqudah, A.M.; Sharma, R.; Pasam, R.K.; Graner, A.; Kilian, B.; Schnurbusch, T. Genetic dissection of photoperiod response based on GWAS of pre-anthesis phase duration in spring barley. PLoS ONE 2014, 9, e113120. [Google Scholar] [CrossRef]
- Romagosa, I.; Fox, P.N. Genotype × environment interaction and adaptation. In Plant Breeding: Principles and Prospects; Plant Breeding. Plant Breeding Series; Hayward, M.D., Bosemark, N.O., Romagosa, I., Eds.; Springer: Dordrecht, The Netherlands, 1993; pp. 373–390. [Google Scholar]
- Molina-Cano, J.L.; Francesch, M.; Perez-Vendrell, A.M.; Ramo, T.; Voltas, J.; Brufau, J.J. Genetic and environmental variation in malting and feed quality of barley. J. Cereal Sci. 1997, 25, 37–47. [Google Scholar] [CrossRef]
- Kaczmarek, Z.; Adamski, T.; Surma, M.; Jezowsk, S.; Leśniewska-Frątczak, M. Genotype-environment interaction of barley doubled haploids with regard to malting quality. Plant Breed. 1999, 118, 243–247. [Google Scholar] [CrossRef]
- Halstead, M.; Morrissy, C.; Fisk, S.; Fox, G.; Hayes, P.; Carrijo, D. Barley grain protein is influenced by genotype, environment, and nitrogen management and is the major driver of malting quality. Crop Sci. 2023, 63, 115–127. [Google Scholar] [CrossRef]
- Waugh, R.; Jannink, J.L.; Muehlbauer, G.J.; Ramsay, L. The emergence of whole genome association scans in barley. Curr. Opin. Plant Biol. 2009, 12, 218–222. [Google Scholar] [CrossRef]
- Muñoz-Amatriaín, M.; Cuesta-Marcos, A.; Hayes, P.M.; Muehlbauer, G.J. Barley genetic variation: Implications for crop improvement. Brief Funct. Genom. 2014, 13, 341–350. [Google Scholar] [CrossRef]
- Massman, J.; Cooper, B.; Horsley, R.; Neate, S.; Dill-Macky, R.; Chao, S.; Dong, Y.; Schwarz, P.; Muehlbauer, G.J.; Smith, K.P. Genome-wide association mapping of Fusarium head blight resistance in contemporary barley breeding germplasm. Mol. Breed. 2011, 27, 439–454. [Google Scholar] [CrossRef]
- Wang, H.; Smith, K.P.; Combs, E.; Blake, T.; Horsley, R.D.; Muehlbauer, G.J. Effect of population size and unbalanced data sets on QTL detection using genome-wide association mapping in barley breeding germplasm. Theor. Appl. Genet. 2012, 124, 111–124. [Google Scholar] [CrossRef]
- Adhikari, A.; Steffenson, B.J.; Smith, K.P.; Smith, M.; Dill-Macky, R. Identification of quantitative trait loci for net form net blotch resistance in contemporary barley breeding germplasm from the USA using genome-wide association mapping. Theor. Appl. Genet. 2020, 133, 1019–1037. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.C.; Flores-Vergara, M.A.; Krasynanski, S.; Kumar, S.; Thompson, W.F. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef] [PubMed]
- The Triticeae Toolbox (T3) Database. Available online: https://triticeaetoolbox.org/barley/ (accessed on 2 February 2023).
- The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 9 December 2022).
- Posit|The Open-Source Data Science Company. Available online: https://posit.co/ (accessed on 9 December 2022).
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 2, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Wirth, T.; Linz, B.; Pritchard, J.K.; Stephens, M.; Kidd, M.; Blaser, M.J.; Graham, D.Y.; Vacher, S.; Perez-Perez, G.I.; et al. Traces of human migrations in Helicobacter pylori populations. Science 2003, 299, 1582–1585. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Segura, V.; Vilhjálmsson, B.J.; Platt, A.; Korte, A.; Seren, Ü.; Long, Q.; Nordborg, M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 2012, 44, 825–830. [Google Scholar] [CrossRef]
- Bayer, M.M.; Rapazote-Flores, P.; Ganal, M.; Hedley, P.E.; Macaulay, M.; Plieske, J.; Ramsay, L.; Russell, J.; Shaw, P.D.; Thomas, W.; et al. Development and evaluation of a barley 50k iSelect SNP array. Front. Plant Sci. 2017, 8, 1792. [Google Scholar] [CrossRef]
- Szűcs, P.; Blake, V.C.; Bhat, P.R.; Chao, S.; Close, T.J.; Cuesta-Marcos, A.; Muehlbauer, G.J.; Ramsay, L.; Waugh, R.; Hayes, P.M. An integrated resource for barley linkage map and malting quality QTL alignment. Plant Genome 2009, 2. [Google Scholar] [CrossRef]
- Tsai, H.-Y.; Janss, L.L.; Andersen, J.R.; Orabi, J.; Jensen, J.D.; Jahoor, A.; Jensen, J. Genomic prediction and GWAS of yield, quality and disease-related traits in spring barley and winter wheat. Sci. Rep. 2020, 10, 3347. [Google Scholar] [CrossRef] [PubMed]
- Alqudah, A.M.; Koppolu, R.; Wolde, G.M.; Graner, A.; Schnurbusch, T. The genetic architecture of barley plant stature. Front. Genet. 2016, 7, 117. [Google Scholar] [CrossRef] [PubMed]
- Genievskaya, Y.; Almerekova, S.; Abugalieva, S.; Chudinov, V.; Blake, T.; Abugalieva, A.; Turuspekov, Y. Identification of SNP Markers Associated with Grain Quality Traits in a Barley Collection (Hordeum vulgare L.) Harvested in Kazakhstan. Agronomy 2022, 12, 2431. [Google Scholar] [CrossRef]
- Close, T.J.; Wanamaker, S.I.; Caldo, R.A.; Turner, S.M.; Ashlock, D.A.; Dickerson, J.A.; Wing, R.A.; Muehlbauer, G.J.; Kleinhofs, A.; Wise, R.P. A new resource for cereal genomics: 22K barley genechip comes of age. Plant Physiol. 2004, 134, 960–968. [Google Scholar] [CrossRef]
- Tibbs Cortes, L.; Zhang, Z.; Yu, J. Status and prospects of genome-wide association studies in plants. Plant Genome 2021, 14, e20077. [Google Scholar] [CrossRef]
- Malysheva-Otto, L.V.; Ganal, M.W.; Röder, M.S. Analysis of molecular diversity, population structure and linkage disequilibrium in a worldwide survey of cultivated barley germplasm (Hordeum vulgare L.). BMC Genet. 2006, 7, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Y.; Marchand, S.; Tinker, N.A.; Belzile, F. Population structure and linkage disequilibrium in barley assessed by DArT markers. Theor. Appl. Genet. 2009, 119, 43–52. [Google Scholar] [CrossRef]
- Hamblin, M.T.; Close, T.J.; Bhat, P.R.; Chao, S.; Kling, J.G.; Abraham, K.J.; Blake, T.; Brooks, W.S.; Cooper, B.; Griffey, C.A.; et al. Population structure and linkage disequilibrium in US barley germplasm: Implications for association mapping. Crop Sci. 2010, 50, 556–566. [Google Scholar] [CrossRef]
- Gryaznov, A.A. Karabalyk Barley (Forage, Groats, Beer); Kustanay Printing House: Kustanay, Kazakhstan, 1996. [Google Scholar]
- Almerekova, S.; Genievskaya, Y.; Abugalieva, S.; Sato, K.; Turuspekov, Y. Population structure and genetic diversity of two-rowed barley accessions from Kazakhstan based on SNP genotyping data. Plants 2021, 10, 2025. [Google Scholar] [CrossRef]
- Rafalski, J.A. Association genetics in crop improvement. Curr. Opin. Plant Biol. 2010, 13, 174–180. [Google Scholar] [CrossRef]
- Samarah, N.H. Effects of drought stress on growth and yield of barley. Agron. Sustain. Dev. 2005, 25, 145–149. [Google Scholar] [CrossRef]
- Haddadin, M.A.F. Assessment of drought tolerant barley varieties under water stress. Int. J. Agric. For. 2015, 5, 131–137. [Google Scholar]
- Gous, P.W.; Gilbert, R.G.; Fox, G.P. Drought-proofing barley (Hordeum vulgare) and its impact on grain quality: A review. J. Inst. Brew. 2015, 121, 19–27. [Google Scholar] [CrossRef]
- Macnicol, P.K.; Jacobsen, J.V.; Keys, M.M.; Stuart, I.M. Effects of heat and water stress on malt quality and grain parameters of Schooner barley grown in cabinets. J. Cereal Sci. 1993, 18, 61–68. [Google Scholar] [CrossRef]
- Cossani, C.M.; Slafer, G.A.; Savin, R. Yield and biomass in wheat and barley under a range of conditions in a Mediterranean site. Field Crops Res. 2009, 112, 205–213. [Google Scholar] [CrossRef]
- Alghabari, F.; Ihsan, M.Z. Effects of drought stress on growth, grain filling duration, yield and quality attributes of barley (Hordeum vulgare L.). Bangladesh J. Bot. 2018, 47, 421–428. [Google Scholar] [CrossRef]
- Savin, R.; Nicolas, M.E. Effects of short periods of drought and high temperature on grain growth and starch accumulation of two malting barley cultivars. Funct. Plant Biol. 1996, 23, 201–210. [Google Scholar] [CrossRef]
- Burton, R.A.; Jobling, S.A.; Harvey, A.J.; Shirley, N.J.; Mather, D.E.; Bacic, A.; Fincher, G.B. The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiol. 2008, 146, 1821–1833. [Google Scholar] [CrossRef]
- Sharma, D.; Tiwari, A.; Sood, S.; Jamra, G.; Singh, N.K.; Meher, P.K.; Kumar, A. Genome wide association mapping of agro-morphological traits among a diverse collection of finger millet (Eleusine coracana L.) genotypes using SNP markers. PLoS ONE 2018, 13, e0199444. [Google Scholar] [CrossRef]
- Yang, Y.; Al-Baidhani, H.H.; Harris, J.; Riboni, M.; Li, Y.; Mazonka, I.; Bazanova, N.; Chirkova, L.; Sarfraz Hussain, S.; Hrmova, M.; et al. DREB/CBF expression in wheat and barley using the stress-inducible promoters of HD-Zip I genes: Impact on plant development, stress tolerance and yield. Plant Biotechnol. J. 2020, 18, 829–844. [Google Scholar] [CrossRef]
- Tommasini, L.; Svensson, J.T.; Rodriguez, E.M.; Wahid, A.; Malatrasi, M.; Kato, K.; Wanamaker, S.; Resnik, J.; Close, T.J. Dehydrin gene expression provides an indicator of low temperature and drought stress: Transcriptome-based analysis of barley (Hordeum vulgare L.). Funct. Integr. Genom. 2008, 8, 387–405. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, R.; Kawahigashi, H.; Ando, T.; Tonooka, T.; Handa, H. Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 2009, 149, 1341–1353. [Google Scholar] [CrossRef]
- Li, Q.; Pan, Z.; Liu, J.; Deng, G.; Long, H.; Zhang, H.; Liang, J.; Zeng, X.; Tang, Y.; Tashi, N.; et al. A mutation in Waxy gene affects amylose content, starch granules and kernel characteristics of barley (Hordeum vulgare). Plant Breed. 2019, 138, 513–523. [Google Scholar] [CrossRef]
- Shen, C.; Yuan, J.; Ou, X.; Ren, X.; Li, X. Genome-wide identification of alcohol dehydrogenase (ADH) gene family under waterlogging stress in wheat (Triticum aestivum). PeerJ 2021, 9, e11861. [Google Scholar] [CrossRef]
- Rabello, A.R.; Guimarães, C.M.; Rangel, P.H.; da Silva, F.R.; Seixas, D.; de Souza, E.; Brasileiro, A.C.; Spehar, C.R.; Ferreira, M.E.; Mehta, Â. Identification of drought-responsive genes in roots of upland rice (Oryza sativa L). BMC Genom. 2008, 9, 485. [Google Scholar] [CrossRef] [PubMed]
- Komatsuda, T.; Pourkheirandish, M.; He, C.; Azhaguvel, P.; Kanamori, H.; Perovic, D.; Stein, N.; Graner, A.; Wicker, T.; Tagiri, A.; et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class. Proc. Natl. Acad. Sci. USA 2007, 104, 1424–1429. [Google Scholar] [CrossRef]
- Turuspekov, Y.; Martin, J.M.; Bowman, J.G.P.; Beecher, B.S.; Giroux, M.J. Associations Between Vrs1 Alleles and Grain Quality Traits in Spring Barley Hordeum vulgare L. Cereal Chem. 2008, 85, 817–823. [Google Scholar] [CrossRef]

| Trait | Year | Range | Median | Mean | SD |
|---|---|---|---|---|---|
| GSC (%) | 2020 | 50.63–62.80 | 61.48 | 61.14 | 1.33 |
| 2021 | 34.86–49.80 | 44.02 | 43.94 | 1.89 | |
| GPC (%) | 2020 | 11.65–16.85 | 13.93 | 13.90 | 0.60 |
| 2021 | 15.15–22.75 | 18.40 | 18.40 | 1.03 | |
| GCC (%) | 2020 | 3.85–6.65 | 5.55 | 5.54 | 0.37 |
| 2021 | 3.50–10.46 | 6.00 | 6.17 | 1.04 | |
| GLC (%) | 2020 | 0.75–3.50 | 2.65 | 2.60 | 0.38 |
| 2021 | 0.60–1.95 | 1.40 | 1.37 | 0.18 | |
| TWL (g/L) | 2020 | 492.5–688.0 | 582.0 | 581.4 | 31.0 |
| 2021 | 419.5–692.0 | 616.0 | 609.7 | 33.6 |
| GSC | |||||
|---|---|---|---|---|---|
| df | SS | MS | p-Value | h2 | |
| G | 406 | 5753 | 14 | <2 × 10−16 | 0.05 |
| E | 1 | 110,551 | 110,551 | <2 × 10−16 | |
| G × E | 387 | 1230 | 3 | 6.26 × 10−14 | |
| Res. | 750 | 1258 | 2 | ||
| GPC | |||||
| df | SS | MS | p-Value | h2 | |
| G | 406 | 1541 | 4 | <2 × 10−16 | 0.15 |
| E | 1 | 7656 | 7656 | <2 × 10−16 | |
| G × E | 387 | 591 | 2 | <2 × 10−16 | |
| Res. | 750 | 377 | 1 | ||
| GCC | |||||
| df | SS | MS | p-Value | h2 | |
| G | 406 | 1298.4 | 3.2 | <2 × 10−16 | 0.52 |
| E | 1 | 141.8 | 141.78 | <2 × 10−16 | |
| G × E | 387 | 444.2 | 1.15 | 5.48 × 10−5 | |
| Res. | 750 | 614.1 | 0.82 | ||
| GLC | |||||
| df | SS | MS | p-Value | h2 | |
| G | 406 | 88.6 | 0.2 | 1.65 × 10−12 | 0.11 |
| E | 1 | 575.2 | 575.2 | <2 × 10−16 | |
| G × E | 387 | 60.4 | 0.2 | 0.00153 | |
| Res. | 750 | 90.4 | 0.1 | ||
| TWL | |||||
| df | SS | MS | p-Value | h2 | |
| G | 406 | 886,157 | 2183 | <2 × 10−16 | 0.34 |
| E | 1 | 324,351 | 324,351 | <2 × 10−16 | |
| G × E | 387 | 739,572 | 1911 | <2 × 10−16 | |
| Res. | 750 | 634,840 | 846 | ||
| Trait | SNP | Chr. | Physical Pos. of SNP (bp) * | QTL Interval (bp) | 2020 | 2021 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p-Value | p-Value (FDR) | PVE (%) | Allele | Effect | p-Value | p-Value (FDR) | PVE (%) | Allele | Effect | |||||
| GSC | 11_21406 | 2H | 718,210,885 | 8.03 × 10−5 | 2.65 × 10−2 | 0.62 | G | 0.41 | ||||||
| GSC | 11_20639 | 3H | 158,707,482 | 158,707,482–226,364,211 | 1.57 × 10−10 | 6.66 × 10−7 | 9.70 | A | 1.33 | 8.88 × 10−6 | 3.66 × 10−3 | 0.00 | G | 0.93 |
| GSC | 12_31484 | 3H | 498,949,534 | 1.43 × 10−6 | 2.96 × 10−3 | 2.90 | A | 0.65 | ||||||
| GSC | 11_20680 | 4H | 19,087,562 | 19,087,562–20,173,462 | 3.10 × 10−7 | 8.44 × 10−4 | 4.30 | A | 1.12 | |||||
| GSC | 11_11473 | 5H | 547,115,792 | 3.12 × 10−6 | 1.72 × 10−3 | 0.26 | C | 0.49 | ||||||
| GSC | 11_20104 | 5H | 624,403,396 | 624,403,396–624,444,586 | 1.79 × 10−4 | 4.91 × 10−2 | 3.90 | G | 0.52 | |||||
| GSC | 12_31042 | 6H | 553,019,586 | 495,778,737–553,203,851 | 5.60 × 10−4 | 2.87 × 10−1 | 0.39 | G | 0.51 | 5.26 × 10−10 | 8.67 × 10−7 | 0.16 | G | 1.08 |
| GSC | 12_30997 | 7H | 130,414,038 | 1.35 × 10−5 | 1.11 × 10−2 | 1.44 | A | 0.57 | ||||||
| GPC | 11_21053 | 1H | 403,309,609 | 403,309,609–481,938,292 | 1.77 × 10−6 | 3.06 × 10−3 | 0.24 | G | 0.46 | 2.74 × 10−4 | 2.25 × 10−1 | 0.18 | A | 0.59 |
| GPC | 12_20632 | 1H | 511,401,867 | 2.16 × 10−5 | 1.95 × 10−2 | 0.11 | A | 0.38 | ||||||
| GPC | 11_20269 | 4H | 72,688,992 | 9.77 × 10−6 | 1.61 × 10−2 | 0.00 | A | 0.28 | ||||||
| GPC | 11_21303 | 4H | 464,028,169 | 459,813,388–464,028,169 | 2.75 × 10−5 | 2.42 × 10−2 | 0.00 | G | 0.28 | |||||
| GPC | 12_31509 | 6H | 203,509,034 | 5.51 × 10−6 | 9.07 × 10−3 | 1.44 | G | 0.51 | ||||||
| GCC | 12_30678 | 2H | UNK | 3.10 × 10−8 | 5.11 × 10−5 | 0.00 | C | 0.22 | ||||||
| GCC | 12_11245 | 5H | 579,324,077 | 6.77 × 10−7 | 1.15 × 10−3 | 0.12 | C | 0.33 | ||||||
| GLC | 11_21057 | 1H | 478,389,125 | 478,389,125–509,511,424 | 1.90 × 10−6 | 3.14 × 10−3 | 0.54 | G | 0.14 | |||||
| GLC | 11_20265 | 5H | 456,062,406 | 2.93 × 10−5 | 4.83 × 10−2 | 1.19 | A | 0.07 | ||||||
| GLC | 11_21528 | 7H | 49,445,658 | 5.50 × 10−5 | 4.53 × 10−2 | 0.80 | T | 0.11 | ||||||
| TWL | 12_30901 | 2H | 652,031,870 | 652,031,870–705,587,677 | 3.84 × 10−5 | 2.11 × 10−2 | 0.18 | G | 9.39 | |||||
| TWL | 12_20274 | 4H | 3,623,098 | 5.40 × 10−10 | 8.90 × 10−7 | 14.79 | G | 56.13 | ||||||
| TWL | 11_20472 | 4H | 494,212,244 | 4.22 × 10−11 | 1.68 × 10−7 | 0.00 | A | 26.79 | ||||||
| TWL | 11_11281 | 5H | 228,224,360 | 9.07 × 10−6 | 1.57 × 10−2 | 0.05 | G | 20.26 | ||||||
| TWL | 12_31034 | 5H | 447,605,783 | 397,043,179–447,605,783 | 3.73 × 10−5 | 2.11 × 10−2 | 1.18 | C | 9.45 | 8.35 × 10−6 | 6.88 × 10−3 | 0.04 | G | 40.35 |
| TWL | 12_21482 | 6H | 351,737,595 | 3.00 × 10−9 | 5.65 × 10−6 | 0.22 | G | 22.87 | ||||||
| TWL | 12_11035 | 7H | 9,613,368 | 2.42 × 10−15 | 3.99 × 10−12 | 0.25 | G | 24.09 | ||||||
| TWL | 11_20060 | 7H | 109,656,682 | 3.97 × 10−6 | 3.70 × 10−3 | 0.04 | A | 9.64 | ||||||
| Trait | Marker | Chr. | Physical Pos. (bp) * | Genetic Pos. (cM) ** | Key Candidate Genes | Candidate QTLs |
| GSC | 11_21406 | 2H | 718,210,885 | 143.1 | ||
| GSC | 11_20639 | 3H | 158,707,482–226,364,211 | 58.3–58.4 | QTL10_SC (51.73–55.77 cM) [34]; qTS-3.1 (176,458,677 bp) [68] | |
| GSC | 12_31484 | 3H | 498,949,534 | - | ||
| GSC | 11_20680 | 4H | 19,087,562–20,173,462 | 31.1–32.4 | ||
| GSC | 11_11473 | 5H | 547,115,792 | 76.3 | CBF4 (559,673,235 bp) dehydration-responsive element-binding protein [66]; CBF5 (560,732,721 bp) dehydration-responsive element-binding protein [66] | qTS-5.1 (536,435,763 bp) [68] |
| GSC | 11_20104 | 5H | 624,403,396–624,444,586 | 144.8–144.9 | Dhn9 (616,115,199 bp) dehydrin [66] | |
| GSC | 12_31042 | 6H | 495,778,737–553,203,851 | 73.8–102.0 | Dhn5 (12_31042, 553,019,586 bp) dehydrin [57]; Amy1 (533,879,986 bp) alpha-amylase [66] | QTL18_SC (71.08 cM) [34] |
| GSC | 12_30997 | 7H | 130,414,038 | 74.8 | CO1 (127,679,215 bp) CONSTANS-like protein [66] | QTL22_SC (78.22 cM) [34] |
| GPC | 11_21053 | 1H | 403,309,609–481,938,292 | 51.9–72.9 | Aglu3 (12_30820, 419,012,101 bp) α-glucosidase [67]; CO9 (60.0 cM) CONSTANS-like protein [69] | QTl1_CPC (55.49 cM) [34] |
| GPC | 12_20632 | 1H | 511,401,867 | - | Adh2 (528,989,695 bp) alcohol dehydrogenase 2 [66]; Ppd-H2 (92.3 cM) pseudo-response regulator PPD-H2 [69] | QTL_Q7 (516,153,706–547,250,913 bp) [70] |
| GPC | 11_20269 | 4H | 72,688,992 | 53.9 | ||
| GPC | 11_21303 | 4H | 459,813,388–464,028,169 | 53.9–54.6 | ||
| GPC | 12_31509 | 6H | 203,509,034 | 58.9 | ndhF (187,155,342 bp) nicotinate dehydrogenase FAD-subunit [66] | QTL_Q24 (12_31509, 203,509,034 bp) [70]; QGpc6H.45 (54.7 cM) [71]; Qcp6a (57.91 cM) [25] |
| GCC | 12_30678 | 2H | UNK | 145.4 | QAX2.S-2H4 (136.0 cM) [44] | |
| GCC | 12_11245 | 5H | 579,324,077 | 109.4 | CBF4 (559,673,235 bp) dehydration-responsive element-binding protein [66]; CBF5 (560,732,721 bp) dehydration-responsive element-binding protein [66] | |
| GLC | 11_21057 | 1H | 478,389,125–509,511,424 | 71.8–90.9 | Ppd-H2 (92.3 cM) pseudo- response regulator PPD-H2 [69] | |
| GLC | 11_20265 | 5H | 456,062,406 | 44.9 | ||
| GLC | 11_21528 | 7H | 49,445,658 | 49.9 | FT1 (39,681,222 bp) flowering locus T [66]; gbp3 (11_21528, 49,445,658 bp) GAMYB-binding protein [57] | |
| TWL | 12_30901 | 2H | 652,031,870–705,587,677 | 90.9–126.6 | Vrs1 (12_30901, 652,031,870 bp) homeodomain leucine zipper protein [57] | QTL_Q10 (641,328,117–652,031,870 bp) [70]; QTw2H.86 (90.99 cM) [71] |
| TWL | 12_20274 | 4H | 3,623,098 | 8.3 | ||
| TWL | 11_20472 | 4H | 494,212,244 | 54.9 | DTDP (12_30839, 494,332,468 bp) d-TDP-glucose dehydratase [67] | QTL_Q14 (11_21303, 464,028,169 bp) [70] |
| TWL | 11_11281 | 5H | 228,224,360 | 45.5 | ||
| TWL | 12_31034 | 5H | 397,043,179–447,605,783 | 44.9–45.0 | Adh3 (12_31034, 447,605,783 bp) alcohol dehydrogenase 3 [57] | |
| TWL | 12_21482 | 6H | 351,737,595 | 58.9 | ||
| TWL | 12_11035 | 7H | 9,613,368 | 6.3 | WAXY (17,091,220 bp) Granule-bound starch synthase 1 [66] | |
| TWL | 11_20060 | 7H | 109,656,682 | 72.8 | B12Dg1 (11_20060, 109,656,682 bp) B12Dg1 protein [57] | QTw7H.70 (71.76 cM) [71] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Genievskaya, Y.; Almerekova, S.; Abugalieva, S.; Abugalieva, A.; Sato, K.; Turuspekov, Y. Identification of SNPs Associated with Grain Quality Traits in Spring Barley Collection Grown in Southeastern Kazakhstan. Agronomy 2023, 13, 1560. https://doi.org/10.3390/agronomy13061560
Genievskaya Y, Almerekova S, Abugalieva S, Abugalieva A, Sato K, Turuspekov Y. Identification of SNPs Associated with Grain Quality Traits in Spring Barley Collection Grown in Southeastern Kazakhstan. Agronomy. 2023; 13(6):1560. https://doi.org/10.3390/agronomy13061560
Chicago/Turabian StyleGenievskaya, Yuliya, Shyryn Almerekova, Saule Abugalieva, Aigul Abugalieva, Kazuhiro Sato, and Yerlan Turuspekov. 2023. "Identification of SNPs Associated with Grain Quality Traits in Spring Barley Collection Grown in Southeastern Kazakhstan" Agronomy 13, no. 6: 1560. https://doi.org/10.3390/agronomy13061560
APA StyleGenievskaya, Y., Almerekova, S., Abugalieva, S., Abugalieva, A., Sato, K., & Turuspekov, Y. (2023). Identification of SNPs Associated with Grain Quality Traits in Spring Barley Collection Grown in Southeastern Kazakhstan. Agronomy, 13(6), 1560. https://doi.org/10.3390/agronomy13061560




