Genotype Selection for Grain Yield of Sorghum through Generalized Linear Mixed Model
Abstract
1. Introduction
2. Methods and Data
2.1. Data and Design Description
2.2. Generalized Linear Mixed Model
3. Results of the Study
3.1. Descriptive Summary
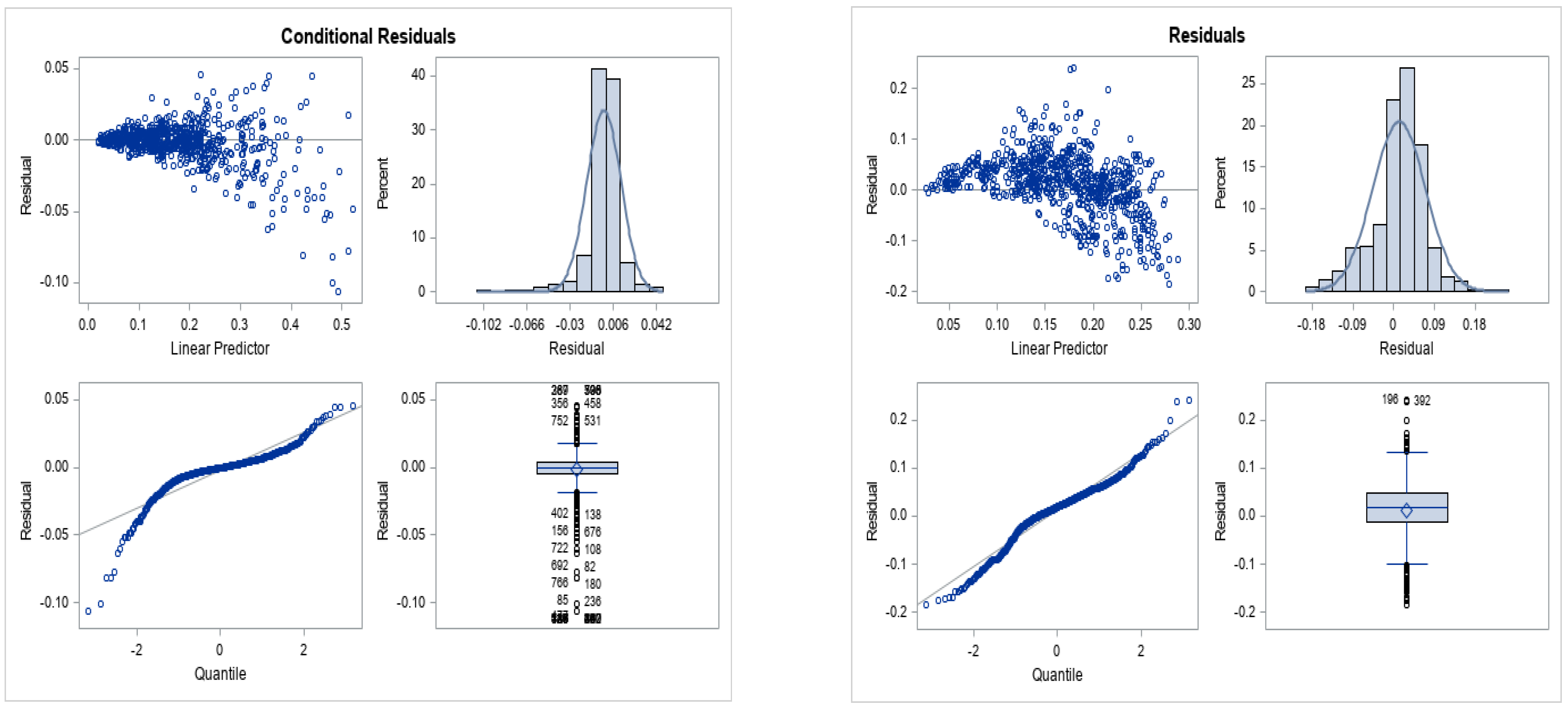
3.2. Results of GLMM
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Amare, K.; Zeleke, H.; Bultosa, G. Variability for yield, yield related traits and association among traits of sorghum (Sorghum bicolor (L.) Moench) varieties in Wollo, Ethiopia. J. Plant Breed. Crop Sci. 2015, 7, 125–133. [Google Scholar]
- Enyew, M.; Feyissa, T.; Carlsson, A.S.; Tesfaye, K.; Hammenhag, C.; Geleta, M. Genetic Diversity and Population Structure of Sorghum [Sorghum bicolor (L.) Moench] Accessions as Revealed by Single Nucleotide Polymorphism Markers. Front. Plant Sci. 2021, 12, e12830. [Google Scholar] [CrossRef] [PubMed]
- Tesfaye, K. Genetic diversity study of sorghum (Sorghum bicolor (L.) Moenc) genotypes, Ethiopia. Acta Univ. Sapientiae Agric. Environ. 2017, 9, 44–54. [Google Scholar] [CrossRef]
- Torres, V.; Medina, Y.; Rodríguez, Y.; Sardiñas, Y.; Herrera, M.; Rodríguez, R. Application of the linear mixed and generalized mixed model as alternatives for analysis in experiments with repeated measures. Cuba. J. Agric. Sci. 2019, 53, 7. [Google Scholar]
- Zhao, Y.; Staudenmayer, J.; Coull, B.A.; Wand, M.P. General design Bayesian generalized linear mixed models. Stat. Sci. 2006, 21, 35–51. [Google Scholar] [CrossRef]
- Yang, R.-C. Towards understanding and use of mixed-model analysis of agricultural experiments. Can. J. Plant Sci. 2010, 90, 605–627. [Google Scholar] [CrossRef]
- Nelder, J.A.; Wedderburn, R.W. Generalized linear models. J. R. Stat. Soc. Ser. A (Gen.) 1972, 135, 370–384. [Google Scholar] [CrossRef]
- Dean, C.; Nielsen, J.D. Generalized linear mixed models: A review and some extensions. Lifetime Data Anal. 2007, 13, 497–512. [Google Scholar] [CrossRef]
- Michel, L.; Brun, F.; Makowski, D. A framework based on generalised linear mixed models for analysing pest and disease surveys. Crop Prot. 2017, 94, 1–12. [Google Scholar] [CrossRef]
- Zhang, Z.; Guillaume, F.; Sartelet, A.; Charlier, C.; Georges, M.; Farnir, F.; Druet, T. Ancestral haplotype-based association mapping with generalized linear mixed models accounting for stratification. Bioinformatics 2012, 28, 2467–2473. [Google Scholar] [CrossRef]
- Jain, S.; Patel, P. Principal component and cluster analysis in sorghum (Sorghum bicolor (L.) Moench). Forage Res. J. 2016, 42, 90–95. [Google Scholar]
- Derese, S.A.; Shimelis, H.; Mwadzingeni, L.; Laing, M. Agro-morphological characterisation and selection of sorghum landraces. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2018, 68, 585–595. [Google Scholar] [CrossRef]
- Bose, R.C. A note on the resolvability of balanced incomplete block designs. Sankhyā Indian J. Stat. 1942, 6, 105–110. [Google Scholar]
- Tuerlinckx, F.; Rijmen, F.; Verbeke, G.; De Boeck, P. Statistical inference in generalized linear mixed models: A review. Br. J. Math. Stat. Psychol. 2006, 59, 225–255. [Google Scholar] [CrossRef] [PubMed]
- Kachman, S.D. An introduction to generalized linear mixed models. In Proceedings of the Symposium at the Organizational Meeting for a NCR Coordinating Committee on “Implementation Strategies for National Beef Cattle Evaluation”, Athens, Greece; pp. 59–73. Available online: https://citeseerx.ist.psu.edu/document?repid=rep1&type=pdf&doi=4f1c417fb9bac04309f0ba9b0ec44d5ef51e36aa (accessed on 1 December 2022).
- Moscatelli, A.; Mezzetti, M.; Lacquaniti, F. Modeling psychophysical data at the population-level: The generalized linear mixed model. J. Vis. 2012, 12, 26. [Google Scholar] [CrossRef]
- Bakbergenuly, I.; Kulinskaya, E. Meta-analysis of binary outcomes via generalized linear mixed models: A simulation study. BMC Med. Res. Methodol. 2018, 18, 70. [Google Scholar] [CrossRef]
- McCulloch, C.E. An Introduction to Generalized Linear Mixed Models. 1997. Available online: https://ecommons.cornell.edu/bitstream/handle/1813/31937/BU-1340-MA.pdf?sequence=1 (accessed on 1 December 2022).
- Capanu, M.; Gönen, M.; Begg, C.B. An assessment of estimation methods for generalized linear mixed models with binary outcomes. Stat. Med. 2013, 32, 4550–4566. [Google Scholar] [CrossRef]
- Breslow, N.E.; Clayton, D.G. Approximate inference in generalized linear mixed models. J. Am. Stat. Assoc. 1993, 88, 9–25. [Google Scholar]
- Vazquez, A.; Bates, D.; Rosa, G.; Gianola, D.; Weigel, K. an R package for fitting generalized linear mixed models in animal breeding. J. Anim. Sci. 2010, 88, 497–504. [Google Scholar] [CrossRef]
- De Resende, M.D.V.; Alves, R.S. Linear, generalized, hierarchical, Bayesian and random regression mixed models in genetics/genomics in plant breeding. Funct. Plant Breed. J. 2020, 2, 121–152. [Google Scholar]
- Groll, A.; Tutz, G. Regularization for generalized additive mixed models by likelihood-based boosting. Methods Inf. Med. 2012, 51, 168–177. [Google Scholar] [PubMed]
- Chen, J.; Zhang, D.; Davidian, M. A Monte Carlo EM algorithm for generalized linear mixed models with flexible random effects distribution. Biostatistics 2002, 3, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Coulibaly, Y.; Mulia, R.; Sanou, J.; Zombré, G.; Bayala, J.; Kalinganire, A.; Van Noordwijk, M. Crop production under different rainfall and management conditions in agroforestry parkland systems in Burkina Faso: Observations and simulation with WaNuLCAS model. Agrofor. Syst. 2014, 88, 13–28. [Google Scholar] [CrossRef]
- Gianinetti, A. Basic features of the analysis of germination data with generalized linear mixed models. Data 2020, 5, 6. [Google Scholar] [CrossRef]
- Baoua, I.; Amadou, L.; Oumarou, N.; Payne, W.; Roberts, J.; Stefanova, K.; Nansen, C. Estimating effect of augmentative biological control on grain yields from individual pearl millet heads. J. Appl. Entomol. 2014, 138, 281–288. [Google Scholar] [CrossRef]
- Gebregziabher, M.; Zhao, Y.; Dismuke, C.; Axon, N.; Hunt, K.; Egede, L. Joint modeling of multiple longitudinal cost outcomes using multivariate generalized linear mixed models. Health Serv. Outcomes Res. Methodol. 2013, 13, 39–57. [Google Scholar] [CrossRef] [PubMed]
- Boykin, D.; Camp, M.J.; Johnson, L.; Kramer, M.; Meek, D.; Palmquist, D.; Vinyard, B.; West, M. Generalized linear mixed model estimation using PROC GLIMMIX: Results from simulations when the data and model match, and when the model is misspecified. In Proceedings of the 22nd Conference on Applied Statistics in Agriculture, Manhattan, KS, USA, 25–28 April 2010; pp. 137–156. [Google Scholar]
- Prasad, P.V.; Boote, K.J.; Allen, L.H., Jr. Adverse high temperature effects on pollen viability, seed-set, seed yield and harvest index of grain-sorghum [Sorghum bicolor (L.) Moench] are more severe at elevated carbon dioxide due to higher tissue temperatures. Agric. For. Meteorol. 2006, 139, 237–251. [Google Scholar] [CrossRef]
- Masasi, B. Evaluating the Impacts of Variable Irrigation Management Strategies on the Performance of Cotton and Grain Sorghum Using Monitoring and Modeling Techniques. Master’s Thesis, Oklahoma State University, Stillwater, OK, USA, 2019. [Google Scholar]
- Assefa, A.; Bezabih, A.; Girmay, G.; Alemayehu, T.; Lakew, A. Evaluation of sorghum (Sorghum bicolor (L.) Moench) variety performance in the lowlands area of wag lasta, north eastern Ethiopia. Cogent Food Agric. 2020, 6, 1778603. [Google Scholar] [CrossRef]
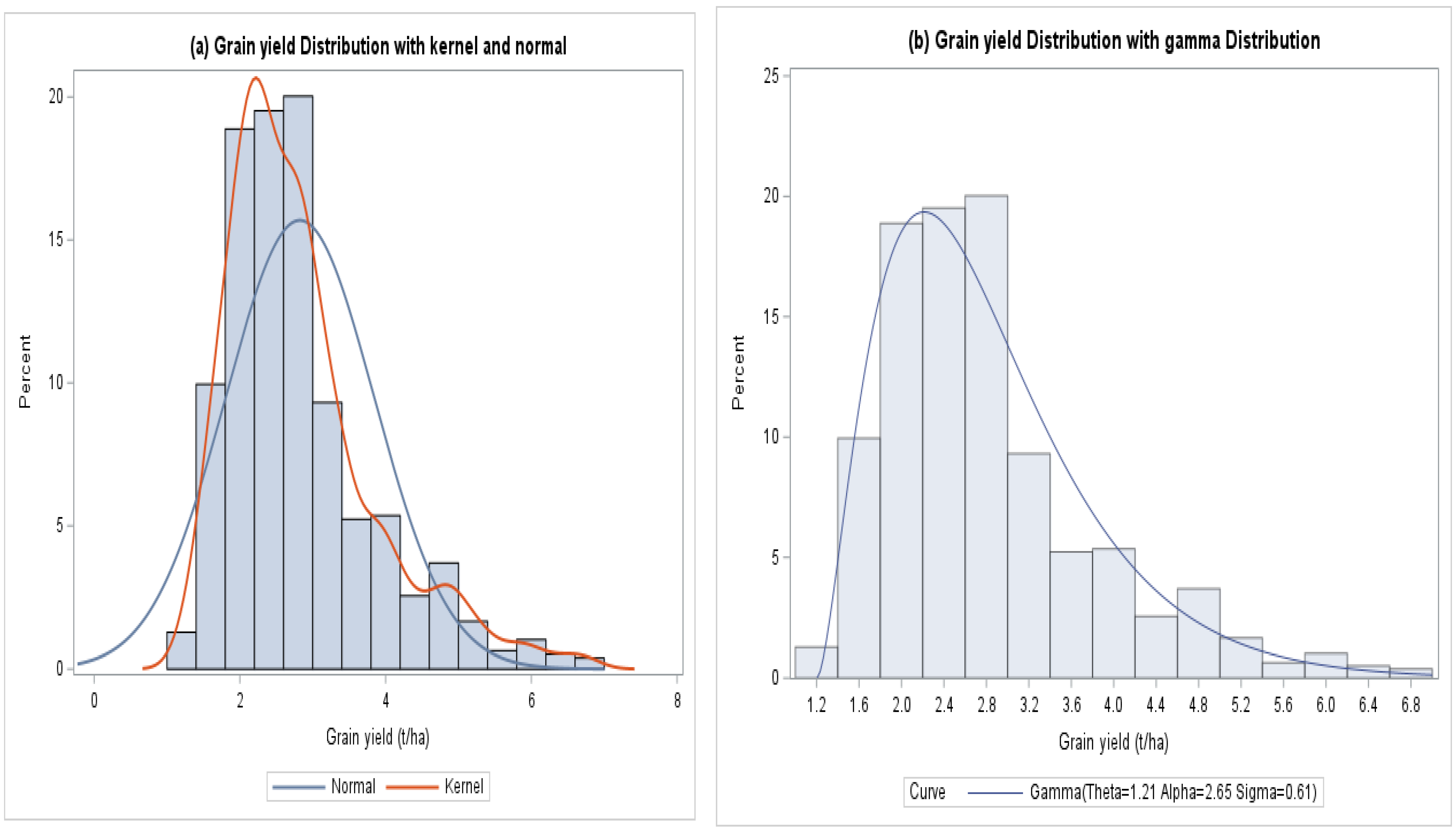
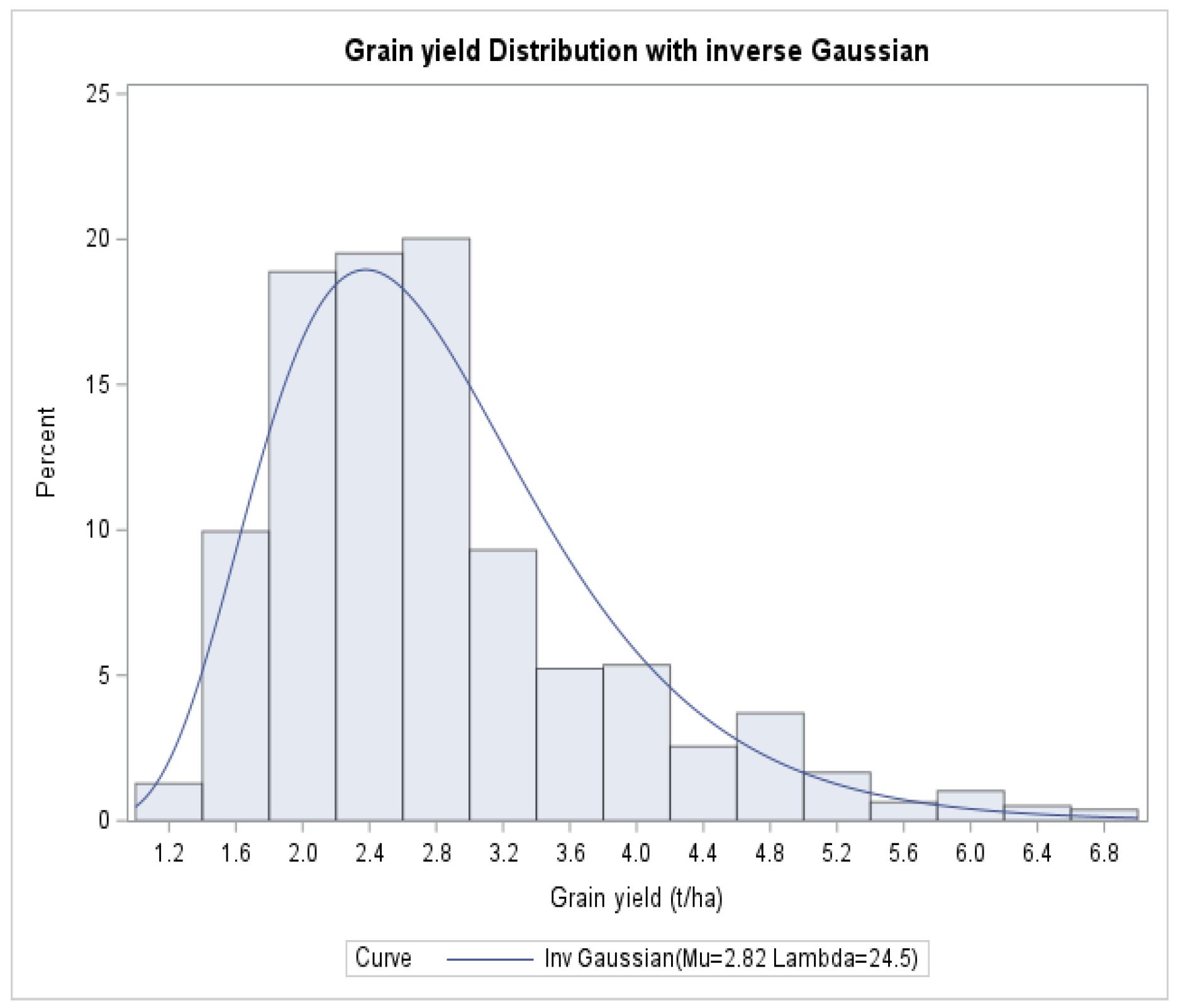

| Distribution | Inverse Gaussian | Gamma | ||||
|---|---|---|---|---|---|---|
| Link Function | Inverse | Inverse Square | Log | Inverse | Inverse Square | Log |
| −2 Res Log PL | −3212.88 | −3285.40 | −1873.19 | −3239.19 | −3324.15 | −1899.14 |
| Pseudo−AIC | −3204.88 | −3277.40 | −1865.19 | −3231.19 | −3316.15 | −1891.14 |
| Pseudo−BIC | −3207.34 | −3279.85 | −1867.64 | −3233.64 | −3318.61 | −1893.60 |
| Pseudo−HQIC | −3210.27 | −3282.79 | −1870.57 | −3236.58 | −3321.54 | −1896.53 |
| Generalized | 0.42 | 0.51 | 0.43 | 1.04 | 1.23 | 1.04 |
| Gen. /DF | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Covariance Parameter | Estimate | Std. Error |
|---|---|---|
| Replication | 0.000034 | 0.000034 |
| Genotype | 0.003071 | 0.000466 |
| Treatment * Genotype | 0.000742 | 0.000101 |
| Residual | 0.001583 | 0.000149 |
| Label | DF | −2 Res Log Like | ChiSq | Pr > ChiSq | Note |
|---|---|---|---|---|---|
| Diagonal G | 0 | −3324.15 | 0.00 | 1.0000 | DF |
| Independence | 3 | −2879.16 | 444.99 | <0.0001 | MI |
| No G-side effects | 3 | −2879.16 | 444.99 | <0.0001 | MI |
| Model Scale | Data Scale | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Treatment | Estimate | St. Err. | DF | t Value | Pr > |t| | LL | UL | Mean | St Err. | LL | UL |
| Irrigated | 0.1447 | 0.00613 | 2 | 23.62 | 0.0018 | 0.1183 | 0.1710 | 2.6293 | 0.05567 | 2.4182 | 2.9075 |
| Stress | 0.1933 | 0.00615 | 2 | 31.44 | 0.0010 | 0.1668 | 0.2197 | 2.2748 | 0.03618 | 2.1335 | 2.4485 |
| Solutions for Fixed Effects | ||||
|---|---|---|---|---|
| Effect | Estimate | Std. Error | t Value | Pr > |t| |
| Intercept | 0.1933 | 0.006147 | 31.44 | 0.0010 |
| Treatment (Irrigation) | −0.0486 | 0.006749 | −7.20 | 0.0187 |
| Treatment (Stress) | 0 | |||
| Prin1 | −0.01723 | 0.001066 | −16.17 | <0.0001 |
| Prin2 | 0.00196 | 0.001148 | 1.71 | 0.0885 |
| Prin3 | −0.00006 | 0.000618 | −0.10 | 0.9213 |
| Overall Tests of the Fixed effects | ||||
| Effect | Num DF | Den DF | F Value | Pr > F |
| Treatment | 1 | 2 | 51.86 | 0.0187 |
| Prin1 | 1 | 387 | 261.50 | <0.0001 |
| Prin2 | 1 | 387 | 2.92 | 0.0885 |
| Prin3 | 1 | 387 | 0.01 | 0.9213 |
| Worst-Performer Genotypes for GLMM | Best-Performer Genotypes for GLMM | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | Estimate | Std.Err.Pred | t Value (Df = 387) | Pr > |t| | Genotype | Estimate | Std.Err.Pred | t Value (Df = 387) | Pr > |t| |
| G196 | −0.08483 | 0.01905 | −4.45 | <0.0001 | G41 | 0.2424 | 0.02547 | 9.52 | <0.0001 |
| G48 | −0.07241 | 0.0191 | −3.79 | 0.0002 | G78 | 0.1772 | 0.02469 | 7.18 | <0.0001 |
| G186 | −0.06368 | 0.01961 | −3.25 | 0.0013 | G66 | 0.1733 | 0.02294 | 7.55 | <0.0001 |
| G40 | −0.06071 | 0.01977 | −3.07 | 0.0023 | G136 | 0.1639 | 0.02383 | 6.88 | <0.0001 |
| G143 | −0.06001 | 0.01906 | −3.15 | 0.0018 | G39 | 0.1588 | 0.0232 | 6.84 | <0.0001 |
| G187 | −0.05132 | 0.01933 | −2.66 | 0.0082 | G108 | 0.1544 | 0.02399 | 6.43 | <0.0001 |
| G25 | −0.05027 | 0.01913 | −2.63 | 0.0089 | G180 | 0.1472 | 0.02287 | 6.44 | <0.0001 |
| G31 | −0.04928 | 0.01972 | −2.5 | 0.0128 | G85 | 0.1244 | 0.02229 | 5.58 | <0.0001 |
| G23 | −0.0479 | 0.01896 | −2.53 | 0.0119 | G56 | 0.122 | 0.02277 | 5.36 | <0.0001 |
| G5 | −0.04771 | 0.01899 | −2.51 | 0.0124 | G137 | 0.1161 | 0.02236 | 5.19 | <0.0001 |
| The Worst-Performer Genotype under Irrigation | The Best-Performer Genotypes under Irrigation | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | Estimate | Std.Err.Pred | t Value (Df = 387) | Pr > |t| | Genotype | Estimate | Std.Err.Pred | t Value (Df = 387) | Pr > |t| |
| G137 | −0.06484 | 0.02134 | −3.04 | 0.0025 | G55 | 0.0658 | 0.02136 | 3.08 | 0.0022 |
| G156 | −0.05602 | 0.02108 | −2.66 | 0.0082 | G41 | 0.0556 | 0.02309 | 2.41 | 0.0166 |
| G157 | −0.05325 | 0.02015 | −2.64 | 0.0085 | G78 | 0.0521 | 0.02262 | 2.3 | 0.0218 |
| G107 | −0.0428 | 0.0196 | −2.18 | 0.0296 | G49 | 0.04216 | 0.02044 | 2.06 | 0.0398 |
| G155 | −0.03719 | 0.02 | −1.86 | 0.0637 | G119 | 0.03617 | 0.02127 | 1.7 | 0.0898 |
| G66 | −0.03569 | 0.02176 | −1.64 | 0.1017 | G36 | 0.03222 | 0.021 | 1.53 | 0.1259 |
| G62 | −0.03505 | 0.01955 | −1.79 | 0.0738 | G2 | 0.03098 | 0.01967 | 1.57 | 0.1161 |
| G167 | −0.03412 | 0.01925 | −1.77 | 0.0771 | G108 | 0.03034 | 0.02226 | 1.36 | 0.1737 |
| G40 | −0.0325 | 0.01915 | −1.7 | 0.0904 | G82 | 0.02641 | 0.02135 | 1.24 | 0.2168 |
| G104 | −0.03238 | 0.01974 | −1.64 | 0.1018 | G185 | 0.02622 | 0.02097 | 1.25 | 0.2119 |
| The Worst-Performer Genotypes under Stress | The Best-Performer Genotypes under Stress | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | Estimate | Std.Err.Pred | t Value (Df = 387) | Pr > |t| | Genotype | Estimate | Std.Err.Pred | t Value (Df = 387) | Pr > |t| |
| G55 | −0.04783 | 0.02113 | −2.26 | 0.0242 | G137 | 0.09289 | 0.02221 | 4.18 | <0.0001 |
| G49 | −0.03849 | 0.0203 | −1.9 | 0.0587 | G66 | 0.07757 | 0.02232 | 3.48 | 0.0006 |
| G8 | −0.03324 | 0.01854 | −1.79 | 0.0737 | G156 | 0.07572 | 0.02171 | 3.49 | 0.0005 |
| G25 | −0.02966 | 0.01841 | −1.61 | 0.108 | G157 | 0.05601 | 0.02067 | 2.71 | 0.007 |
| G196 | −0.02804 | 0.01842 | −1.52 | 0.1287 | G147 | 0.04183 | 0.02137 | 1.96 | 0.051 |
| G26 | −0.02736 | 0.01933 | −1.42 | 0.1577 | G155 | 0.03839 | 0.0204 | 1.88 | 0.0606 |
| G7 | −0.02665 | 0.01851 | −1.44 | 0.1507 | G107 | 0.03838 | 0.02002 | 1.92 | 0.056 |
| G23 | −0.02588 | 0.01843 | −1.4 | 0.1611 | G104 | 0.03144 | 0.02007 | 1.57 | 0.1181 |
| G16 | −0.02532 | 0.01853 | −1.37 | 0.1726 | G136 | 0.03137 | 0.02242 | 1.4 | 0.1625 |
| G3 | −0.02514 | 0.01874 | −1.34 | 0.1806 | G138 | 0.03123 | 0.02124 | 1.47 | 0.1422 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tesfa, M.; Zewotir, T.; Derese, S.A.; Belay, D.B.; Laing, M. Genotype Selection for Grain Yield of Sorghum through Generalized Linear Mixed Model. Agronomy 2023, 13, 852. https://doi.org/10.3390/agronomy13030852
Tesfa M, Zewotir T, Derese SA, Belay DB, Laing M. Genotype Selection for Grain Yield of Sorghum through Generalized Linear Mixed Model. Agronomy. 2023; 13(3):852. https://doi.org/10.3390/agronomy13030852
Chicago/Turabian StyleTesfa, Mulugeta, Temesgen Zewotir, Solomon Assefa Derese, Denekew Bitew Belay, and Mark Laing. 2023. "Genotype Selection for Grain Yield of Sorghum through Generalized Linear Mixed Model" Agronomy 13, no. 3: 852. https://doi.org/10.3390/agronomy13030852
APA StyleTesfa, M., Zewotir, T., Derese, S. A., Belay, D. B., & Laing, M. (2023). Genotype Selection for Grain Yield of Sorghum through Generalized Linear Mixed Model. Agronomy, 13(3), 852. https://doi.org/10.3390/agronomy13030852





