Composition and Diversity of LTR Retrotransposons in the Coffee Leaf Rust Genome (Hemileia vastatrix)
Abstract
1. Introduction
2. Materials and Methods
2.1. Genomic Data Used
2.2. Identification and Annotation of TEs
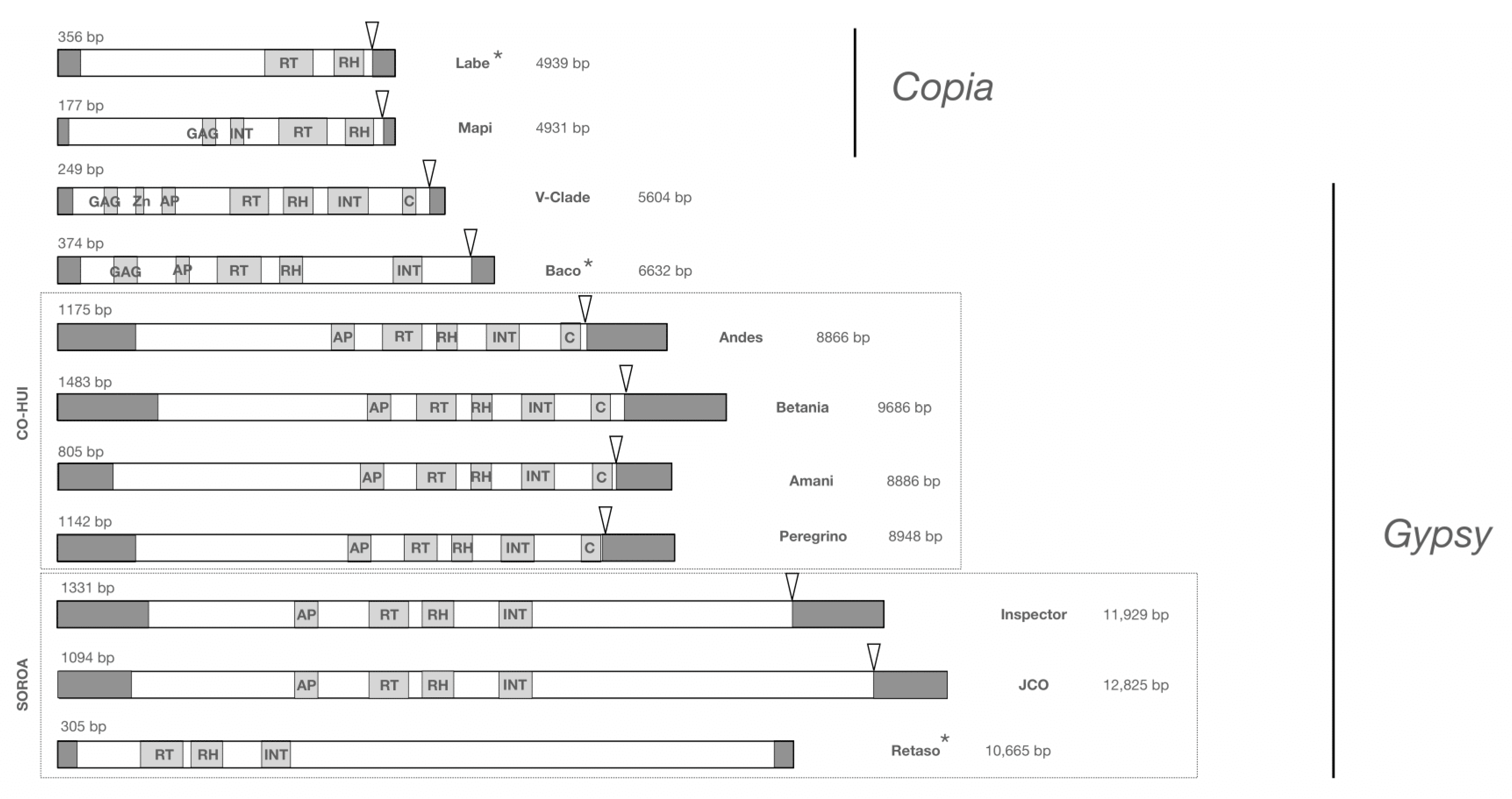
2.3. Structural Characterization of the LTR-RT Lineages
2.4. Expression Analysis and Relationship with Coding Sequences
2.5. LTR-RT Analysis in Other Pucciniales Genomes
2.6. Setting Up a Reference Library of Domains and Complete LTR-RT Sequences of the Pucciniales Order (PucciDB)
3. Results
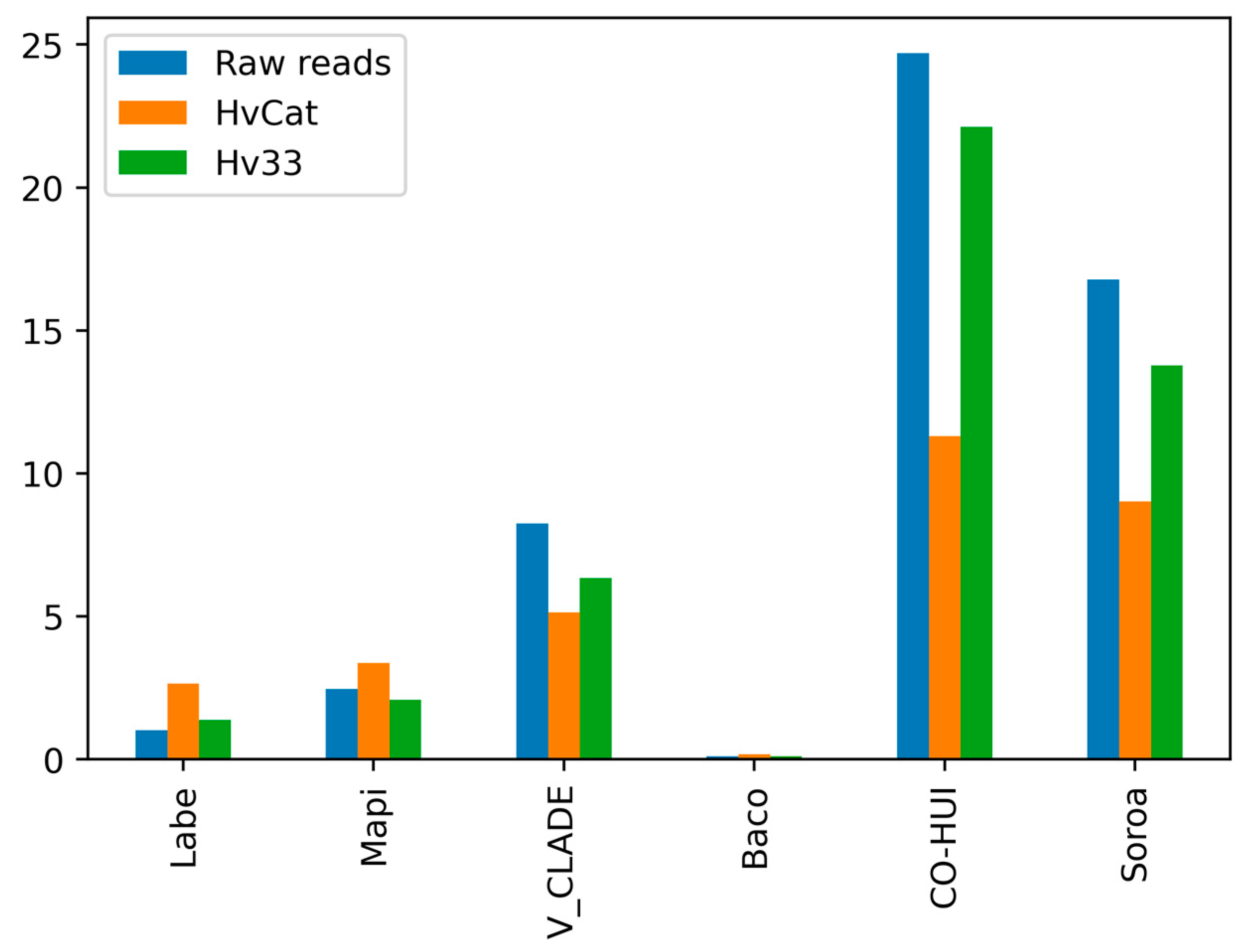
3.1. Identification and Annotation of Transposable Elements in the Hva Genome Assemblies
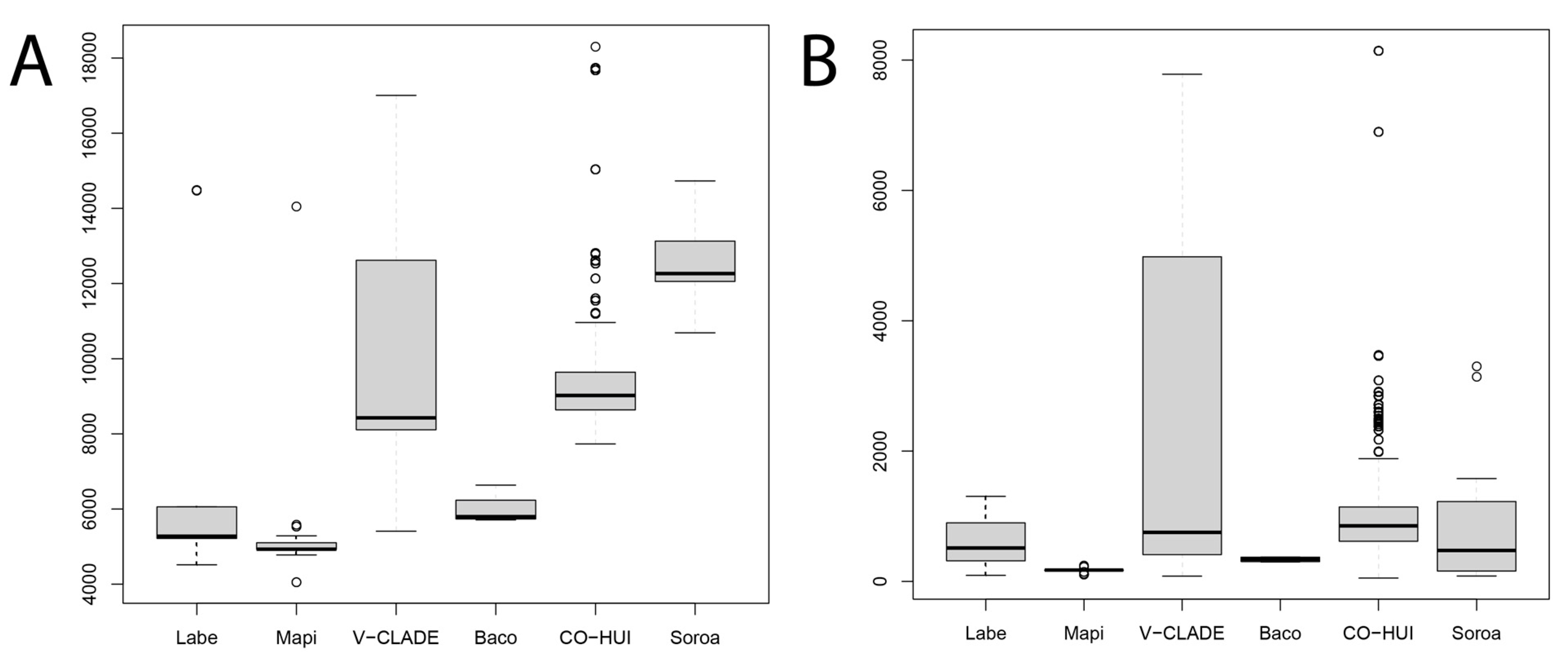
3.2. Copy Number Estimation and the Contribution of TEs to the Hva Genome Size
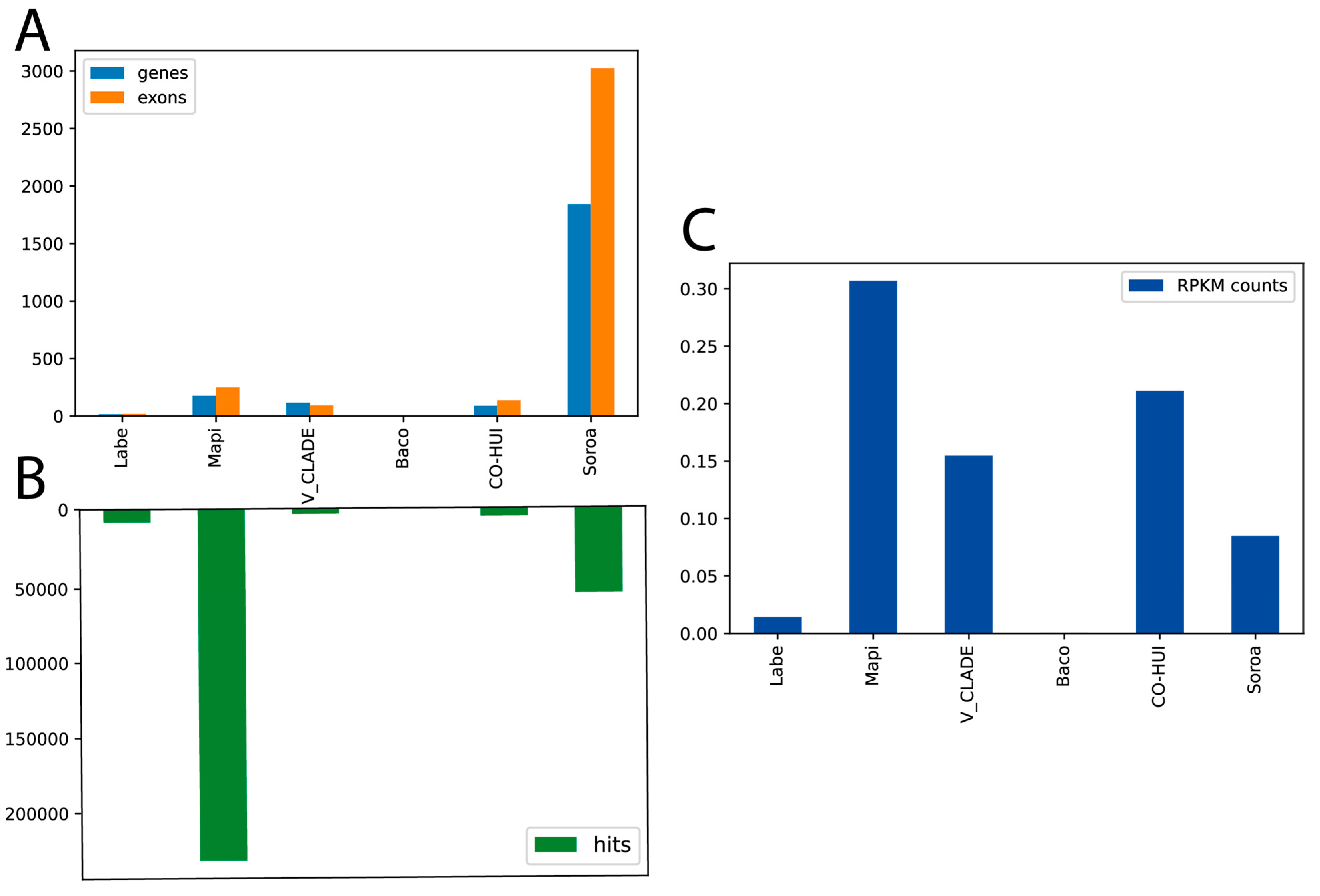
3.3. Transcriptional Analysis of the LTR-RT Lineages and Families Using RNA-Seq, and Their Relationships with Coding Genes
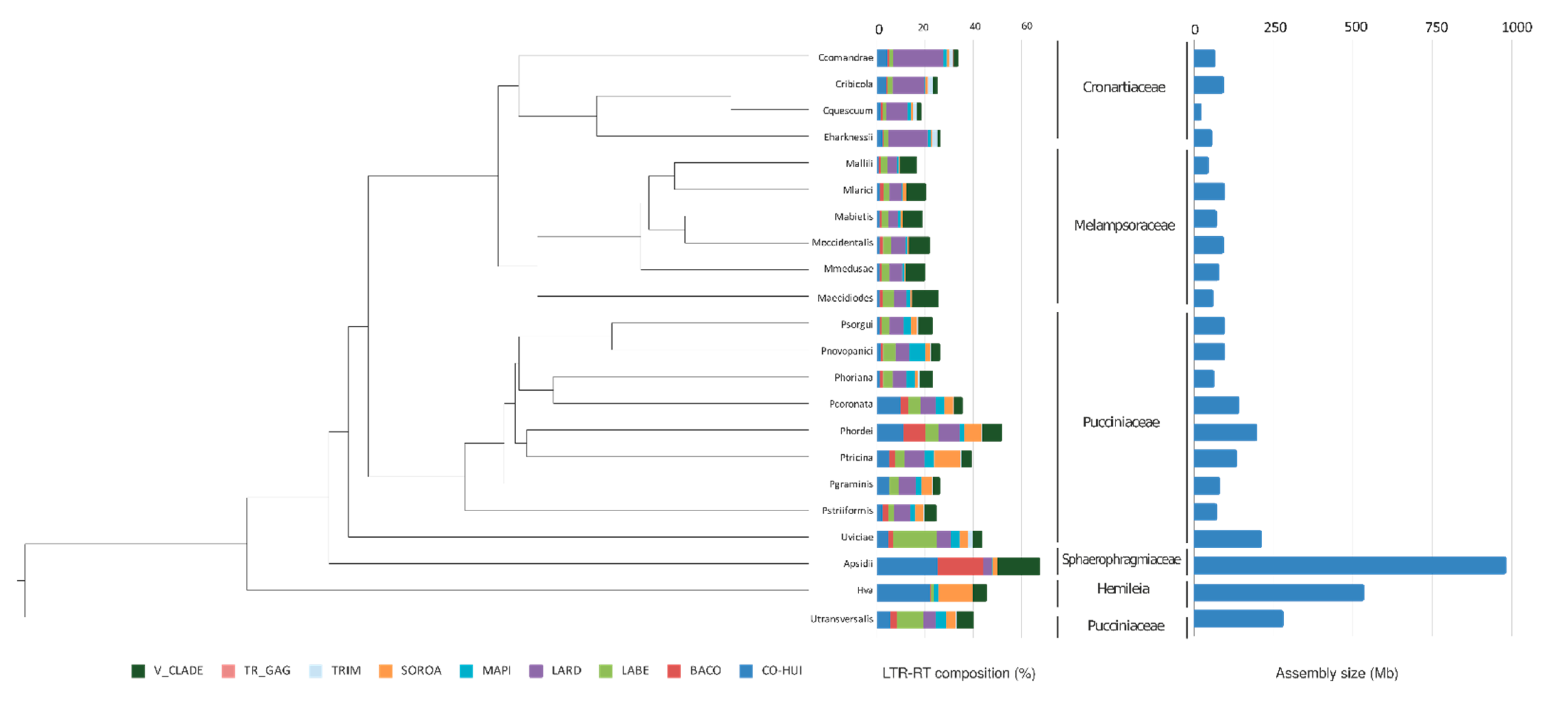
3.4. Characterization of the Conserved LTR-RT Lineages in the Pucciniales Genomes
4. Discussion
4.1. Identification and Annotation of the Transposable Elements in Available Hva Genomes: A Challenge
4.2. The Hva Genome Is Mainly Composed of Three LTR-RT Lineages
4.3. Pattern of LTR-RT Fractions in the Pucciniales Genomes
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lopez, O.L.O.; Alvarez-Herrera, L.M. Tendencia de la producción y el consumo del café en Colombia. Apunt. Cenes 2017, 36, 139–166. [Google Scholar] [CrossRef]
- Cristancho-Ardila, M.A.; Escobar-Ochoa, C.; Dickson Ocampo-Muñoz, J. Evolución de razas de Hemileia vastatrix en Colombia. Cenicafé 2007, 58, 340–359. [Google Scholar]
- Rhiney, K.; Guido, Z.; Knudson, C.; Avelino, J.; Bacon, C.M.; Leclerc, G.; Aime, M.C.; Bebber, D.P. Epidemics and the future of coffee production. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Talhinhas, P.; Batista, D.; Diniz, I.; Vieira, A.; Silva, D.N.; Loureiro, A.; Tavares, S.; Pereira, A.P.; Azinheira, H.G.; Guerra-Guimarães, L.; et al. The coffee leaf rust pathogen Hemileia vastatrix: One and a half centuries around the tropics. Mol. Plant Pathol. 2017, 18, 1039–1051. [Google Scholar] [CrossRef] [PubMed]
- Monaco, L.C. Consequences of the introduction of coffee rust into Brazil. Ann. N. Y. Acad. Sci. 1977, 287, 57–71. [Google Scholar] [CrossRef]
- McCook, S. Global rust belt: Hemileia vastatrix and the ecological integration of world coffee production since 1850. J. Glob. Hist. 2006, 1, 177–195. [Google Scholar] [CrossRef]
- Silva, M.D.C.; Guerra-Guimarães, L.; Diniz, I.; Loureiro, A.; Azinheira, H.; Pereira, A.P.; Tavares, S.; Batista, D.; Várzea, V. An Overview of the Mechanisms Involved in Coffee-Hemileia vastatrix Interactions: Plant and Pathogen Perspectives. Agronomy 2022, 12, 326. [Google Scholar] [CrossRef]
- Gouveia, M.M.C.; Ribeiro, A.; Várzea, V.M.; Rodrigues, C.J., Jr. Genetic diversity in Hemileia vastatrix based on RAPD markers. Mycologia 2017, 97, 396–404. [Google Scholar] [CrossRef]
- Cabral, P.G.C.; Maciel-Zambolim, E.; Oliveira, S.A.S.; Caixeta, E.; Zambolim, L. Genetic diversity and structure of Hemileia vastatrix populations on Coffea spp. Plant Pathol. 2016, 65, 196–204. [Google Scholar] [CrossRef]
- Santana, M.F.; Zambolim, E.M.; Caixeta, E.T.; Zambolim, L. Population genetic structure of the coffee pathogen Hemileia vastatrix in Minas Gerais, Brazil. Trop. Plant Pathol. 2018, 43, 473–476. [Google Scholar] [CrossRef]
- Nemri, A.; Saunders, D.G.O.; Anderson, C.; Upadhyaya, N.M.; Win, J.; Lawrence, G.; Jones, D.; Kamoun, S.; Ellis, J.; Dodds, P. The genome sequence and effector complement of the flax rust pathogen Melampsora lini. Front. Plant Sci. 2014, 5, 98. [Google Scholar] [CrossRef] [PubMed]
- Schirawski, J.; Mannhaupt, G.; Münch, K.; Brefort, T.; Schipper, K.; Doehlemann, G.; Di Stasio, M.; Rössel, N.; Mendoza-Mendoza, A.; Pester, D.; et al. Pathogenicity Determinants in Smut Fungi Revealed by Genome Comparison. Science 2010, 330, 1546–1548. [Google Scholar] [CrossRef] [PubMed]
- Toome, M.; Ohm, R.A.; Riley, R.W.; James, T.Y.; Lazarus, K.L.; Henrissat, B.; Albu, S.; Boyd, A.; Chow, J.; Clum, A.; et al. Genome sequencing provides insight into the reproductive biology, nutritional mode and ploidy of the fern pathogen Mixia osmundae. New Phytol. 2014, 202, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Tavares, S.; Ramos, A.P.; Pires, A.S.; Gil Azinheira, H.; Caldeirinha, P.; Link, T.; Abranches, R.; Silva, M.D.C.; Voegele, R.T.; Loureiro, J.; et al. Genome size analyses of Pucciniales reveal the largest fungal genomes. Front. Plant Sci. 2014, 5, 422. [Google Scholar] [CrossRef]
- Aime, M.C.; McTaggart, A.R.; Mondo, S.J.; Duplessis, S. Phylogenetics and Phylogenomics of Rust Fungi. Adv. Genet. 2017, 100, 267–307. [Google Scholar] [CrossRef]
- Silva, D.N.; Várzea, V.; Paulo, O.S.; Batista, D. Population genomic footprints of host adaptation, introgression and recombination in coffee leaf rust. Mol. Plant Pathol. 2018, 19, 1742–1753. [Google Scholar] [CrossRef]
- Rodrigues, A.S.; Silva, D.N.; Várzea, V.; Paulo, O.S.; Batista, D. Worldwide population structure of the coffee rust fungus Hemileia vastatrix is strongly shaped by local adaptation and breeding history. Phytopathology 2022. [Google Scholar] [CrossRef]
- Duplessis, S.; Cuomo, C.A.; Lin, Y.-C.; Aerts, A.; Tisserant, E.; Veneault-Fourrey, C.; Joly, D.; Hacquard, S.; Amselem, J.; Cantarel, B.L.; et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA 2011, 108, 9166–9171. [Google Scholar] [CrossRef]
- Kiran, K.; Rawal, H.C.; Dubey, H.; Jaswal, R.; Devanna, B.; Gupta, D.K.; Bhardwaj, S.C.; Prasad, P.; Pal, D.; Chhuneja, P.; et al. Draft Genome of the Wheat Rust Pathogen (Puccinia triticina) Unravels Genome-Wide Structural Variations during Evolution. Genome Biol. Evol. 2016, 8, 2702–2721. [Google Scholar] [CrossRef]
- Cantu, D.; Segovia, V.; MacLean, D.; Bayles, R.; Chen, X.; Kamoun, S.; Dubcovsky, J.; Saunders, D.G.; Uauy, C. Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. triticireveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genom. 2013, 14, 270. [Google Scholar] [CrossRef]
- McTaggart, A.R.; A Duong, T.; Le-Quy, V.; Shuey, L.S.; Smidt, W.; Naidoo, S.; Wingfield, M.J.; Wingfield, B.D. Chromium sequencing: The doors open for genomics of obligate plant pathogens. BioTechniques 2018, 65, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Cristancho, M.A.; Botero-Rozo, D.O.; Giraldo, W.; Tabima, J.; RiañO-Pachón, D.M.; Escobar, C.; Rozo, Y.; Rivera, L.F.; Durán, A.; Restrepo, S.; et al. Annotation of a hybrid partial genome of the coffee rust (Hemileia vastatrix) contributes to the gene repertoire catalog of the Pucciniales. Front. Plant Sci. 2014, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Porto, B.N.; Caixeta, E.T.; Mathioni, S.M.; Vidigal, P.M.P.; Zambolim, L.; Zambolim, E.M.; Donofrio, N.; Polson, S.W.; Maia, T.; Chen, C.; et al. Genome sequencing and transcript analysis of Hemileia vastatrix reveal expression dynamics of candidate effectors dependent on host compatibility. PLoS ONE 2019, 14, e0215598. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, G.M.A.; Carvalho, C.R.; Barreto, R.W.; Evans, H.C. Coffee rust genome measured using flow cytometry: Does size matter? Plant Pathol. 2013, 63, 1022–1026. [Google Scholar] [CrossRef]
- Cristancho, M.; Giraldo, W.; Botero, D.; Tabima, J.; Ortiz, D.; Peralta, A.; Gaitán, À.; Restrepo, S.; Riaño, D. Application of Genome Studies of Coffee Rust. Adv. Comput. Biol. 2014, 133–139. [Google Scholar] [CrossRef]
- Orozco-Arias, S.; Isaza, G.; Guyot, R. Retrotransposons in Plant Genomes: Structure, Identification, and Classification through Bioinformatics and Machine Learning. Int. J. Mol. Sci. 2019, 20, 3837. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 1–12. [Google Scholar] [CrossRef]
- Boeke, J.D.; Garfinkel, D.J.; Styles, C.A.; Fink, G.R. Ty elements transpose through an RNA intermediate. Cell 1985, 40, 491–500. [Google Scholar] [CrossRef]
- Feschotte, C.; Pritham, E.J. DNA Transposons and the Evolution of Eukaryotic Genomes. Annu. Rev. Genet. 2007, 41, 331–368. [Google Scholar] [CrossRef]
- Han, Y.; Qin, S.; Wessler, S.R. Comparison of class 2 transposable elements at superfamily resolution reveals conserved and distinct features in cereal grass genomes. BMC Genom. 2013, 14, 71. [Google Scholar] [CrossRef] [PubMed]
- Choulet, F.; Alberti, A.; Theil, S.; Glover, N.; Barbe, V.; Daron, J.; Pingault, L.; Sourdille, P.; Couloux, A.; Paux, E.; et al. Structural and functional partitioning of bread wheat chromosome 3B. Science 2014, 345, 1249721. [Google Scholar] [CrossRef] [PubMed]
- Orozco-Arias, S.; Isaza, G.; Guyot, R.; Tabares-Soto, R. A systematic review of the application of machine learning in the detection and classification of transposable elements. PeerJ 2019, 7, e8311. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Gu, X.; Peterson, T. TIR-Learner, a New Ensemble Method for TIR Transposable Element Annotation, Provides Evidence for Abundant New Transposable Elements in the Maize Genome. Mol. Plant 2019, 12, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 2014, 345, 1181–1184. [Google Scholar] [CrossRef]
- Seidl, M.F.; Thomma, B.P. Transposable Elements Direct The Coevolution between Plants and Microbes. Trends Genet. 2017, 33, 842–851. [Google Scholar] [CrossRef]
- Nunes, R.D.C.; Orozco-Arias, S.; Crouzillat, D.; Mueller, L.A.; Strickler, S.R.; Descombes, P.; Fournier, C.; Moine, D.; de Kochko, A.; Yuyama, P.M.; et al. Structure and Distribution of Centromeric Retrotransposons at Diploid and Allotetraploid Coffea Centromeric and Pericentromeric Regions. Front. Plant Sci. 2018, 9, 175. [Google Scholar] [CrossRef]
- Ayala-Usma, D.A.; Cardenas, M.; Guyot, R.; Mares, M.C.D.; Bernal, A.; Munoz, A.R.; Restrepo, S. A whole genome duplication drives the genome evolution of Phytophthora betacei, a closely related species to Phytophthora infestans. BMC Genom. 2021, 22, 795. [Google Scholar] [CrossRef]
- Castanera, R.; López-Varas, L.; Borgognone, A.; LaButti, K.; Lapidus, A.; Schmutz, J.; Grimwood, J.; Pérez, G.; Pisabarro, A.G.; Grigoriev, I.V.; et al. Transposable Elements versus the Fungal Genome: Impact on Whole-Genome Architecture and Transcriptional Profiles. PLoS Genet. 2016, 12, e1006108. [Google Scholar] [CrossRef]
- Belyayev, A. Bursts of transposable elements as an evolutionary driving force. J. Evol. Biol. 2014, 27, 2573–2584. [Google Scholar] [CrossRef]
- Mat Razali, N.; Cheah, B.H.; Nadarajah, K. Transposable Elements Adaptive Role in Genome Plasticity, Pathogenicity and Evolution in Fungal Phytopathogens. Int. J. Mol. Sci. 2019, 20, 3597. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Chen, M.; Zhong, Z.; Tang, W.; Lin, L.; Zhang, X.; Jiang, H.; Zhang, D.; Miao, C.; Tang, H.; et al. PacBio Sequencing Reveals Transposable Elements as a Key Contributor to Genomic Plasticity and Virulence Variation in Magnaporthe oryzae. Mol. Plant 2017, 10, 1465–1468. [Google Scholar] [CrossRef] [PubMed]
- Hua-Van, A.; Le Rouzic, A.; Boutin, T.S.; Filée, J.; Capy, P. The struggle for life of the genome’s selfish architects. Biol. Direct 2011, 6, 19. [Google Scholar] [CrossRef]
- Plissonneau, C.; Benevenuto, J.; Mohd-Assaad, N.; Fouché, S.; Hartmann, F.E.; Croll, D. Using Population and Comparative Genomics to Understand the Genetic Basis of Effector-Driven Fungal Pathogen Evolution. Front. Plant Sci. 2017, 8, 119. [Google Scholar] [CrossRef]
- Flutre, T.; Duprat, E.; Feuillet, C.; Quesneville, H. Considering Transposable Element Diversification in De Novo Annotation Approaches. PLoS ONE 2011, 6, e16526. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, E.M.; McDonald, J.F. LTR_STRUC: A novel search and identification program for LTR retrotransposons. Bioinformatics 2003, 19, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.; Su, W.; Liao, Y.; Chougule, K.; Agda, J.R.A.; Hellinga, A.J.; Lugo, C.S.B.; Elliott, T.A.; Ware, D.; Peterson, T.; et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 2019, 20, 275. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, H. LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007, 35, W265–W268. [Google Scholar] [CrossRef]
- Orozco-Arias, S.; Liu, J.; Tabares-Soto, R.; Ceballos, D.; Domingues, D.S.; Garavito, A.; Ming, R.; Guyot, R. Inpactor, Integrated and Parallel Analyzer and Classifier of LTR Retrotransposons and Its Application for Pineapple LTR Retrotransposons Diversity and Dynamics. Biology 2018, 7, 32. [Google Scholar] [CrossRef]
- Llorens, C.; Futami, R.; Covelli, L.; Dominguez-Escriba, L.; Viu, J.M.; Tamarit, D.; Aguilar-Rodríguez, J.; Vicente-Ripolles, M.; Fuster, G.; Bernet, G.P.; et al. The Gypsy Database (GyDB) of mobile genetic elements: Release 2.0. Nucleic Acids Res. 2011, 39, D70–D74. [Google Scholar] [CrossRef]
- Hoede, C.; Arnoux, S.; Moissette, M.; Chaumier, T.; Inizan, O.; Jamilloux, V.; Quesneville, H. PASTEC: An Automatic Transposable Element Classification Tool. PLoS ONE 2014, 9, e91929. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Boratyn, G.M.; Thierry-Mieg, J.; Thierry-Mieg, D.; Busby, B.; Madden, T.L. Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinform. 2019, 20, 405. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Sundaravaradan, V.; Hahn, T.; Ahmad, N. Conservation of functional domains and limited heterogeneity of HIV-1 reverse transcriptase gene following vertical transmission. Retrovirology 2005, 2, 1–17. [Google Scholar] [CrossRef][Green Version]
- Witte, C.-P.; Le, Q.H.; Bureau, T.; Kumar, A. Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc. Natl. Acad. Sci. USA 2001, 98, 13778–13783. [Google Scholar] [CrossRef]
- Kalendar, R.; Vicient, C.M.; Peleg, O.; Anamthawat-Jonsson, K.; Bolshoy, A.; Schulman, A.H. Large Retrotransposon Derivatives: Abundant, Conserved but Nonautonomous Retroelements of Barley and Related Genomes. Genetics 2004, 166, 1437–1450. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, C.; Gayraud, T.; De Souza, R.F.; Domingues, U.S.; Akaffou, S.; Vanzela, A.L.L.; De Kochko, A.; Rigoreau, M.; Crouzillat, M.; Hamon, S.; et al. Terminal-Repeat Retrotransposons with GAG Domain in Plant Genomes: A New Testimony on the Complex World of Transposable Elements. Genome Biol. Evol. 2015, 7, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; VanBuren, R.; Wai, C.M.; Tang, H.; Schatz, M.C.; E Bowers, J.; Lyons, E.; Wang, M.-L.; Chen, J.; Biggers, E.; et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015, 47, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, S.; Kiss, E.; Kuo, A.; Drula, E.; Kohler, A.; Sánchez-García, M.; Morin, E.; Andreopoulos, B.; Barry, K.W.; Bonito, G.; et al. Large-scale genome sequencing of mycorrhizal fungi provides insights into the early evolution of symbiotic traits. Nat. Commun. 2020, 11, 5125. [Google Scholar] [CrossRef]
- Castanera, R.; Borgognone, A.; Pisabarro, A.G.; Ramírez, L. Biology, dynamics, and applications of transposable elements in basidiomycete fungi. Appl. Microbiol. Biotechnol. 2017, 101, 1337–1350. [Google Scholar] [CrossRef]
- Muszewska, A.; Hoffman-Sommer, M.; Grynberg, M. LTR Retrotransposons in Fungi. PLoS ONE 2011, 6, e29425. [Google Scholar] [CrossRef]
- Marín, I.; Lloréns, C. Ty3/Gypsy Retrotransposons: Description of New Arabidopsis thaliana Elements and Evolutionary Perspectives Derived from Comparative Genomic Data. Mol. Biol. Evol. 2000, 17, 1040–1049. [Google Scholar] [CrossRef]
- Hess, J.; Skrede, I.; Wolfe, B.E.; LaButti, K.; Ohm, R.A.; Grigoriev, I.V.; Pringle, A. Transposable Element Dynamics among Asymbiotic and Ectomycorrhizal Amanita Fungi. Genome Biol. Evol. 2014, 6, 1564–1578. [Google Scholar] [CrossRef]
- Devos, K.M.; Brown, J.K.M.; Bennetzen, J.L. Genome Size Reduction through Illegitimate Recombination Counteracts Genome Expansion in Arabidopsis. Genome Res. 2002, 12, 1075–1079. [Google Scholar] [CrossRef]
- SanMiguel, P.; Tikhonov, A.; Jin, Y.-K.; Motchoulskaia, N.; Zakharov, D.; Melake-Berhan, A.; Springer, P.S.; Edwards, K.J.; Lee, M.; Avramova, Z.; et al. Nested Retrotransposons in the Intergenic Regions of the Maize Genome. Science 1996, 274, 765–768. [Google Scholar] [CrossRef]
- Piegu, B.; Guyot, R.; Picault, N.; Roulin, A.; Saniyal, A.; Kim, H.; Collura, K.; Brar, D.S.; Jackson, S.; Wing, R.A.; et al. Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006, 16, 1262–1269. [Google Scholar] [CrossRef] [PubMed]
- Horns, F.; Petit, E.; Hood, M.E. Massive Expansion of Gypsy-like Retrotransposons in Microbotryum Fungi. Genome Biol. Evol. 2017, 9, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Fowler, T.J.; Mitton, M.F. Scooter, a New Active Transposon in Schizophyllum commune, Has Disrupted Two Genes Regulating Signal Transduction. Genetics 2000, 156, 1585–1594. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.-K.; Chen, Z.; Wilkins, M.R.; Collins, D.; Englezou, A. A brief overview of the size and composition of the myrtle rust genome and its taxonomic status. Mycology 2014, 5, 52–63. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orozco-Arias, S.; Candamil, M.S.; Jaimes, P.A.; Cristancho, M.; Tabares-Soto, R.; Guyot, R. Composition and Diversity of LTR Retrotransposons in the Coffee Leaf Rust Genome (Hemileia vastatrix). Agronomy 2022, 12, 1665. https://doi.org/10.3390/agronomy12071665
Orozco-Arias S, Candamil MS, Jaimes PA, Cristancho M, Tabares-Soto R, Guyot R. Composition and Diversity of LTR Retrotransposons in the Coffee Leaf Rust Genome (Hemileia vastatrix). Agronomy. 2022; 12(7):1665. https://doi.org/10.3390/agronomy12071665
Chicago/Turabian StyleOrozco-Arias, Simon, Mariana S. Candamil, Paula A. Jaimes, Marco Cristancho, Reinel Tabares-Soto, and Romain Guyot. 2022. "Composition and Diversity of LTR Retrotransposons in the Coffee Leaf Rust Genome (Hemileia vastatrix)" Agronomy 12, no. 7: 1665. https://doi.org/10.3390/agronomy12071665
APA StyleOrozco-Arias, S., Candamil, M. S., Jaimes, P. A., Cristancho, M., Tabares-Soto, R., & Guyot, R. (2022). Composition and Diversity of LTR Retrotransposons in the Coffee Leaf Rust Genome (Hemileia vastatrix). Agronomy, 12(7), 1665. https://doi.org/10.3390/agronomy12071665







