Molecular Markers for Identifying Resistance Genes in Brassica napus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genotypic Characterization of Resistance Genes
2.2. Phenotypic Characterization of Resistance Genes
2.3. KASP Marker Development and Protocols
3. Results
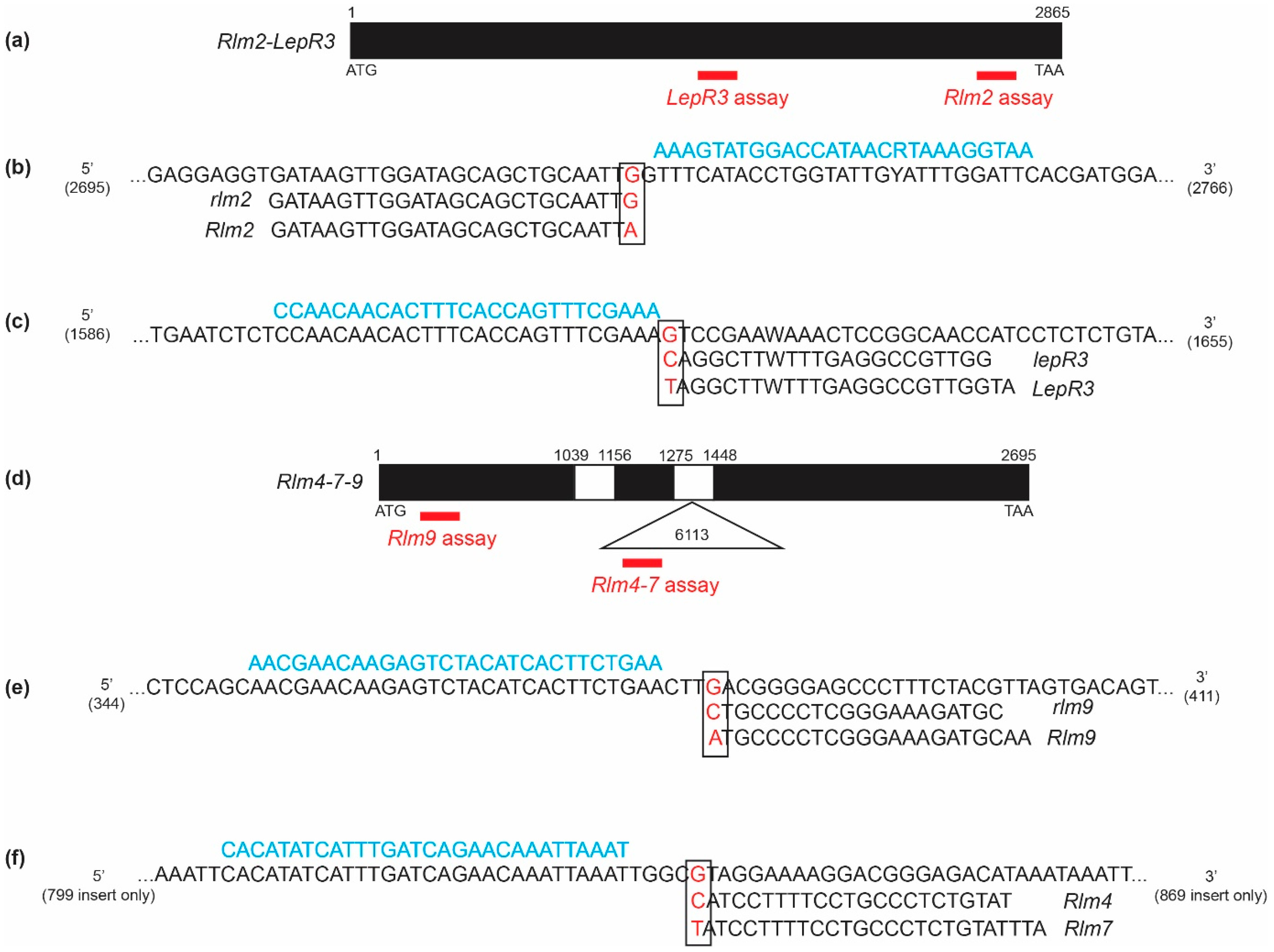
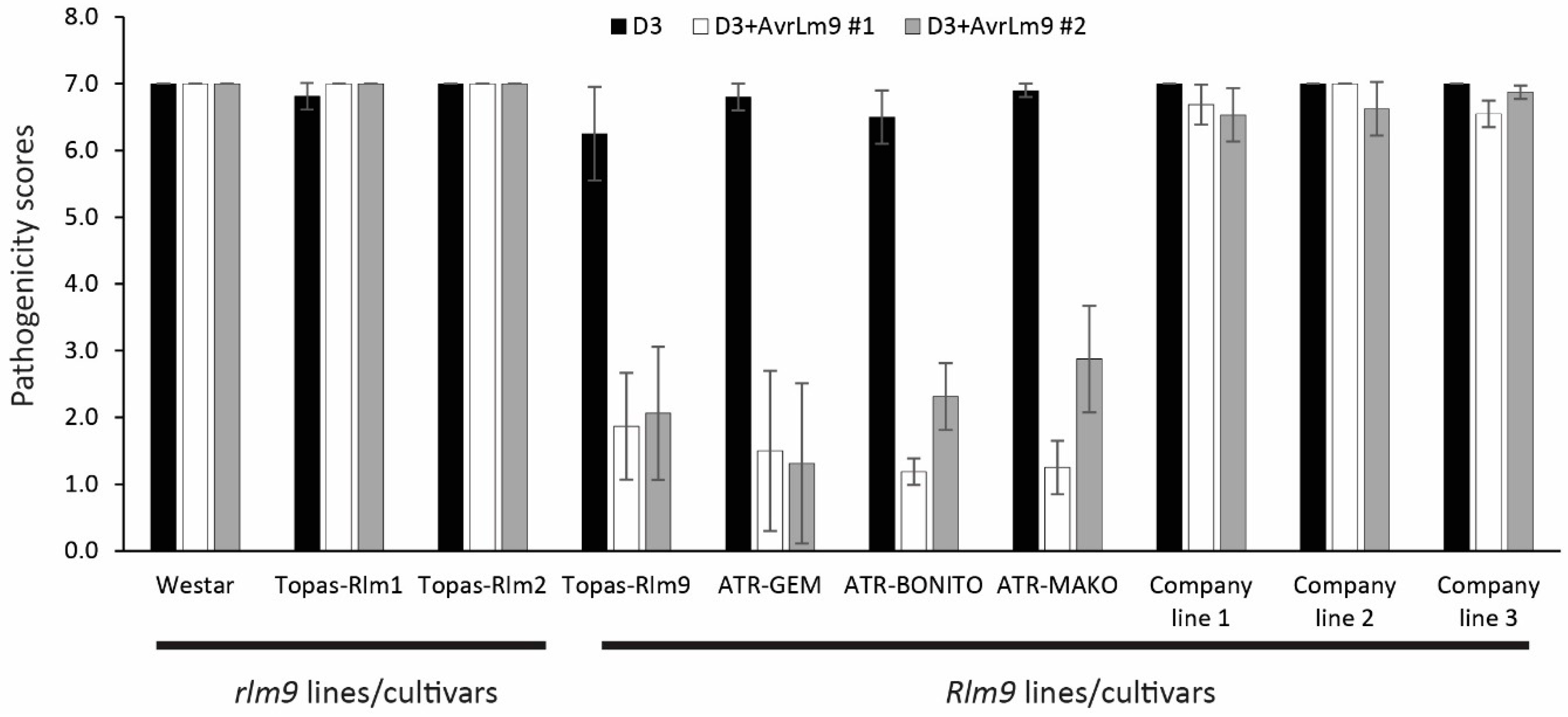
3.1. Identification of Resistance and Susceptible Alleles at the LepR3-Rlm2 and Rlm4-7-9 Gene Loci
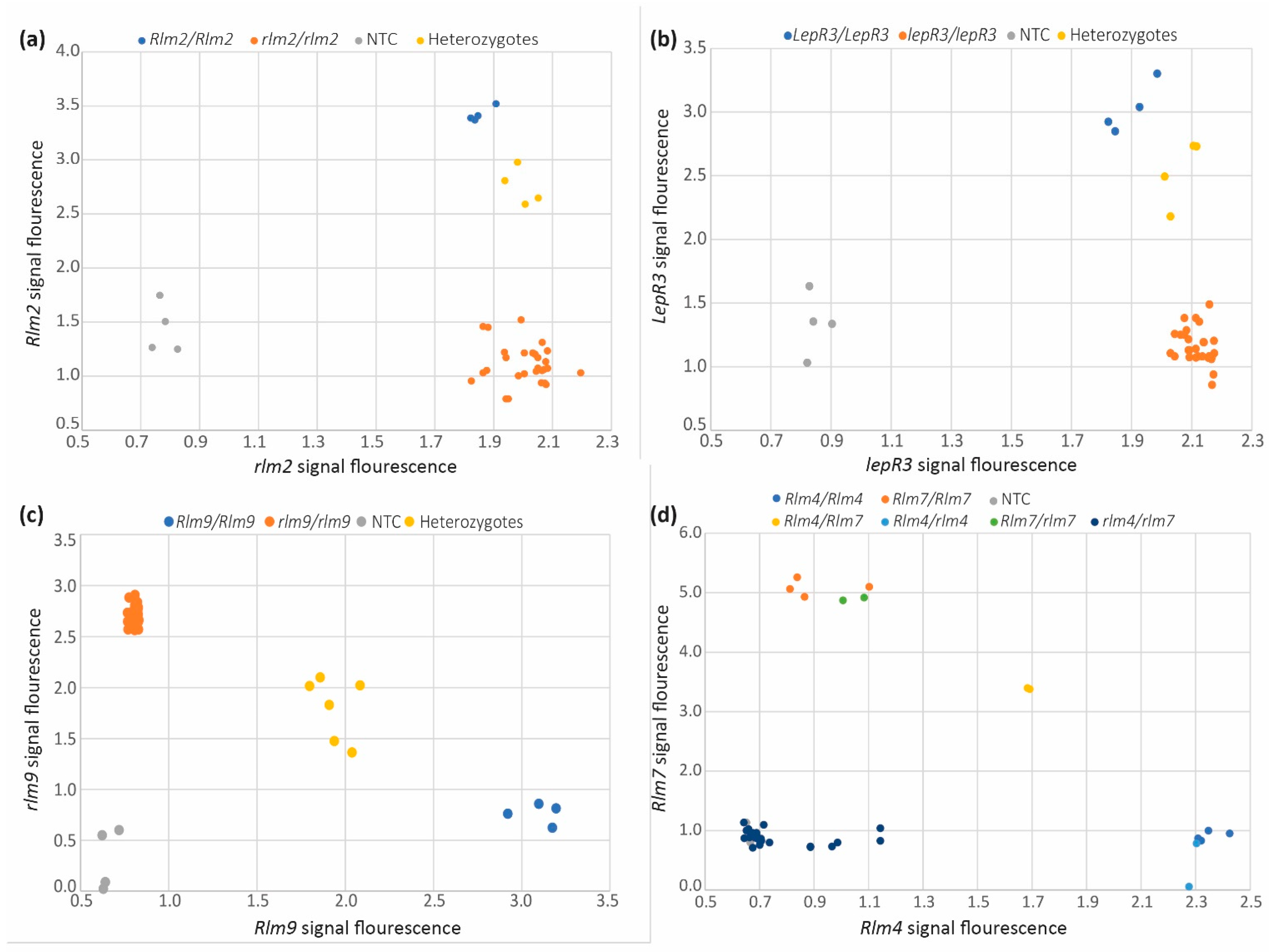
3.2. Rlm2 Molecular Marker
3.3. LepR3 Molecular Marker
3.4. Rlm9 Molecular Marker
3.5. Rlm4 and Rlm7 Molecular Marker
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stotz, H.U.; Mitrousia, G.K.; de Wit, P.J.; Fitt, B.D.L. Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci. 2014, 19, 491–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitt, B.D.L.; Fraaije, B.A.; Chandramohan, P.; Shaw, M.W. Impacts of changing air composition on severity of arable crop disease epidemics. Plant Pathol. 2011, 60, 44–53. [Google Scholar] [CrossRef] [Green Version]
- Van de Wouw, A.P.; Marcroft, S.J.; Sprague, S.J.; Scanlan, J.L.; Vesk, P.A.; Idnurm, A. Epidemiology and management of blackleg of canola in response to changing farming practices in Australia. Australas. Plant Pathol. 2021, 50, 137–149. [Google Scholar] [CrossRef]
- Yang, Y.; Marcroft, S.J.; Forsyth, L.M.; Zhao, J.; Ziqin, L.; Van de Wouw, A.P.; Idnurm, A. Sterol demethylation inhibitor fungicide resistance in Leptosphaeria maculans is caused by modifications in the regulatory region of ERG11. Plant Dis. 2020, 104, 1280–1290. [Google Scholar] [CrossRef] [PubMed]
- Delourme, R.; Chèvre, A.M.; Brun, H.; Rouxel, T.; Balesdent, M.H.; Dias, J.S.; Salisbury, P.; Renard, M.; Rimmer, S.R. Major gene and polygenic resistance to Leptosphaeria maculans in oilseed rape (Brassica napus). Eur. J. Plant Pathol. 2006, 114, 41–52. [Google Scholar] [CrossRef]
- Stuthmann, D.D.; Leonard, J.J.; Miller-Garvin, J. Breeding crops for durable resistance to disease. Adv. Agron. 2007, 95, 319–367. [Google Scholar] [CrossRef]
- Marcroft, S.J.; Elliott, V.L.; Cozijnsen, A.J.; Salisbury, P.A.; Howlett, B.J.; Van de Wouw, A.P. Identifying resistance genes to Leptosphaeria maculans in Australian Brassica napus cultivars based on reactions to isolates with known avirulence genotypes. Crop Pasture Sci. 2012, 63, 338–350. [Google Scholar] [CrossRef]
- Schnippenkoetter, W.; Hoque, S.; Maher, R.; Van de Wouw, A.P.; Marcroft, S.J.; Hands, P.; Rolland, V.; Barrett, L.; Sprague, S.J. Comparison of non-subjective relative fungal biomass measurements to quantify the Leptosphaeria maculans-Brassica napus interaction. BMC Plant Methods 2021, 17, 122. [Google Scholar] [CrossRef]
- Balesdent, M.H.; Barbetti, M.J.; Li, H.; Sivasithamparam, K.; Gout, L.; Rouxel, T. Analysis of Leptosphaeria maculans race structure in a worldwide collection of isolates. Phytopathology 2005, 95, 1061–1071. [Google Scholar] [CrossRef] [Green Version]
- Delourme, R.; Pilet-Nayel, M.L.; Archipiano, M.; Horvais, R.; Tanguy, X.; Rouxel, T.; Brun, H.; Renard, M.; Balesdent, M.H. A cluster of major specific resistance genes to Leptosphaeria maculans in Brassica napus. Phytopathology 2004, 94, 578–583. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Lydiate, D.J.; Rimmer, S.R. Identification of two novel genes for blackleg resistance in Brassica napus. Theor. Appl. Genet. 2005, 110, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Gugel, R.K.; Kutcher, R.; Peng, G.; Rimmer, S.R. Identification and mapping of a novel blackleg resistance locus LepR4 in the progenies from Brassica napus × B. rapa subsp. sylvestris. Theor. Appl. Genet. 2013, 126, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Raman, H.; Raman, R.; Qiu, Y.; Zhang, Y.; Batley, J.; Liu, S. The Rlm13 gene, a new player of Brassica napus-Leptosphaeria maculans interaction maps on chromosome C03 in canola. Front. Plant Sci. 2021, 12, 654604. [Google Scholar] [CrossRef]
- Larkan, N.J.; Ma, L.; Haddadi, P.; Buchwaldt, M.; Parkin, I.A.; Djavaheri, M.; Borhan, H. The Brassica napus wall-associated kinase-like (WAKL) gene Rlm9 provides race-specific blackleg resistance. Plant J. 2020, 104, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Haddadi, P.; Larkan, N.J.; Van de Wouw, A.P.; Zhang, Y.; Xiang Neik, T.; Beynon, E.; Bayer, P.E.; Edwards, D.; Batley, J.; Borhan, H. Brassica napus genes Rlm4 and Rlm7, conferring resistance to Leptosphaeria maculans, are alleles of the Rlm9 wall-associated kinase-like resistance locus. Plant Biotechnol. J. 2022, in press. [Google Scholar] [CrossRef]
- Larkan, N.J.; Lydiate, D.J.; Parkin, I.A.; Nelson, M.N.; Epp, D.J.; Cowling, W.A.; Rimmer, S.R.; Borhan, M.H. The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 2013, 197, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Larkan, N.J.; Ma, L.; Borhan, H. The Brassica napus receptor-like protein RLM2 is encoded by a second allele of the LepR3/Rlm2 blackleg resistance locus. Plant Biotechnol. J. 2015, 13, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Bohar, R.; Chitkineni, A.; Varshney, R.K. Genetic molecular markers to accelerate genetic gains in crops. BioTechniques 2020, 69, 158–160. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Fernando, D.W.G. Insights into fighting against blackleg disease of Brassica napus in Canada. Crop Pasture Sci. 2018, 69, 40–47. [Google Scholar] [CrossRef]
- Van de Wouw, A.P.; Elliott, C.E.; Howlett, B.J. Transformation of fungal isolates with avirulence genes provides tools for identification of corresponding resistance genes in the host plant. Eur. J. Plant Pathol. 2014, 140, 875–882. [Google Scholar] [CrossRef]
- Van de Wouw, A.P.; Marcroft, S.J.; Barbetti, M.J.; Hua, L.; Salisbury, P.A.; Gout, L.; Rouxel, T.; Howlett, B.J.; Balesdent, M.H. Dual control of avirulence in Leptosphaeria maculans towards a Brassica napus cultivar with ’sylvestris-derived’ resistance suggests involvement of two resistance genes. Plant Pathol. 2009, 58, 305–313. [Google Scholar] [CrossRef]
- Parlange, F.; Daverdin, G.; Fudal, I.; Kuhn, M.L.; Balesdent, M.H.; Blaise, F.; Grezes-Besset, B.; Rouxel, T. Leptosphaeria maculans avirulence gene AvrLm4-7 confers a dual recognition specificity by the Rlm4 and Rlm7 resistance genes of oilseed rape, and circumvents Rlm4-mediated recognition through a single amino acid change. Mol. Microbiol. 2009, 71, 851–863. [Google Scholar] [CrossRef] [PubMed]
- Plissonneau, C.; Daverdin, G.; Ollivier, B.; Blaise, F.; Degrave, A.; Fudal, I.; Rouxel, T.; Balesdent, M. A game of hide and seek between avirulence genes AvrLm4-7 and AvrLm3 in Leptosphaeria maculans. New Phytol. 2016, 209, 1613–1624. [Google Scholar] [CrossRef] [Green Version]
- Ghanbarnia, K.; Ma, L.; Larkan, N.J.; Haddadi, P.; Fernando, D.W.G.; Borhan, M.H. Leptosphaeria maculans AvrLm9: A new player in the game of hide and seek with AvrLm4-7. Mol. Plant Pathol. 2018, 19, 1754–1764. [Google Scholar] [CrossRef] [Green Version]
- Ravi, R.K.; Walton, K.; Khosroheidari, M. MiSeq: A next generation sequencing platform for genomic analysis. Methods Mol. Biol. 2018, 1706, 223–232. [Google Scholar]
- Cheng, F.; Wu, J.; Cai, C.; Fu, L.; Liang, J.; Borm, T.; Zhuang, M.; Zhang, Y.; Zhang, F.; Bonnema, G.; et al. Genome requencing and comparative variome analysis in a Brassica rapa and Brassica oleracea collection. Sci. Data 2016, 20, 160119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouxel, T.; Willner, E.; Coudard, L.; Balesdent, M.H. Screening and identification of resistance to Leptosphaeria maculans (stem canker) in Brassica napus accessions. Euphytica 2003, 133, 219–231. [Google Scholar] [CrossRef]
- Larkan, N.J.; Yu, F.; Lydiate, D.J.; Rimmer, S.R.; Borhan, M.H. Single R gene introgression lines for accurate dissection of the Brassica-Leptosphaeria pathosystem. Front. Plant Sci. 2016, 7, 1771. [Google Scholar] [CrossRef] [Green Version]
- Elliott, C.E.; Howlett, B.J. Overexpression of a 3-ketoacyl-CoA thiolase in Leptosphaeria maculans causes reduced pathogenicity on Brassica napus. Mol. Plant Microbe Interact. 2006, 19, 588–596. [Google Scholar] [CrossRef] [Green Version]
- Gardiner, D.M.; Howlett, B.J. Negative selection using thymidine kinase increases the efficiency of recovery of transformants with targeted genes in the filamentous fungus Leptosphaeria maculans. Curr. Genet. 2004, 45, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Dogan, Y.; Comertpay, G.; Yildiz, M.; Hatipoglu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef] [Green Version]
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcroft, S.J.; Van de Wouw, A.P.; Salisbury, P.A.; Potter, T.D.; Howlett, B.J. Rotation of canola (Brassica napus) cultivars with different complements of blackleg resistance genes decreases disease severity. Plant Pathol. 2012, 61, 934–944. [Google Scholar] [CrossRef]
- Van de Wouw, A.P.; Marcroft, S.J.; Ware, A.; Lindbeck, K.; Khangura, R.; Howlett, B.J. Breakdown of resistance to the fungal disease, blackleg, is averted in commercial canola (Brassica napus) crops in Australia. Field Crops Res. 2014, 166, 144–151. [Google Scholar] [CrossRef]
- Van de Wouw, A.P.; Marcroft, S.J.; Howlett, B.J. Blackleg disease of canola in Australia. Crop Pasture Sci. 2016, 67, 273–282. [Google Scholar] [CrossRef]
- Van de Wouw, A.P.; Howlett, B.J.; Idnurm, A. Changes in allele frequencies of avirulence genes in the blackleg fungus, Leptosphaeria maculans, over two decades in Australia. Crop Pasture Sci. 2018, 69, 20–29. [Google Scholar] [CrossRef]
| Cultivar/Line | Resistance Gene(s) | Reference |
|---|---|---|
| Westar | None | [27] |
| Topas-DH16516 | None | [28] |
| Topas-Rlm1 | Rlm1 | [28] |
| Topas-Rlm2 | Rlm2 | [28] |
| Topas-Rlm3 | Rlm3 | [28] |
| Topas-Rlm4 | Rlm4 | [28] |
| Topas-Rlm7 | Rlm7 | [28] |
| Topas-Rlm9 | Rlm9 | [28] |
| Topas-LepR1 | LepR1 | [28] |
| Topas-LepR2 | LepR2 | [28] |
| Topas-LepR3 | LepR3 | [28] |
| Express | Rlm2 | [27] |
| Surpass501TT | LepR3, RlmS | [16,21] |
| BASF3000TR | Rlm4 | [7] |
| Caiman | Rlm7 | [7] |
| ATR-Gem | Rlm1, Rlm9 | [7] |
| Gene | Primer Name | Primer Sequence | SNP | Allele Detected | Fluorescence |
|---|---|---|---|---|---|
| Rlm2 | Rlm2_AlleleX | GAAGGTGACCAAGTTCATGCTGATAAGTTGGATAGCAGCTGCAATTA | A | Rlm2 | FAM |
| Rlm2_AlleleY | GAAGGTCGGAGTCAACGGATTGATAAGTTGGATAGCAGCTGCAATTG | G | rlm2 | HEX | |
| Rlm2_Common | AATCCAAATRCAATACCAGGTATGAAA | ||||
| LepR3 | LepR3_AlleleX | GAAGGTGACCAAGTTCATGCTATGGTTGCCGGAGTTTWTTCGGAT | A | LepR3 | FAM |
| LepR3_AlleleY | GAAGGTCGGAGTCAACGGATTGGTTGCCGGAGTTTWTTCGGAC | G | lepR3 | HEX | |
| LepR3_Common | CCAACAACACTTTCACCAGYTTCGAAA | ||||
| Rlm9 | Rlm9_AlleleX | GAAGGTGACCAAGTTCATGCTCGTAGAAAGGGCTCCCCGTC | G | rlm9 | FAM |
| Rlm9_AlleleY | GAAGGTCGGAGTCAACGGATTAACGTAGAAAGGGCTCCCCGTA | T | Rlm9 | HEX | |
| Rlm9_Common | AACGAACAAGAGTCTACATCACTTCTGAA | ||||
| Rlm4 and Rlm7 1 | Rlm47_AlleleX | GAAGGTGACCAAGTTCATGCTATTTATGTCTCCCGTCCTTTTCCTAT | A | Rlm7 | FAM |
| Rlm47_AlleleY | GAAGGTCGGAGTCAACGGATTTATGTCTCCCGTCCTTTTCCTAC | G | Rlm4 | HEX | |
| Rlm47_Common | CACATATCATTTGATCAGAACAAATTAAAT |
| Cycles | Temp | Time | Type of Cycling |
|---|---|---|---|
| 1× | 94 °C | 15 min | Denature |
| 10× | 94 °C | 20 s | PCR |
| 61–55 °C (decreasing temp each cycle) | 60 s | ||
| 26× | 94 °C | 20 s | PCR |
| 55 °C | 60 s | ||
| 1× | 25 °C | 1 min | Cooling |
| 1× | 25 °C | 30 s | Acquire data |
| 3× | 94 °C | 20 s | Recycling (if required) 1 |
| 57 °C | 60 s | ||
| 1× | 25 °C | 1 min | |
| 25 °C | 30 s | Second data acquisition |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van de Wouw, A.P.; Zhang, Y.; Mohd Saad, N.S.; Yang, H.; Sheedy, E.; Elliott, C.E.; Batley, J. Molecular Markers for Identifying Resistance Genes in Brassica napus. Agronomy 2022, 12, 985. https://doi.org/10.3390/agronomy12050985
Van de Wouw AP, Zhang Y, Mohd Saad NS, Yang H, Sheedy E, Elliott CE, Batley J. Molecular Markers for Identifying Resistance Genes in Brassica napus. Agronomy. 2022; 12(5):985. https://doi.org/10.3390/agronomy12050985
Chicago/Turabian StyleVan de Wouw, Angela P., Yueqi Zhang, Nur Shuhadah Mohd Saad, Hua Yang, Elizabeth Sheedy, Candace E. Elliott, and Jacqueline Batley. 2022. "Molecular Markers for Identifying Resistance Genes in Brassica napus" Agronomy 12, no. 5: 985. https://doi.org/10.3390/agronomy12050985
APA StyleVan de Wouw, A. P., Zhang, Y., Mohd Saad, N. S., Yang, H., Sheedy, E., Elliott, C. E., & Batley, J. (2022). Molecular Markers for Identifying Resistance Genes in Brassica napus. Agronomy, 12(5), 985. https://doi.org/10.3390/agronomy12050985







