Antagonism of Bacillus velezensis Isolate from Anaerobically Digested Dairy Slurry against Fusarium Wilt of Spinach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and Characteristics of ADSs
2.2. Antifungal Activity of Raw and Filtrate ADSs
2.3. Isolation of Bacteria from Five ADSs
2.4. Antifungal Activity of Bacterial Isolates from ADSs
2.5. Pot Experiment
2.5.1. Preparation of Fos Inoculum
2.5.2. Preparation of AD-3
2.5.3. Pot Assay
2.6. Genome Sequencing
2.7. Genome Analysis
3. Results
3.1. Antagonistic Activities of Raw and Filtrate ADSs In Vitro
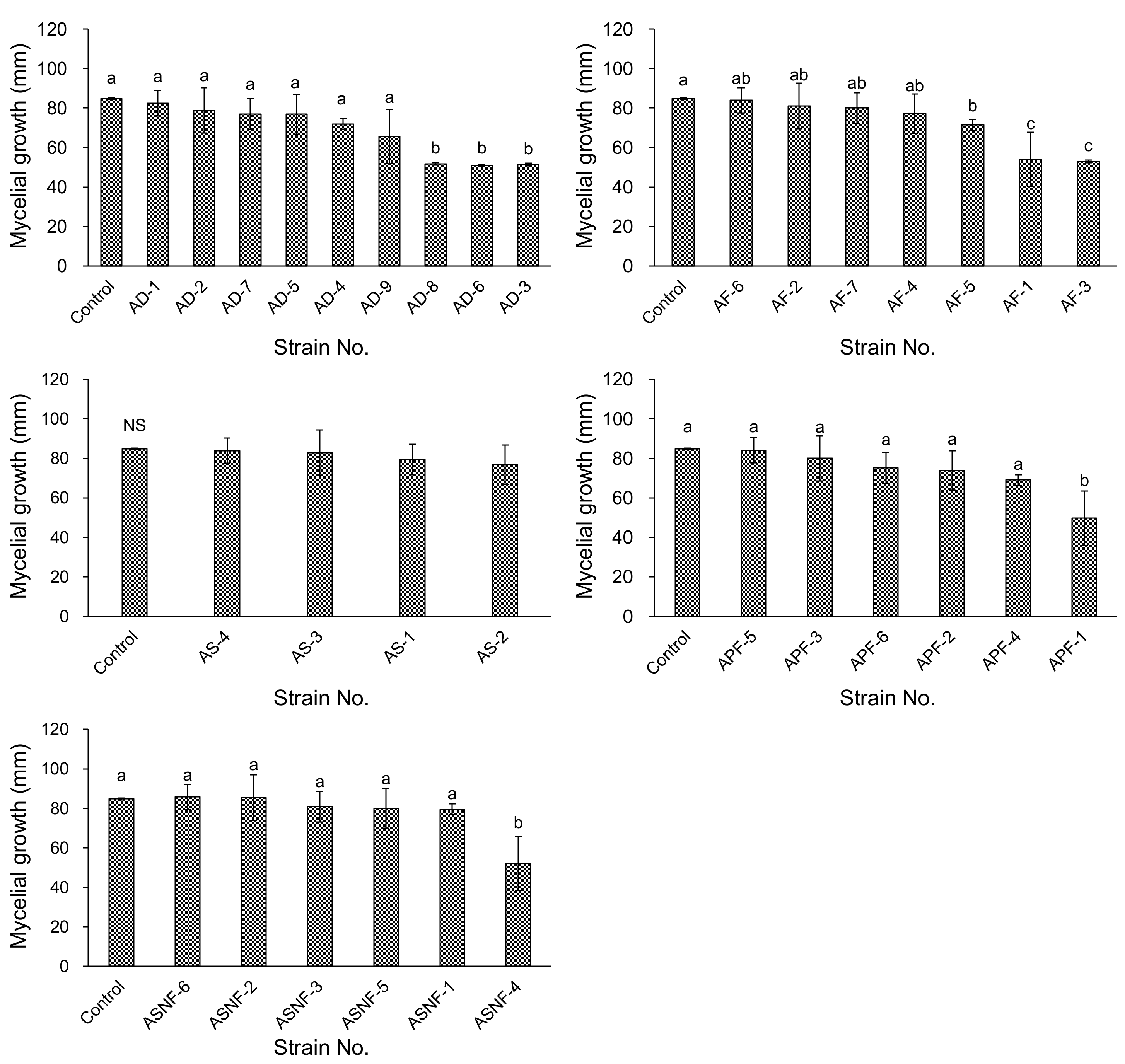
3.2. Antagonistic Activities of Bacterial Isolates from ADSs In Vitro
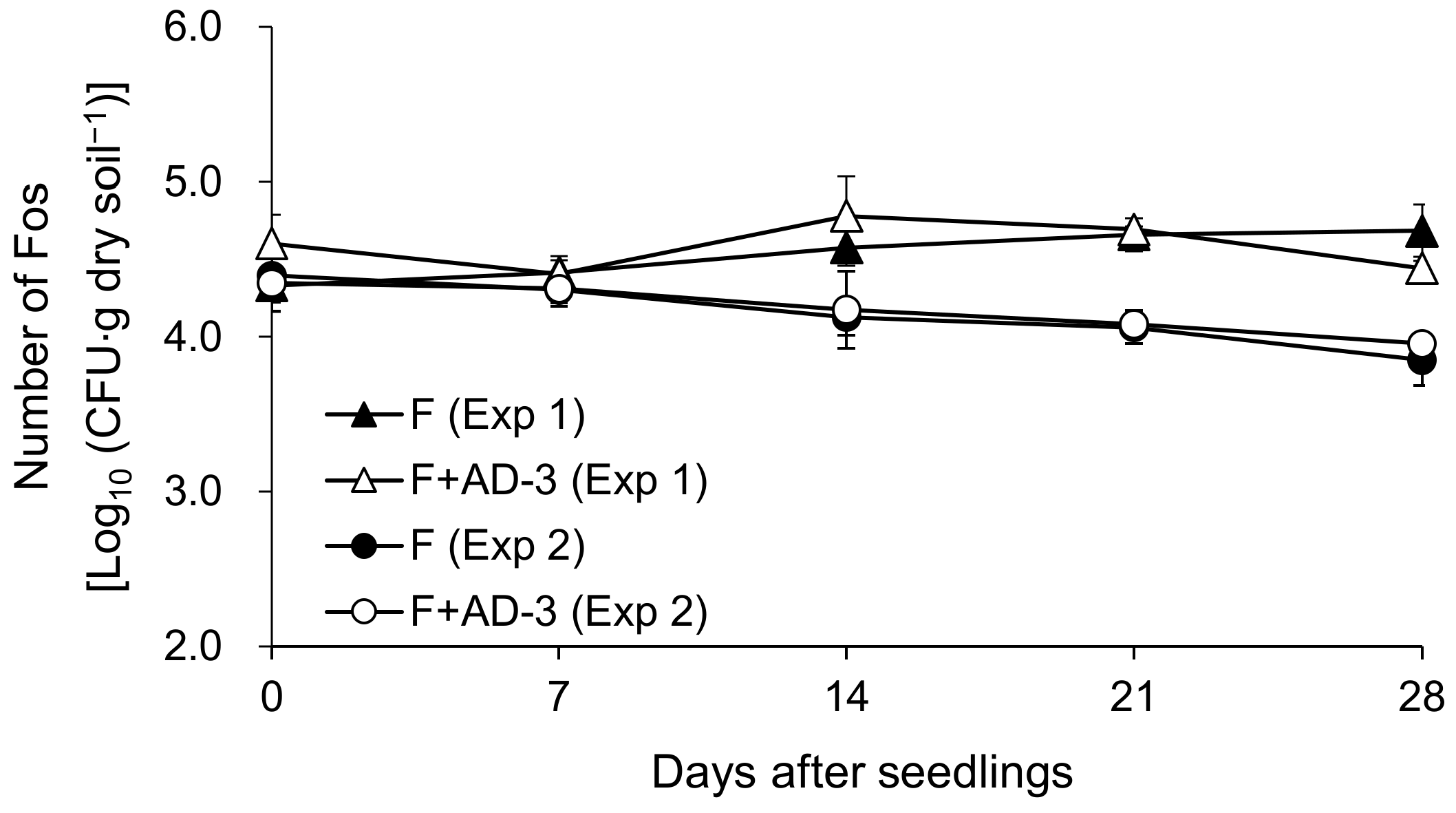
3.3. Effects of AD-3 on Fusarium Wilt of Spinach
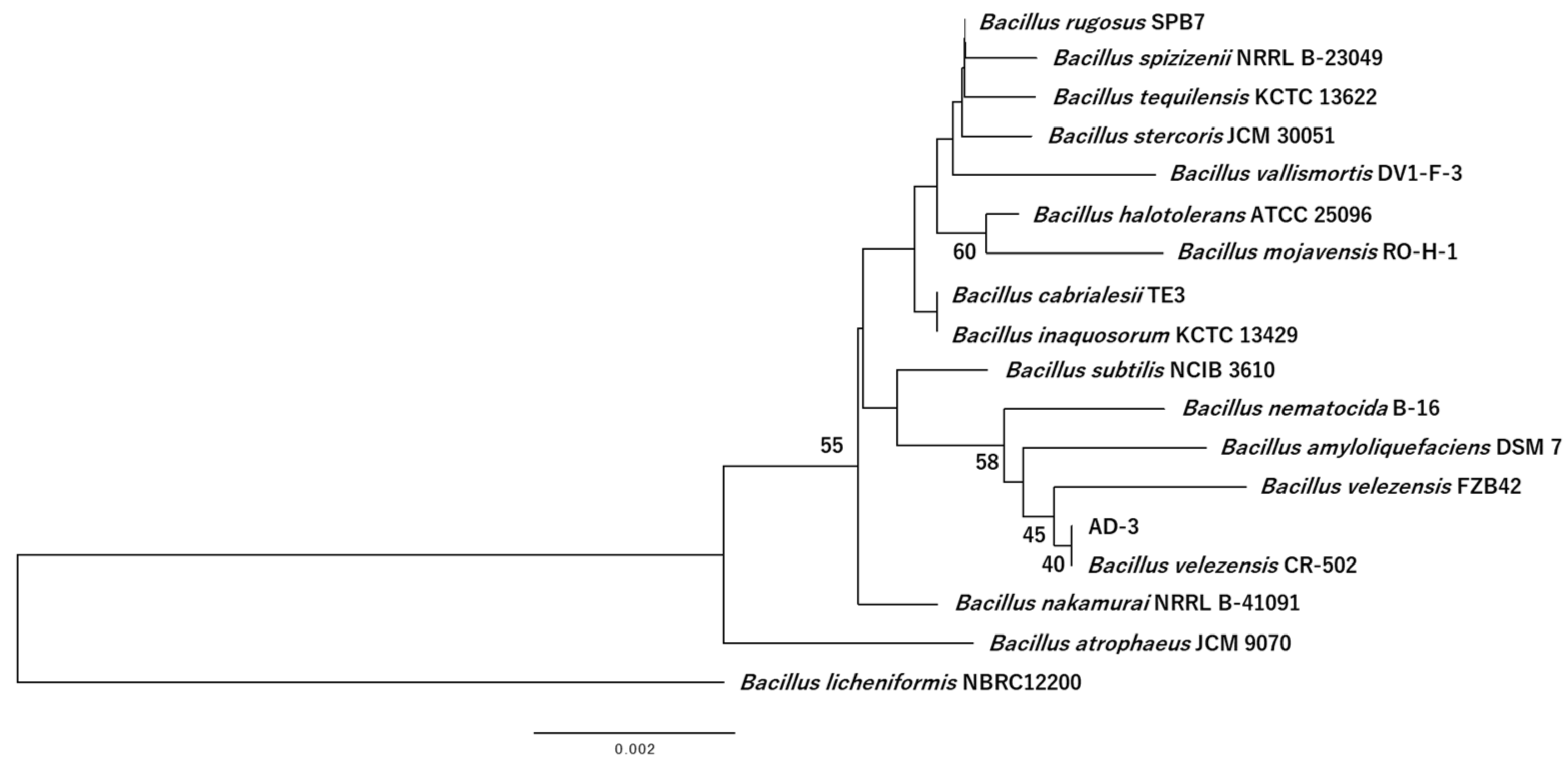
3.4. Identification of AD-3
3.5. Genomic Characterization of AD-3
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fravel, D.; Olivain, C.; Alabouvette, C. Fusarium oxysporum and its biocontrol. New Phytologist. 2003, 157, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond−Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panth, M.; Hassler, S.C.; Baysal-Gurel, F. Methods for management of soilborne diseases in crop production. Agriculture 2020, 10, 16. [Google Scholar] [CrossRef] [Green Version]
- Bonanomi, G.; Zotti, M.; Idbella, M.; Cesarano, G.; Al-Rowaily, S.L.; Abd-ElGawad, A.M. Mixtures of organic amendments and biochar promote beneficial soil microbiota and affect Fusarium oxysporum f. sp. lactucae, Rhizoctonia solani and Sclerotinia minor disease suppression. Plant Pathol. 2022, 71, 818–829. [Google Scholar] [CrossRef]
- Vida, C.; de Vicente, A.; Cazorla, F.M. The role of organic amendments to soil for crop protection: Induction of suppression of soilborne pathogens. Ann. Appl. Biol. 2020, 176, 1–15. [Google Scholar] [CrossRef]
- Banegas, V.; Moreno, J.L.; Moreno, J.I.; García, C.; León, G.; Hernández, T. Composting anaerobic and aerobic sewage sludges using two proportions of sawdust. Waste Manag. 2007, 27, 1317–1327. [Google Scholar] [CrossRef]
- Bonanomi, G.; Antignani, V.; Pane, C.; Scala, F. Suppression of soil-borne fungal diseases with organic amendments. J. Plant Pathol. 2007, 89, 311–324. [Google Scholar]
- Bonanomi, G.; Ippolito, F.; Scala, F. A “black” future for plant pathology? Biochar as a new soil amendment for controlling plant diseases. J. Plant Pathol. 2015, 97, 223–234. [Google Scholar]
- Neshat, S.A.; Mohammadi, M.; Najafpour, G.D.; Lahijani, P. Anaerobic co-digestion of animal manures and lignocellulosic residues as a potent approach for sustainable biogas production. Renew. Sustain. Energy Rev. 2017, 79, 308–322. [Google Scholar]
- Mao, C.; Feng, Y.; Wang, X.; Ren, G. Review on research achievements of biogas from anaerobic digestion. Renew. Sustain. Energy Rev. 2015, 45, 540–555. [Google Scholar] [CrossRef]
- Baştabak, B.; Koçar, G. A review of the biogas digestate in agricultural framework. J. Mater. Cycles Waste Manag. 2020, 22, 1318–1327. [Google Scholar] [CrossRef]
- Alburquerque, J.A.; de la Fuente, C.; Ferre-Costa, A.; Carrasco, L.; Cegarra, J.; Abad, M.; Bernal, M.P. Assessment of the fertiliser potential of digestates from farm and agro-industrial residues. Biomass Bioenerg. 2012, 40, 181–189. [Google Scholar] [CrossRef]
- Provenzano, M.R.; Iannuzi, G.; Fabbri, C.; Senesi, N. Qualitative characterization and differentiation of digestates from different biowastes using FTIR and fluorescence spectroscopies. J. Environ. Prot. 2011, 2, 83–89. [Google Scholar] [CrossRef] [Green Version]
- Monlau, F.; Sambusiti, C.; Ficara, E.; Aboulkas, A.; Barakat, A.; Carrere, H. New opportunities for agricultural digestate valorization, current situation and perspectives. Energ. Environ. Sci. 2015, 8, 2600–2621. [Google Scholar] [CrossRef]
- Tambone, F.; Scaglia, B.; D’Imporzano, G.; Schievano, A.; Salati, V.; Adani, F. Assessing amendment and fertilizing properties of digestates from anaerobic digestion through a comparative study with digested sludge and compost. Chemosphere 2010, 81, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Weiland, P. Biogas production: Current state and perspectives. Appl. Microbiol. Biotechnol. 2010, 85, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Xian, Y.; Wu, J.; Gu, Y.; Yang, G.; Zhang, X.; Peng, H.; Yu, X.; Xiao, Y.; Li, L. Effect of biogas slurry addition on soil properties, yields, and bacterial composition in the rice-rape rotation ecosystem over 3 years. J. Soils Sediments 2019, 19, 2534–2542. [Google Scholar] [CrossRef]
- Yu, F.B.; Luo, X.P.; Song, C.F.; Shan, S.D. Concentrated biogas slurry enhanced soil fertility and tomato quality. Acta Agric. Scand. Sect. B Soil Plant Sci. 2010, 60, 262–268. [Google Scholar] [CrossRef]
- Abubaker, J.; Risberg, K.; Pell, M. Biogas residues as fertilisers-effects on wheat growth and soil microbial activities. Appl. Energy 2012, 99, 126–134. [Google Scholar] [CrossRef]
- Theuerl, S.; Herrmann, C.; Heiermann, M.; Grundmann, P.; Landwehr, N.; Kreidenweis, U.; Prochnow, A. The Future Agricultural Biogas Plant in Germany: A Vision. Energies 2019, 12, 396. [Google Scholar] [CrossRef] [Green Version]
- Tao, X.; Shang, B.; Dong, H.; Chen, Y.; Huang, H. Effects of anaerobically digested pig slurry on in vitro growth crop pathogenic fungi. Animal production technology. In Proceedings of the International Conference of Agricultural Engineering-CIGR-AgEng 2012: Agriculture and Engineering for a Healthier Life, Valencia, Spain, 8–12 July 2012. [Google Scholar]
- Amari, M.; Toyota, K.; Islam, M.S.; Masuda, K.; Kuroda, T.; Watanabe, A. Effect of the addition of anaerobically digested slurry to soil and hydroponics on soil-borne plant disease. Soil Microorg. 2008, 62, 106–113. (In Japanese) [Google Scholar]
- Cao, Y.; Chang, Z.; Wang, J.; Ma, Y.; Yang, H.; Fu, G. Potential use of anaerobically digested manure slurry to suppress Phytophthora root rot of chilli pepper. Sci. Hortic. 2014, 168, 124–131. [Google Scholar] [CrossRef]
- Katsube, K. Studies on Fusarium Wilt of Spinach [Spinacia oleracea], caused by Fusarium oxysporum f. sp. spinaciae. Bull. Iwate Agric. Res. Cent. 2001. Available online: https://agris.fao.org/agris-search/search.do?recordID=JP2002006095 (accessed on 20 March 2022).
- Komada, H. Development of a selective medium for quantitative isolation of Fusarium oxysporum from natural soil. Rev. Plant Prot. Res. 1975, 8, 114–124. [Google Scholar]
- Wick, R.R.; Louise, J.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanizawa, Y.; Fujisawa, T.; Nakamura, Y. DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 2018, 34, 1037–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Wu, H.; Yan, S.; Guo, D.; Wang, G.; Ma, Y. Soil chemical and microbial responses to biogas slurry amendment and its effect on Fusarium wilt suppression. Appl. Soil Ecol. 2016, 107, 116–123. [Google Scholar] [CrossRef]
- Moreno-Velandia, C.A.; Izquierdo-García, L.F.; Ongena, M.; Kloepper, J.W.; Cotes, A.M. Soil sterilization, pathogen and antagonist concentration affect biological control of Fusarium wilt of cape gooseberry by Bacillus velezensis Bs006. Plant Soil 2019, 435, 39–55. [Google Scholar] [CrossRef] [Green Version]
- Dunlap, C.A.; Kim, S.J.; Kwon, S.W.; Rooney, A.P. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and Bacillus oryzicola are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int. J. Syst. Evol. Microbiol. 2016, 66, 1212–1217. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Wang, C.; Song, X.; Ding, X.; Wu, L.; Wu, H.; Gao, X.; Borriss, R. Bacillus velezensis FZB42 in 2018: The gram-positive model strain for plant growth promotion and biocontrol. Front. Microbiol. 2018, 9, 2491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Y.; Zhu, H.; Luo, S.; Yang, W.; Zhang, L.; Li, S.; Jin, Q.; Cao, Q.; Sun, S.; Xiao, M. Role of maize root exudates in promotion of colonization of Bacillus velezensis strain S3-1 in rhizosphere soil and root tissue. Curr. Microbiol. 2019, 76, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shan, Y.; Li, Y.; Li, Q.; Wu, C. The soil nutrient environment determines the strategy by which Bacillus velezensis HN03 suppresses Fusarium wilt in banana plants. Front. Plant Sci. 2020, 11, 1801. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-García, C.; Béjar, V.; Martínez-Checa, F.; Llamas, I.; Quesada, E. Bacillus velezensis sp. nov., a surfactant-producing bacterium isolated from the river Vélez in Málaga, southern Spain. Int. J. Syst. Evol. Microbiol. 2005, 55, 191–195. [Google Scholar] [CrossRef] [Green Version]
- Min, S.C.; Yong, J.J.; Bo, K.K.; Yu, K.P.; Kim, C.K.; Dong, S.P. Understanding the ontogeny and succession of Bacillus velezensis and B. subtilis subsp. subtilis by focusing on kimchi fermentation. Sci. Rep. 2018, 8, 7045. [Google Scholar]
- Cao, Y.; Pi, H.; Chandrangsu, P.; Li, Y.; Wang, Y.; Zhou, H.; Xiong, H.; Helmann, J.D.; Cai, Y. Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci. Rep. 2018, 8, 4360. [Google Scholar] [CrossRef]
- Xu, Z.; Shao, J.; Li, B.; Yan, X.; Shen, Q.; Zhang, R. Contribution of bacillomycin D in Bacillus amyloliquefaciens SQR9 to antifungal activity and biofilm formation. Appl. Environ. Microbiol. 2013, 79, 808–815. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, S.P.; Dietel, K.; Rändler, M.; Schmid, M.; Junge, H.; Borriss, R.; Hartman, A.; Grosch, R. Effects of Bacillus amyloliquefaciens FZB42 on lettuce growth and health under pathogen pressure and its impact on the rhizosphere bacterial community. PLoS ONE 2013, 8, e68818. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.C.; Liu, C.H.; Wang, B.T.; Xue, Y.R. Genomic and metabolic traits endow Bacillus velezensis CC09 with a potential biocontrol agent in control of wheat powdery mildew disease. Microbiol. Res. 2017, 196, 89–94. [Google Scholar] [CrossRef]
- Koumoutsi, A.; Chen, X.H.; Henne, A.; Liesegang, H.; Hitzeroth, G.; Franke, P.; Vater, J.; Borriss, R. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB4. J. Bacteriol. 2004, 186, 1084–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Schneider, K.; Vater, J.; Süssmuth, R.; Piel, J.; Borriss, R. Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J. Biotechnol. 2009, 140, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Palazzini, J.M.; Dunlap, C.A.; Bowman, M.J.; Chulze, S.N. Bacillus velezensis RC 218 as a biocontrol agent to reduce Fusarium head blight and deoxynivalenol accumulation: Genome sequencing and secondary metabolite cluster profiles. Microbiol. Res. 2016, 192, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Yokota, K.; Hayakawa, H. Impact of antimicrobial lipopeptides from Bacillus sp. on suppression of Fusarium yellows of tatsoi. Microbes Environ. 2015, 30, 281–283. [Google Scholar] [CrossRef] [Green Version]
- Ling, N.; Xue, C.; Huang, Q.; Yang, X.; Xu, Y.; Shen, Q. Development of a mode of application of bioorganic fertilizer for improving the biocontrol efficacy to Fusarium wilt. Biocontrol 2010, 55, 673–683. [Google Scholar] [CrossRef]
- El-Hassan, S.; Gowen, S. Formulation and delivery of the bacterial antagonist Bacillus subtilis for management of lentil vascular wilt caused by Fusarium oxysporum f.sp. lentis. J. Phytopathol. 2006, 154, 148–155. [Google Scholar] [CrossRef]
| Sample 1 | Rate of Source Materials | Processing Condition | Microbial Immobilization Method | Pre-Treatment | Facility Location | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dairy Cow Manure | Sewage Sludge | Food Garbage | Pig Manure | Night Soil Sludge | Treatment Temperature | HRT 2 | ||||
| (%) | (%) | (%) | (%) | (%) | (°C) | (Days) | ||||
| AD | 100 | 35–36 | 45 | Anaerobic fluidized bed process | Solid liquid separation | Tochigi prefecture | ||||
| AS | 100 | 35–36 | 30 | Anaerobic contact process | Gravitation enrichment | Yamagata prefecture | ||||
| AF | 100 | 35–36 | 20 | Anaerobic fluidized bed process | Acid fermentation | Hokkaido | ||||
| APF | 10 | 90 | 35–36 | 30 | Anaerobic fluidized bed process | Crush | Okayama prefecture | |||
| ASNF | 74 | 9 | 17 | 35–36 | 30 | Anaerobic fluidized bed process | Acid fermentation | Fukuoka prefecture | ||
| Sample * | Mycelial Growth (mm) | |||
|---|---|---|---|---|
| Raw | Filtrate | |||
| AD | 56.1 | b | 85.6 | NS |
| AS | 55.9 | b | 85.2 | |
| AF | 60.8 | b | 85.5 | |
| APF | 57.6 | b | 86.3 | |
| ASNF | 53.5 | b | 86.4 | |
| Control | 82.9 | a | 83.3 | |
| Treatment | IE (%) 2 | ||||||
|---|---|---|---|---|---|---|---|
| F | F+AD-3 | Untreated | |||||
| Exp 1 1 | 77.8 | a | 27.8 | b | 0.0 | c | 64.3 |
| Exp 2 | 95.8 | a | 53.6 | b | 0.0 | c | 44.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sugiyama, T.; Natsuaki, K.T.; Tanaka, N.; Shiwa, Y.; Irie, M. Antagonism of Bacillus velezensis Isolate from Anaerobically Digested Dairy Slurry against Fusarium Wilt of Spinach. Agronomy 2022, 12, 1058. https://doi.org/10.3390/agronomy12051058
Sugiyama T, Natsuaki KT, Tanaka N, Shiwa Y, Irie M. Antagonism of Bacillus velezensis Isolate from Anaerobically Digested Dairy Slurry against Fusarium Wilt of Spinach. Agronomy. 2022; 12(5):1058. https://doi.org/10.3390/agronomy12051058
Chicago/Turabian StyleSugiyama, Tomomi, Keiko T. Natsuaki, Naoto Tanaka, Yuh Shiwa, and Mami Irie. 2022. "Antagonism of Bacillus velezensis Isolate from Anaerobically Digested Dairy Slurry against Fusarium Wilt of Spinach" Agronomy 12, no. 5: 1058. https://doi.org/10.3390/agronomy12051058
APA StyleSugiyama, T., Natsuaki, K. T., Tanaka, N., Shiwa, Y., & Irie, M. (2022). Antagonism of Bacillus velezensis Isolate from Anaerobically Digested Dairy Slurry against Fusarium Wilt of Spinach. Agronomy, 12(5), 1058. https://doi.org/10.3390/agronomy12051058





