Does the Use of an Intercropping Mixture Really Improve the Biology of Monocultural Soils?—A Search for Bacterial Indicators of Sensitivity and Resistance to Long-Term Maize Monoculture
Abstract
:1. Introduction
2. Materials and Methods
2.1. Description of the Study Area and Soil Sampling Procedure
2.2. Soil Chemical Features
2.3. Soil Biological Activities
2.4. Next Generation Sequencing and Bioinformatic Analysis
3. Results
3.1. Soil Chemical Characteristics
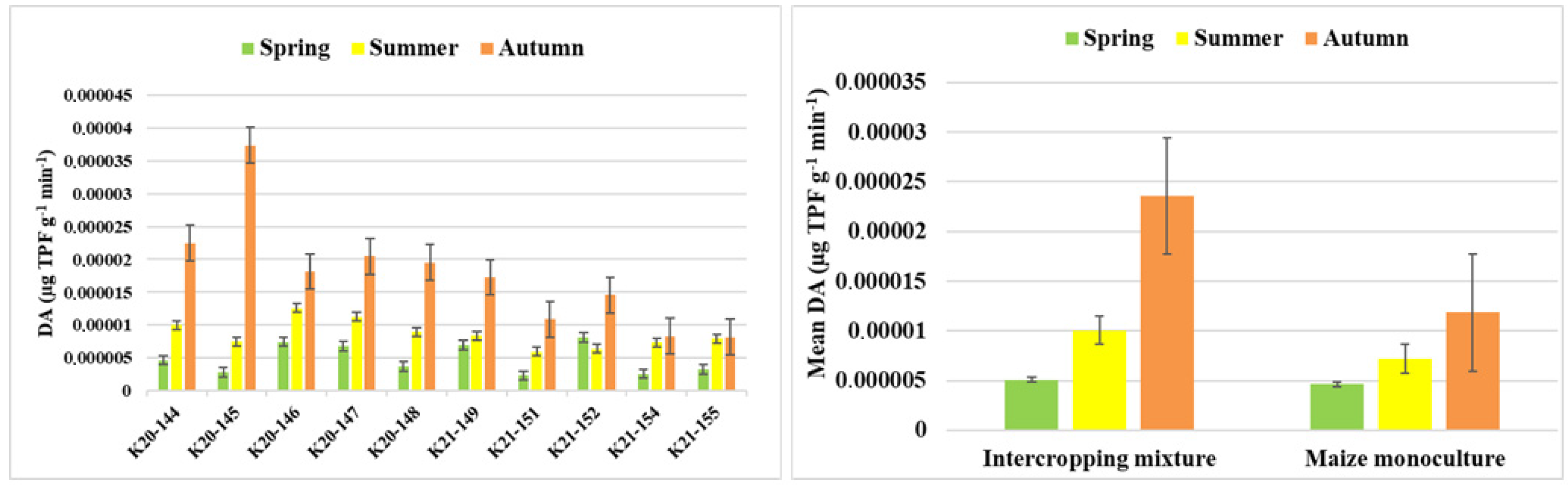
3.2. Soil Biological Activities
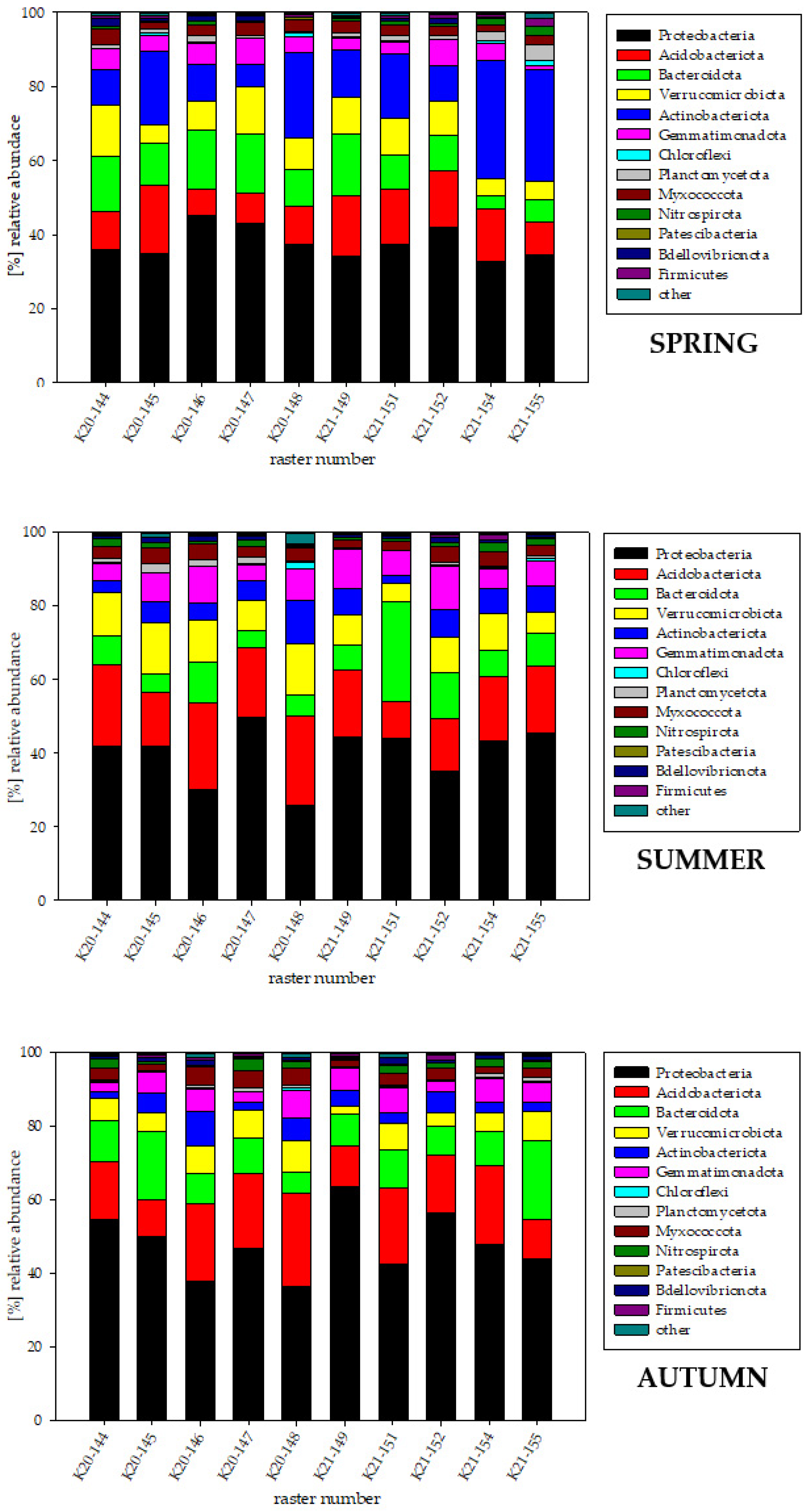
3.3. Bacterial Biodiversity at the Phylum Level
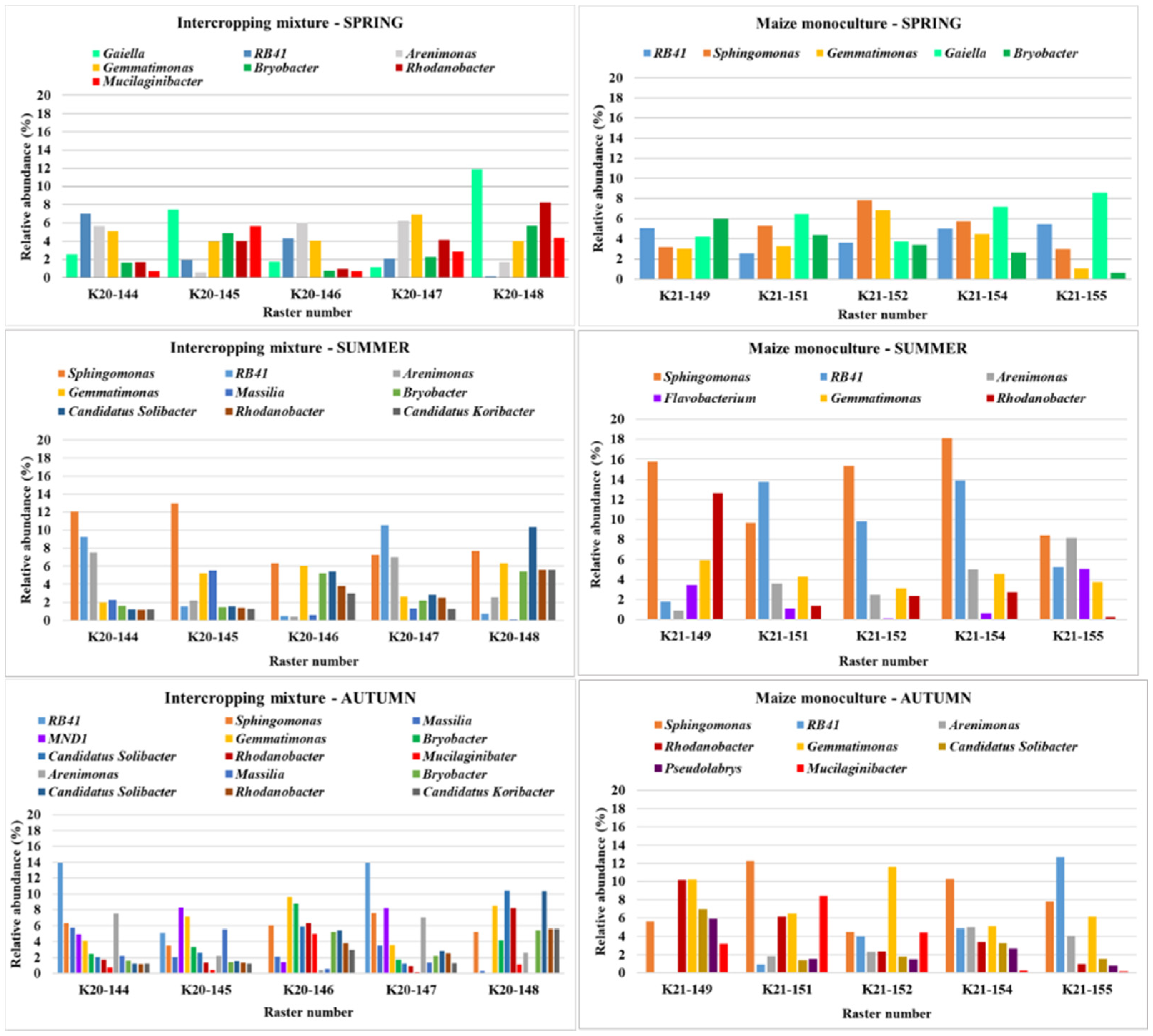
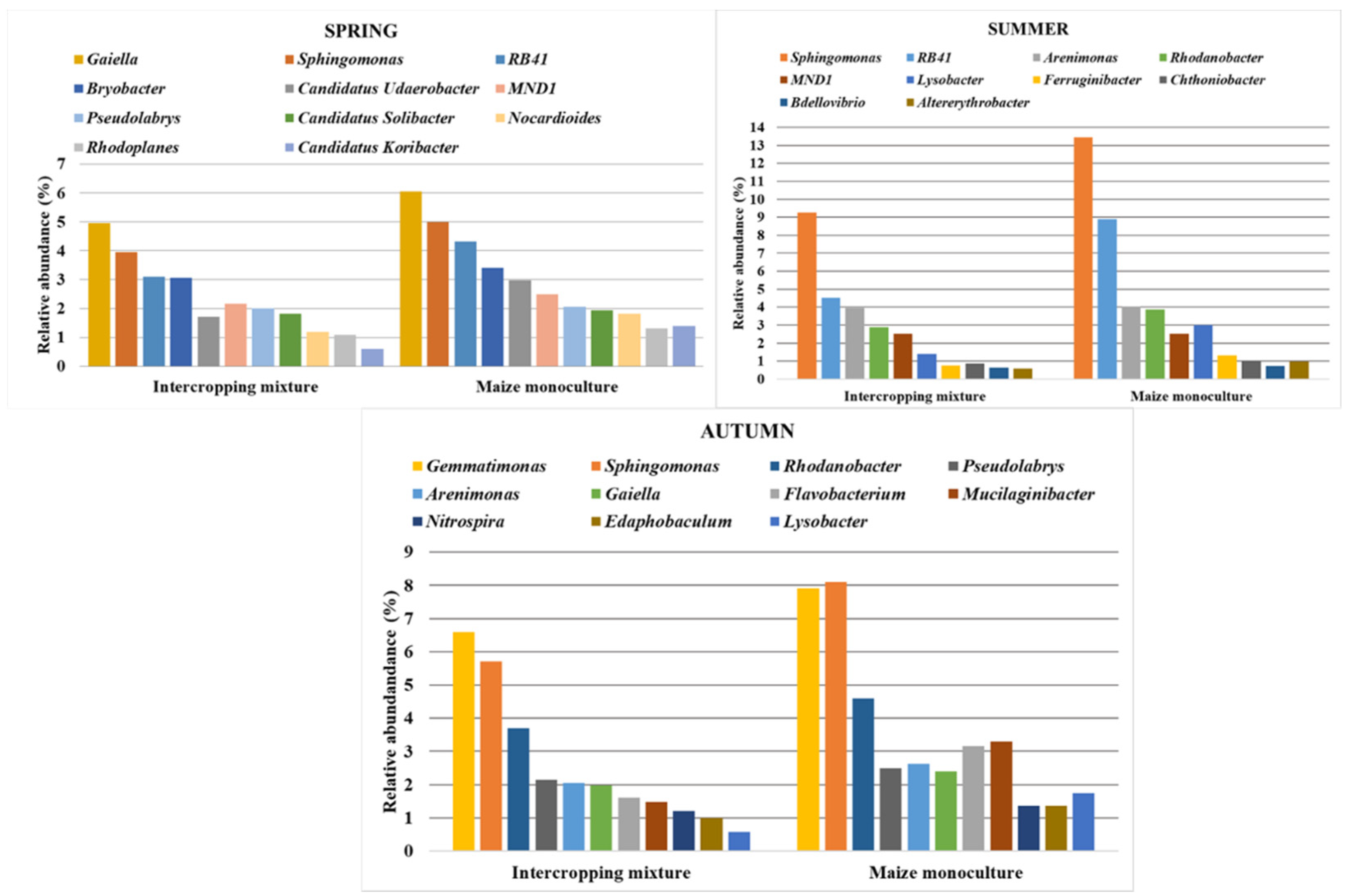
3.4. Bacterial Biodiversity at the Genus Level
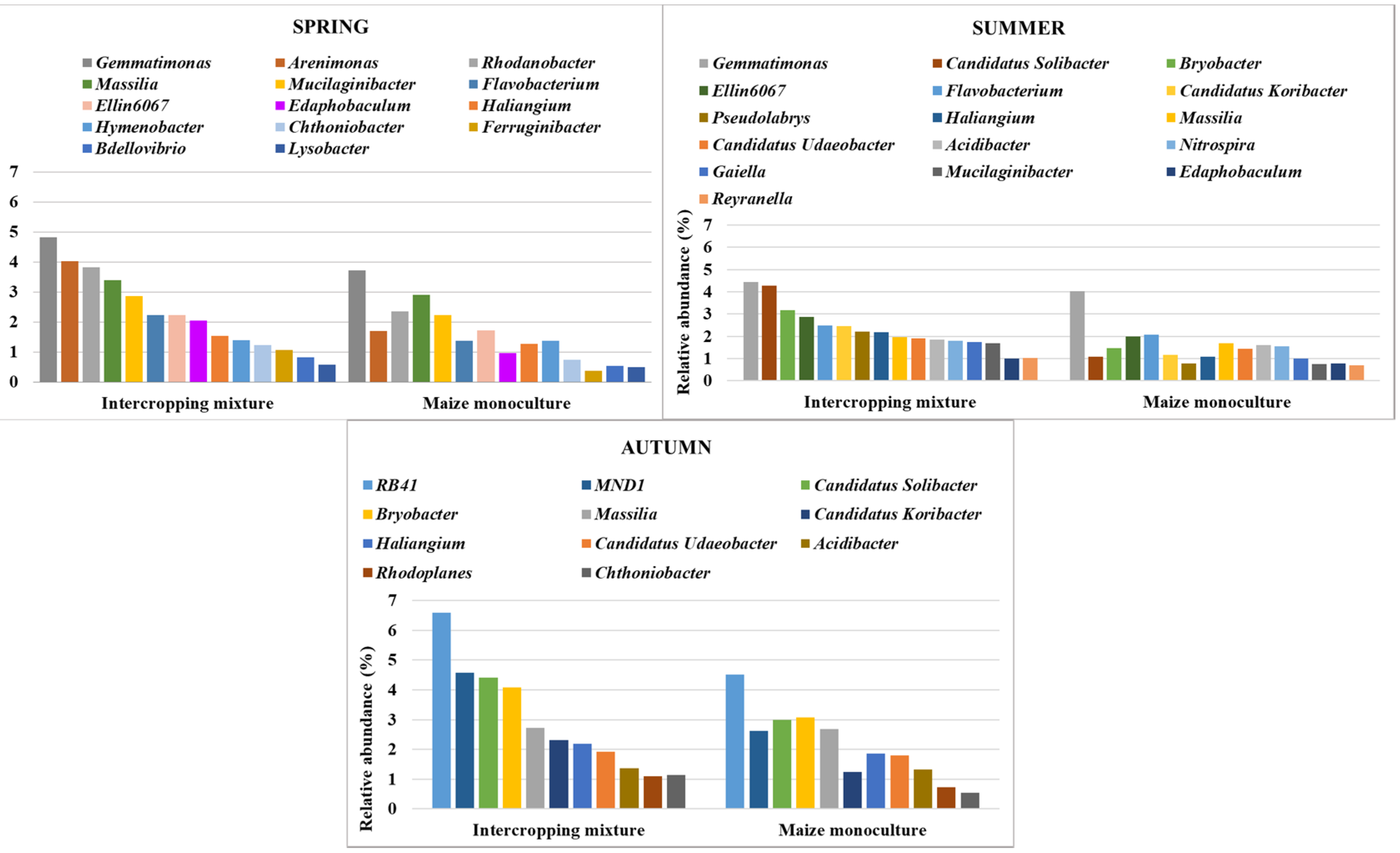
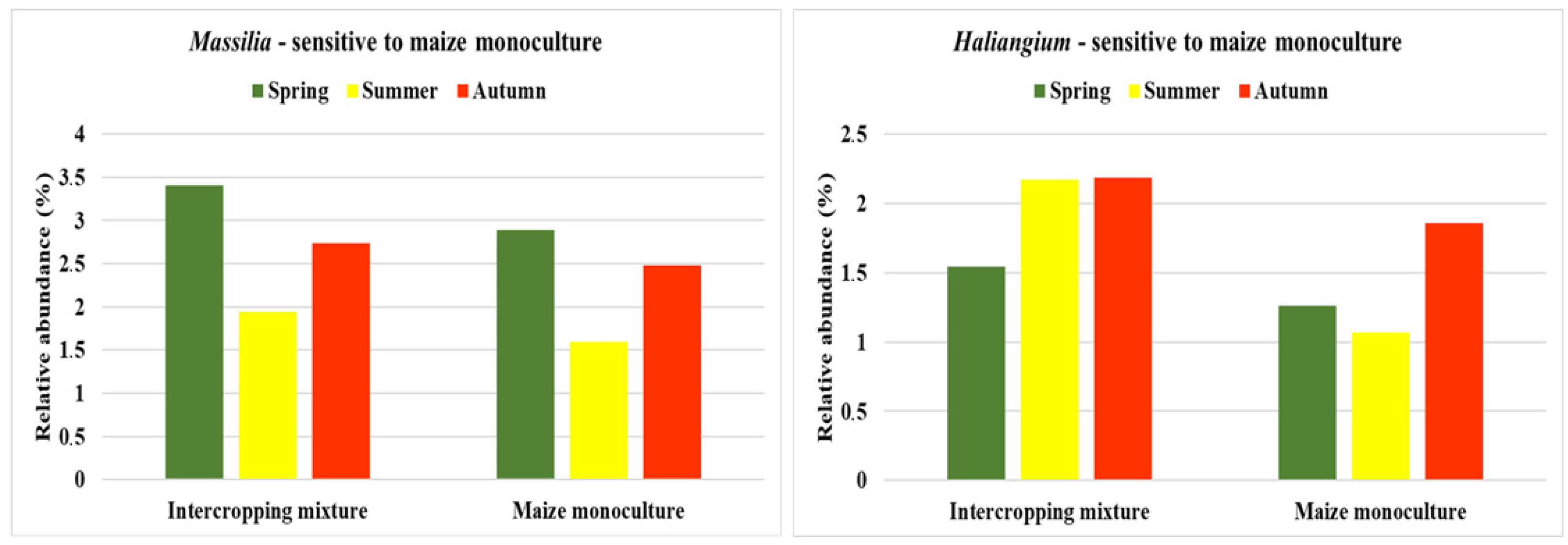
3.5. Bacterial Indicators of Sensitivity and Resistance to Maize Monoculture
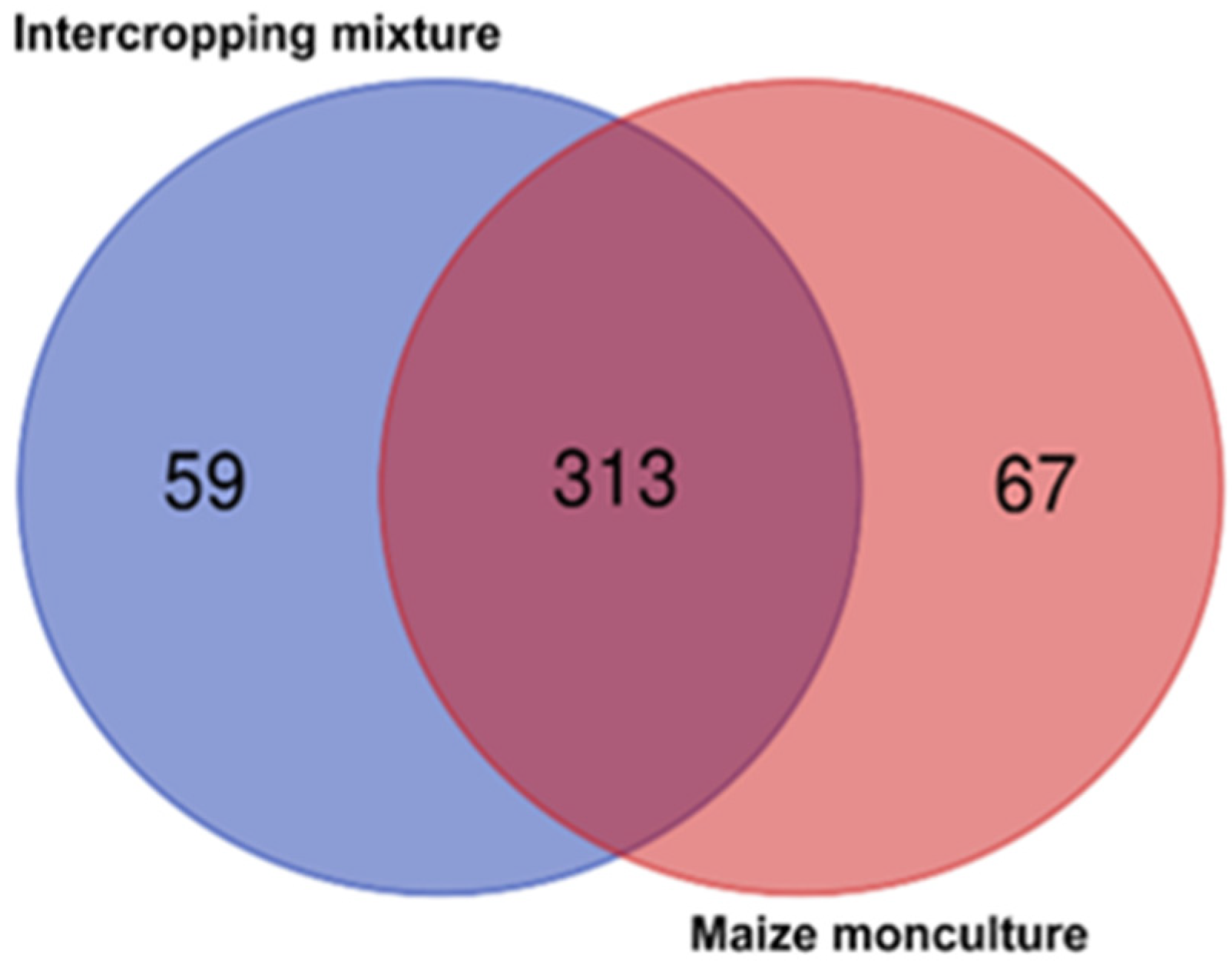
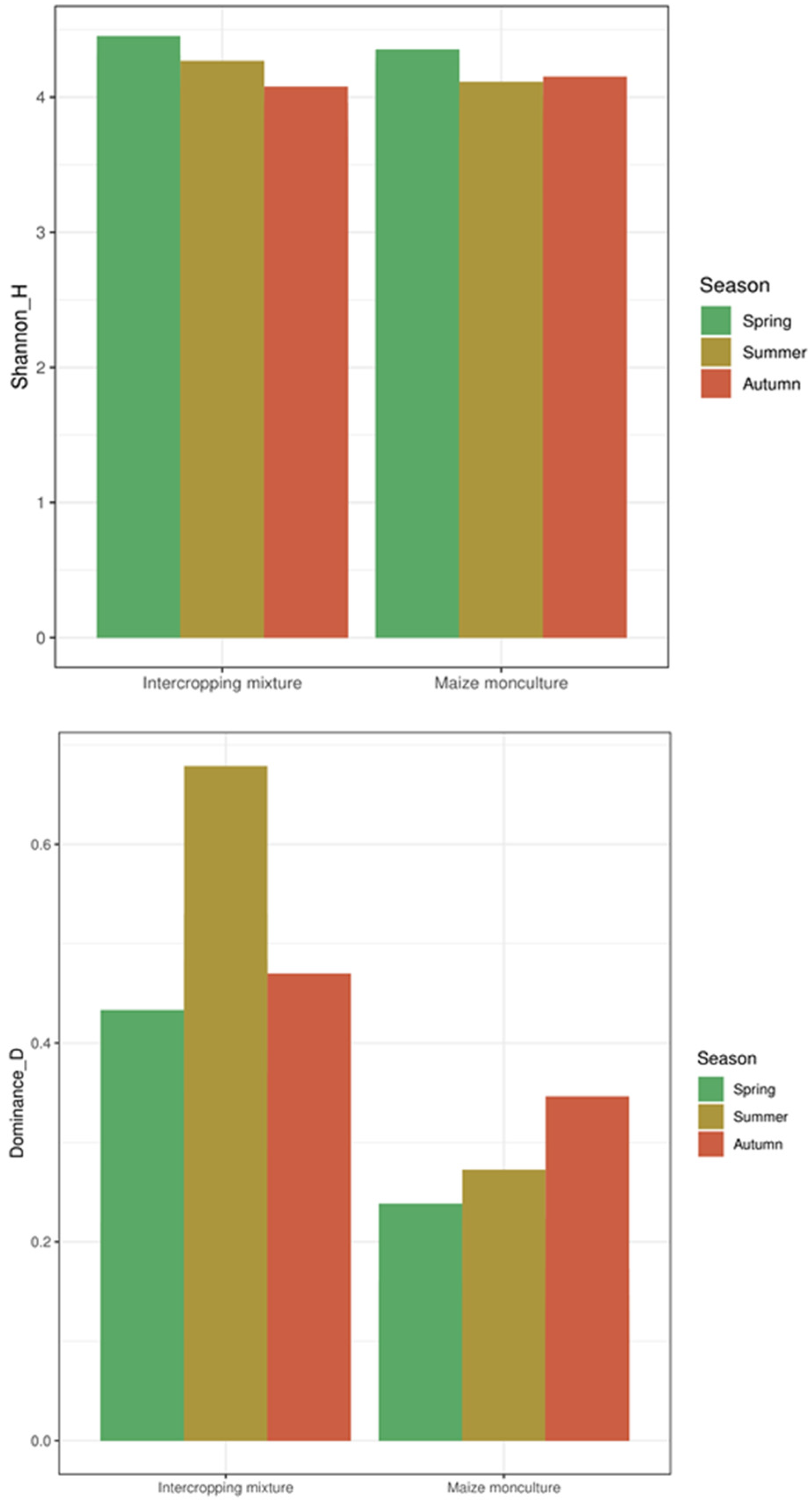
3.6. Biodiversity Indices and Beta-Diversity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, X.; Wang, Y.; Sun, L.; Qi, X.; Song, F.; Zhu, X. Impact of maize–mushroom intercropping on the soil bacterial community composition in Northeast China. Agronomy 2020, 10, 1526. [Google Scholar] [CrossRef]
- Zhu, X.C.; Sun, L.Y.; Song, F.B.; Liu, S.Q.; Liu, F.L.; Li, X.N. Soil microbial community and activity are affected by integrated agricultural practices in China. Eur. J. Soil Sci. 2018, 69, 924–935. [Google Scholar] [CrossRef]
- Ullah, S.; Ai, C.; Huang, S.; Song, D.; Abbas, T.; Zhang, J.; Zhou, W.; He, P. Substituting ecological intensification of agriculture for conventional agricultural practices increased yield and decreased nitrogen losses in North China. Appl. Soil Ecol. 2020, 147, 103395. [Google Scholar] [CrossRef]
- Uchida, S.; Hayashi, K. Comparative life cycle assessment of improved and conventional cultivation practices for energy crops in Japan. Biomass Bioenergy 2012, 36, 302–315. [Google Scholar] [CrossRef]
- Heinze, S.; Vogel, A. Reversion from organic to conventional agriculture in Germany. Build. Org. Brid. 2014, 20, 347–350. [Google Scholar]
- Stroud, J.L. Soil health pilot study in England: Outcomes from an on-farm earthworm survey. PLoS ONE 2019, 14, e0203909. [Google Scholar] [CrossRef] [Green Version]
- Domagała–Świątkiewicz, I.; Gąstoł, M. Soil chemical properties under organic and conventional crop management systems in south Poland. Biol. Agric. Hortic. 2013, 29, 12–28. [Google Scholar] [CrossRef]
- Munkholm, L.J.; Heck, R.J.; Deen, B. Long-term rotation and tillage effects on soil structure and crop yield. Soil Tillage Res. 2013, 127, 85–91. [Google Scholar] [CrossRef]
- Doula, M.K.; Sarris, A. Chapter 4-Soil environment. In Environment and Development; Poulopoulos, S.G., Inglezakis, V.J., Eds.; Elsevier: Amsterdam, The Netherland, 2016; pp. 213–286. [Google Scholar]
- Zhang, B.; Li, Y.; Ren, T.; Tian, Z.; Wang, G.; He, X.; Tian, C. Short-term effect of tillage and crop rotation on microbial community structure and enzyme activities of a clay loam soil. Biol. Fertil. Soils 2014, 50, 1077–1085. [Google Scholar] [CrossRef]
- Chen, H.; Hou, R.; Gong, Y.; Li, H.; Fan, M.; Kuzyakov, Y. Effects of 11 years of conservation tillage on soil organic matter fractions in wheat monoculture in Loess Plateau of China. Soil Tillage Res. 2009, 106, 85–94. [Google Scholar] [CrossRef]
- Griffiths, B.S.; Philippot, L. Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol. Rev. 2013, 37, 112–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coudrain, V.; Hedde, M.; Chauvat, M.; Maron, P.; Bourgeois, E.; Mary, B.; Léonard, J.; Ekelund, F.; Villenave, C.; Recous, S. Temporal differentiation of soil communities in response to arable crop managenment strategies. Agric. Ecosyst. Environ. 2016, 225, 12–21. [Google Scholar] [CrossRef]
- Li, L.; Zhang, L.; Zhang, F. Crop mixtures and the mechanisms of overyielding. Encycl. Biodivers. 2013, 2, 382–395. [Google Scholar]
- Askari, M.S.; Holden, N.M. Indices for quantitative evaluation of soil quality under grassland management. Geoderma 2014, 230–231, 131–142. [Google Scholar] [CrossRef]
- Wolińska, A.; Rekosz–Burlaga, H.; Goryluk–Salmonowicz, A.; Błaszczyk, M.K.; Stępniewska, Z. Bacterial Abundance and Dehydrogenase Activity in Selected Agricultural Soils from Lublin Region. Pol. J. Environ. Stud. 2015, 24, 2677–2682. [Google Scholar] [CrossRef]
- Brooker, R.W.; Bennett, A.E.; Cong, W.F.; Daniell, T.J.; George, T.S.; Hallett, P.D.; Hawes, C.; Iannetta, P.P.M.; Jones, H.G.; Karley, A.J.; et al. Improving intercropping: A synthesis of research in agronomy, plant physiology and ecology. New Phytol. 2015, 206, 107–117. [Google Scholar] [CrossRef]
- Zhao, Q.; Xiong, W.; Xing, Y.; Sun, Y.; Lin, X.; Dong, Y. Long–Term Coffee Monoculture Alters Soil Chemical Properties and Microbial Communities. Sci. Rep. 2018, 8, 6116. [Google Scholar] [CrossRef]
- Stomph, T.; Dordas, C.; Baranger, A.; Bedoussac, L.; de Rijk, J.; Dong, B.; Evers, J.; Gu, C.; Li, L.; Simon, J.; et al. Designing intercrops for high yield, yield stability and efficient use of resources: Are there principles? In Advances in Agronomy; Academic Press: Cambridge, MA, USA, 2020; Volume 160, pp. 1–50. [Google Scholar]
- Zhu, J.; van der Werf, W.; Anten, N.P.R.; Vos, J.; Evers, J.B. The contribution of phenotypic plasticity to complementary light capture in plant mixtures. New Phytol. 2015, 207, 1213–1222. [Google Scholar] [CrossRef]
- Zaeem, M.; Nadeem, M.; Pham, T.H.; Ashiq, W.; Ali, W.; Gilani, S.S.M.; Elavarthi, S.; Kavanagh, V.; Cheema, M.; Galagedara, L.; et al. The potential of corn-soybean intercropping to improve the soil health status and biomass production in cool climate boreal ecosystems. Sci. Rep. 2019, 9, 13148. [Google Scholar] [CrossRef] [Green Version]
- Du, Q.; Zhou, L.; Chen, P.; Liu, X.; Song, C.; Yang, F.; Wang, X.; Liu, W.; Sun, X.; Du, J.; et al. Relay-intercropping soybean with maize maintains soil fertility and increases nitrogen recovery efficiency by reducing nitrogen input. Crop J. 2020, 8, 140–152. [Google Scholar] [CrossRef]
- Bedoussac, L.; Journet, E.P.; Hauggaard–Nielsen, H.; Naudin, C.; Corre–Hellou, G.; Jensen, E.S.; Prieur, L.; Justes, E. Ecological principles under lying the increase of productivity achieved by cereal—Grain legume intercrops in organic farming. A review. Agron. Sustain. Dev. 2015, 35, 911–935. [Google Scholar] [CrossRef]
- Lithourgidis, A.; Dordas, C.; Damalas, C.; Vlachostergios, D. Annual Intercrops: An Alternative Pathway for Sustainable Agriculture. Aust. J. Crop Sci. 2011, 5, 396–410. [Google Scholar]
- Pankou, C.; Lithourgidis, A.; Dordas, C. Effect of Irrigation on Intercropping Systems of Wheat (Triticum aestivum L.) with Pea (Pisum sativum L.). Agronomy 2021, 11, 283. [Google Scholar] [CrossRef]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of Climate Change on Crops Adaptation and Strategies to Tackle Its Outcome: A Review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [Green Version]
- Bedoussac, L.; Journet, E.-P.; Hauggaard-Nielsen, H.; Naudin, C.; Corre-Hellou, G.; Jensen, E.S.; Justes, E. Grainlegume—cerealinter cropping systems. In Achieving Sustainable Cultivation of Grain Legumes Volume 1: Advances in Breeding and Cultivation Techniques; Sivasankar, S., Ed.; Burleigh Dodds Science Publishing: Cambridge, UK, 2018. [Google Scholar]
- Tang, X.; Zhang, C.; Yu, Y.; Shen, J.; van der Werf, W.; Zhang, F. Intercropping legumes and cereals increases phosphorus use efficiency; a meta-analysis. Plant Soil 2021, 460, 89–104. [Google Scholar] [CrossRef]
- Xu, Y.; Feng, J.; Li, H. How intercropping and mixed systems reduce cadmium concentration in rice grains and improve grain yields. J. Hazard. Mater. 2021, 402, 123762. [Google Scholar] [CrossRef]
- Schulz, V.S.; Schumann, C.; Weisenburger, S.; Müller-Lindenlauf, M.; Stolzenburg, K.; Möller, K. Row Intercropping Maize (Zea mays L.) with Biodiversity Enhancing Flowering-Partners—Effect on Plant Growth, Silage Yield, and Composition of Harvest Material. Agriculture 2020, 10, 524. [Google Scholar] [CrossRef]
- Szafranek-Nakonieczna, A.; Stępniewska, Z. Aerobic and anaerobic respiration in profiles of Polesie Lubelskie peatlands. Int. Agrophysics 2014, 28, 219–229. [Google Scholar] [CrossRef]
- Casida, L.E.; Klein, D.A.; Santoro, T. Soil dehydrogenase activity. Soil Sci. 1964, 98, 371–376. [Google Scholar] [CrossRef]
- Wolińska, A.; Kuźniar, A.; Gałązka, A. Biodiversity in the rhizosphere of selected winter wheat (Triticum aestivum L.) cultivars—genetic and catabolic fingerprinting. Agronomy 2020, 10, 953. [Google Scholar] [CrossRef]
- Thijs, S.; Op De Beeck, M.; Beckers, B.; Truyens, S.; Stevens, V.; Van Hamme, J.D.; Weyens, N.; Vangronsveld, J. Comparative evaluation of four bacteria-specific primer pairs for 16S rRNA gene surveys. Front. Microbiol. 2017, 8, 494. [Google Scholar] [CrossRef] [PubMed]
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Banach, A.; Izak, D.; Stępniewska, Z.; Błaszczyk, M. Metagenomic analysis of some potential nitrogen-fixing bacteria in arable soils at different formation processes. Microb. Ecol. 2017, 73, 162–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuźniar, A.; Włodarczyk, K.; Grządziel, J.; Goraj, W.; Gałązka, A.; Wolińska, A. Culture-independent analysis of an endophytic core microbiome in two species of wheat: Triticum aestivum L. (cv. ‘Hondia’) and the first report of microbiota in Triticum spelta L. (cv. ‘Rokosz’). Syst. Appl. Microbiol. 2020, 43, 126025. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Wright, E.S. Using DECIPHER v2.0 to Analyze Biological Sequence Data in R. R J. 2016, 8, 352–359. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Reeve, J.R.; Hoagland, L.A.; Villalba, J.J.; Carr, P.M.; Atucha, A.; Cambardella, C.; Davis, D.R.; Delate, K. Chapter Six—Organic Farming, Soil Health, and Food Quality: Considering Possible Links. In Advances in Agronomy; Academic Press: Cambridge, MA, USA, 2016; Volume 137, pp. 319–367. [Google Scholar]
- Lemaire, G.; Franzluebbers, A.; Carvalho, P.C.D.F.; Dedieu, B. Integrated crop-livestock systems: Strategies to achieve synergy between agricultural production and environmental quality. Agric. Ecosyst. Environ. 2014, 190, 4–8. [Google Scholar] [CrossRef]
- Vukicevich, E.; Lowery, T.; Bowen, P.; Úrbez-Torres, J.R.; Hart, M. Cover crops to increase soil microbial diversity and mitigate decline in perennial agriculture. A review. Agron. Sustain. Dev. 2016, 36, 48. [Google Scholar] [CrossRef] [Green Version]
- Bukovsky-Reyes, S.; Isaac, M.E.; Blesh, J. Effects of intercropping and soil properties on root functional traits of cover crops. Agric. Ecosyst. Environ. 2019, 285, 106614. [Google Scholar] [CrossRef]
- Hinsinger, P.; Betencourt, E.; Bernard, L.; Brauman, A.; Plassard, C.; Shen, J.; Tang, X.; Zhang, F. P for two, sharing a scarce resource: Soil phosphorus acquisition in the rhizosphere of intercropped species. Plant Physiol. 2011, 156, 1078–1086. [Google Scholar] [CrossRef] [Green Version]
- Sun, F.; Pan, K.; Olatunji, O.A.; Li, Z.; Chen, W.; Zhang, A.; Song, D.; Sun, X.; Huang, D.; Tan, X. Specific legumes allay drought effects on soil microbial food web activities of the focal species in agroecosystem. Plant Soil 2019, 437, 455–471. [Google Scholar] [CrossRef]
- Lopes, A.R.; Faria, C.; Prieto-Fernandez, A.; Trasar-Cepeda, C.; Manaia, C.M.; Nunes, O.C. Comparative study of the microbial diversity of bulk paddy soil of two rice fields subjected to organic and conventional farming. Soil Biol. Biochem. 2011, 43, 115–125. [Google Scholar] [CrossRef] [Green Version]
- Wolińska, A.; Szafranek-Nakonieczna, A.; Banach, A.; Rekosz-Burlaga, H.; Goryluk-Salmonowicz, A.; Błaszczyk, M.; Stępniewska, Z.; Górski, A. Biological degradation of arable soils from Lublin region (SE Poland). Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 558–571. [Google Scholar]
- Uzarowicz, Ł.; Wolińska, A.; Błońska, E.; Szafranek-Nakonieczna, A.; Kuźniar, A.; Słodczyk, Z.; Kwasowski, W. Technogenic soils (Technosols) developed from mine spoils containing Fe sulphides: Microbiological activity as an indicator of soil development following land reclamation. Appl. Soil Ecol. 2020, 156, 103699. [Google Scholar] [CrossRef]
- Grządziel, J.; Furtak, K.; Gałązka, A. Community-level physiological profiles of microorganisms from different types of soils that are characteristic to Poland-a long term microplot experiment. Sustainability 2019, 11, 56. [Google Scholar] [CrossRef] [Green Version]
- Czerwonka, G.; Konieczna, I.; Żarnowiec, P.; Zieliński, A.; Malinowska-Gniewosz, A.; Gałuszka, A.; Migaszewski, Z.; Szarlip, P. Characterization of microbial communities in acidified, Sulphur containing soils. Pol. J. Microbiol. 2017, 66, 509–517. [Google Scholar] [CrossRef] [Green Version]
- Wolińska, A.; Górniak, D.; Zielenkiewicz, U.; Goryluk-Salmonowicz, A.; Kuźniar, A.; Stępniewska, Z.; Błaszczyk, M. Microbial biodiversity in arable soils is affected by agricultural practices. Int. Agrophysics 2017, 31, 259–271. [Google Scholar] [CrossRef] [Green Version]
- Minasny, B.; Hong, S.Y.; Hartemink, A.E.; Kim, Y.H.; Kang, S.S. Soil pH increase under paddy in South Korea between 2000 and 2012. Agric. Ecosyst. Environ. 2016, 221, 205–213. [Google Scholar] [CrossRef]
- Neina, D. The Role of Soil pH in Plant Nutrition and Soil Remediation. Appl. Environ. Soil Sci. 2019, 2019, 9. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Lauber, C.L.; Weintraub, M.N.; Ahmed, B.; Allison, S.D.; Crenshaw, C.; Contosta, A.R.; Cusack, D.; Frey, S.; Gallo, M.E.; et al. Stoichiometry of soil enzyme activity at global scale. Ecol. Lett. 2008, 11, 1252–1264. [Google Scholar] [CrossRef]
- Pawar, R.M. The effect of soil pH on bioremediation of polycyclic aromatic hydrocarbons (PAHS). J. Bioremediation Biodegrad. 2015, 6, 291–304. [Google Scholar] [CrossRef]
- Husson, O. Redox potential (Eh) and pH as drivers of soil/plant/microorganism systems: A transdisciplinary overview pointing to integrative opportunities for agronomy. Plant Soil 2012, 362, 389–417. [Google Scholar] [CrossRef] [Green Version]
- Błońska, E. Enzyme activity in forest peat soils. Folia For. Pol. 2010, 52, 20. [Google Scholar]
- Brzezińska, M.; Stępniewska, Z.; Stępniewski, W. Soil oxygen status and dehydrogenase activity. Soil Biol. Biochem. 1998, 30, 1783. [Google Scholar] [CrossRef]
- Włodarczyk, T. Some aspects of dehydrogenase activity in soils. Int. Agrophysics 2000, 14, 365–376. [Google Scholar]
- Wu, C.S.; Liu, G.H.; Ma, G.X.; Liu, Q.S.; Yu, F.; Huang, C.; Zhao, Z.; Liang, L. Study of the differences in soil properties between the dry season and rainy season in the Mun River basin. Catena 2019, 182, 104103. [Google Scholar] [CrossRef]
- Upton, R.N.; Bach, E.M.; Hofmockel, K.S. Spatio-temporal microbial community dynamics within soil aggregates. Soil Biol. Biochem. 2019, 132, 58–68. [Google Scholar] [CrossRef]
- Zhang, P.; Cui, Z.; Guo, M.; Xi, R. Characteristics of the soil microbial community in the forestland of Camellia oleifera. PeerJ 2019, 8, e9117. [Google Scholar] [CrossRef]
- Wolińska, A.; Włodarczyk, K.; Kuźniar, A.; Marzec-Grządziel, A.; Grządziel, J.; Gałązka, A.; Uzarowicz, Ł. Soil Microbial Community Profiling and Bacterial Metabolic Activity of Technosols as an Effect of Soil Properties following Land Reclamation: A Case Study from the Abandoned Iron Sulphide and Uranium Mine in Rudki (South-Central Poland). Agronomy 2020, 10, 1795. [Google Scholar] [CrossRef]
- Sadaf, K.; Anirban, B.; Iqbal, A.; Sayyed, R.Z.; El-Enshasy, H.A.; Dailin, D.J.; Suriani, N.L. Recent Understanding of Soil Acidobacteria and Their Ecological Significance: A Critical Review. Front. Microbiol. 2020, 11, 2712. [Google Scholar]
- Männistö, M.K.; Tiirola, M.; Haggblom, M.M. Bacterial communities in Arctic fields of Finnish Lapland are stable but highly pH-dependent. FEMS Microbiol. Ecol. 2007, 59, 452–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lladó, S.; López-Mondéjar, R.; Baldrian, P. Drivers of microbial community structure in forest soils. Appl. Microbiol. Biotechnol. 2018, 102, 4331–4338. [Google Scholar] [CrossRef] [PubMed]
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Banach, A.; Błaszczyk, M. Indicators of arable soils fatigue—Bacterial families and genera: A metagenomic approach. Ecol. Indic. 2018, 93, 490–500. [Google Scholar] [CrossRef]
- Wolińska, A. Metagenomic Achievements in Microbial Diversity Determination in Croplands: A Review. In Microbial Diversity in the Genomic Era; Academic Press: Cambridge, MA, USA, 2019; Volume 2, pp. 15–35. [Google Scholar]
- Xiao, X.M.; Cheng, Z.H.; Meng, H.W.; Liu, L.H.; Li, H.Z.; Dong, Y.X. Intercropping of green garlic (Allium sativum L.) induces nutrient concentration changes in the soil and plants in continuously cropped cucumber (Cucumis sativus L.) in a plastic tunnel. PLoS ONE 2013, 8, 62173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Yu, G.; Wu, F. Effects of intercropping cucumber with onion or garlic on soil enzyme activities, microbial communities and cucumber yield. Eur. J. Soil Biol. 2011, 47, 279–287. [Google Scholar] [CrossRef]
- Bargaz, A.; Noyce, G.L.; Fulthorpe, R.; Carlsson, G.; Furze, J.R.; Jensen, E.S.; Dhiba, D.; Isaac, M.E. Species interactions enhance root allocation, microbial diversity and P acquisition in intercropped wheat and soybean under P deficiency. Appl. Soil Ecol. 2017, 120, 179–188. [Google Scholar] [CrossRef]
- Wang, G.; Bei, S.; Li, J.; Bao, X.; Zhang, J.; Schultz, P.A.; Li, H.; Li, L.; Zhang, F.; Bever, J.D.; et al. Soil microbial legacy drives crop diversity advantage: Linking ecological plant–soil feedback with agricultural intercropping. J. Appl. Ecol. 2020, 186, 281. [Google Scholar] [CrossRef]
- Dang, K.; Gong, X.; Zhao, G.; Wang, H.; Ivanistau, A.; Feng, B. Intercropping alters the soil microbial diversity and community to facilitate Nitrogen assimilation: A potential mechanism for increasing pros millet grain yield. Front. Microbiol. 2020, 11, 2975. [Google Scholar] [CrossRef]
- Li, Q.; Chen, J.; Wu, L.; Luo, X.; Li, N.; Arafat, Y.; Lin, S.; Lin, W. Belowground Interactions Impact the Soil Bacterial Community, Soil Fertility, and Crop Yield in Maize/Peanut Intercropping Systems. Int. J. Mol. Sci. 2018, 19, 622. [Google Scholar] [CrossRef] [Green Version]
- Correa-Galeote, D.; Bedmar, E.J.; Arone, G.J. Maize endophytic bacterial diversity as affected by soil cultivation history. Front. Microbiol. 2018, 9, 484. [Google Scholar] [CrossRef]
- Liu, Y.; Zuo, S.; Xu, L.; Zou, Y.; Song, W. Study on diversity of endophytic bacterial communities in seeds of hybrid maize and their parental lines. Arch. Microbiol. 2012, 194, 1001–1012. [Google Scholar] [CrossRef] [PubMed]



| No of Raster | pH(H2O) | Eh (mV) | TOC (%) | Moisture (%) |
|---|---|---|---|---|
| SPRING | ||||
| K20-144 | 6.07 ± 0.04 | 509.23 ± 0.12 | 0.71 ± 0.01 | 4.59 ± 0.07 |
| K20-145 | 6.07 ± 0.03 | 526.50 ± 0.85 | 0.82 ± 0.04 | 5.89 ± 0.11 |
| K20-146 | 7.90 ± 0.03 | 493.60 ± 0.08 | 0.61 ± 0.03 | 7.56 ± 0.05 |
| K20-147 | 6.21 ± 0.03 | 514.30 ± 0.16 | 1.16 ± 0.04 | 9.53 ± 0.08 |
| K20-148 | 6.07 ± 0.04 | 525.60 ± 0.21 | 1.34 ± 0.03 | 8.92 ± 0.06 |
| K21-149 | 6.02 ± 0.01 | 514.70 ± 0.04 | 0.52 ± 0.03 | 6.20 ± 0.03 |
| K21-151 | 6.40 ± 0.01 | 509.60 ± 1.60 | 0.59 ± 0.03 | 7.36 ± 0.06 |
| K21-152 | 5.82 ± 0.04 | 520.20 ± 0.08 | 0.56 ± 0.01 | 8.51 ± 0.12 |
| K21-154 | 6.83 ± 0.02 | 507.00 ± 0.16 | 0.67 ± 0.05 | 7.40 ± 0.06 |
| K21-155 | 7.93 ± 0.03 | 499.70 ± 0.21 | 1.02 ± 0.02 | 8.41 ± 0.05 |
| SUMMER | ||||
| K20-144 | 6.63 ± 0.05 | 534.40 ± 0.20 | 0.85 ± 0.02 | 10.26 ± 0.03 |
| K20-145 | 6.20 ± 0.05 | 534.33 ± 0.26 | 0.79 ± 0.02 | 7.70 ± 0.06 |
| K20-146 | 5.95 ± 0.01 | 533.86 ± 0.25 | 0.77 ± 0.03 | 9.13 ± 0.07 |
| K20-147 | 6.29 ± 0.03 | 527.07 ± 0.20 | 0.77 ± 0.04 | 10.92 ± 0.04 |
| K20-148 | 5.70 ± 0.02 | 534.57 ± 0.05 | 1.14 ± 0.04 | 14.64 ± 0.08 |
| K21-149 | 5.31 ± 0.02 | 561.53 ± 0.70 | 0.71 ± 0.05 | 10.24 ± 0.03 |
| K21-151 | 5.44 ± 0.01 | 567.60 ± 0.17 | 0.45 ± 0.01 | 9.09 ± 0.08 |
| K21-152 | 5.46 ± 0.01 | 568.03 ± 0.59 | 0.62 ± 0.04 | 8.84 ± 0.06 |
| K21-154 | 6.08 ± 0.02 | 542.67 ± 0.37 | 0.77 ± 0.03 | 10.73 ± 0.09 |
| K21-155 | 6.60 ± 0.04 | 537.73 ± 0.95 | 0.70 ± 0.04 | 10.44 ± 0.05 |
| AUTUMN | ||||
| K20-144 | 6.17 ± 0.03 | 491.07 ± 0.28 | 0.58 ± 0.02 | 13.05 ± 0.06 |
| K20-145 | 6.60 ± 0.01 | 417.73 ± 3.02 | 0.52 ± 0.03 | 10.88 ± 0.08 |
| K20-146 | 6.94 ± 0.01 | 424.83 ± 0.12 | 0.42 ± 0.01 | 12.23 ± 0.09 |
| K20-147 | 6.61 ± 0.03 | 469.67 ± 0.25 | 0.51 ± 0.05 | 12.01 ± 0.11 |
| K20-148 | 5.99 ± 0.16 | 499.13 ± 0.76 | 0.87 ± 0.01 | 16.32 ± 0.04 |
| K21-149 | 5.99 ± 0.02 | 496.53 ± 0.33 | 0.23 ± 0.02 | 11.87 ± 0.06 |
| K21-151 | 5.84 ± 0.01 | 515.30 ± 0.21 | 0.35 ± 0.01 | 9.66 ± 0.03 |
| K21-152 | 6.33 ± 0.01 | 506.57 ± 0.23 | 0.36 ± 0.02 | 9.61 ± 0.05 |
| K21-154 | 6.55 ± 0.02 | 484.07 ± 0.17 | 0.39 ± 0.03 | 13.35 ± 0.02 |
| K21-155 | 6.99 ± 0.04 | 426.17 ± 0.12 | 0.40 ± 0.03 | 11.88 ± 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolińska, A.; Kruczyńska, A.; Podlewski, J.; Słomczewski, A.; Grządziel, J.; Gałązka, A.; Kuźniar, A. Does the Use of an Intercropping Mixture Really Improve the Biology of Monocultural Soils?—A Search for Bacterial Indicators of Sensitivity and Resistance to Long-Term Maize Monoculture. Agronomy 2022, 12, 613. https://doi.org/10.3390/agronomy12030613
Wolińska A, Kruczyńska A, Podlewski J, Słomczewski A, Grządziel J, Gałązka A, Kuźniar A. Does the Use of an Intercropping Mixture Really Improve the Biology of Monocultural Soils?—A Search for Bacterial Indicators of Sensitivity and Resistance to Long-Term Maize Monoculture. Agronomy. 2022; 12(3):613. https://doi.org/10.3390/agronomy12030613
Chicago/Turabian StyleWolińska, Agnieszka, Anna Kruczyńska, Jacek Podlewski, Andrzej Słomczewski, Jarosław Grządziel, Anna Gałązka, and Agnieszka Kuźniar. 2022. "Does the Use of an Intercropping Mixture Really Improve the Biology of Monocultural Soils?—A Search for Bacterial Indicators of Sensitivity and Resistance to Long-Term Maize Monoculture" Agronomy 12, no. 3: 613. https://doi.org/10.3390/agronomy12030613
APA StyleWolińska, A., Kruczyńska, A., Podlewski, J., Słomczewski, A., Grządziel, J., Gałązka, A., & Kuźniar, A. (2022). Does the Use of an Intercropping Mixture Really Improve the Biology of Monocultural Soils?—A Search for Bacterial Indicators of Sensitivity and Resistance to Long-Term Maize Monoculture. Agronomy, 12(3), 613. https://doi.org/10.3390/agronomy12030613









