Multi-Scale and Multi-Match for Few-Shot Plant Disease Image Semantic Segmentation
Abstract
1. Introduction
- Multi-scale and multi-prototypes match is proposed for few-shot plant disease semantic segmentation. This method generates multiple prototypes and multiple query feature maps at different scales, and then the relationships between prototypes and query feature maps are established through the similarity measure method. Finally, the relationships at different scales are fused. With this approach, our network can more precisely identify plant disease signatures.
- The mixed similarity is designed as the weighted sum of cosine similarity and euclidean distance. When the similarity of the direction and the actual distance between two vectors are jointly considered, more accurate similarity can be obtained.
- A CBAM attention module is added to our network to make the network pay attention to the important plant disease feature and ignore interference information, which is beneficial to improve accuracy.
- To accomplish the few-shot semantic segmentation task, we constructed a plant disease dataset(PDID-5i) that is suitable for the task. Experiments on the dataset show that the model we designed is very effective.
2. Related Work
2.1. Semantic Segmentation of Plant Disease Images
2.2. Few-Shot Semantic Segmentation
3. The Proposed Method
3.1. Problem Setting
3.2. Evaluation Indicators
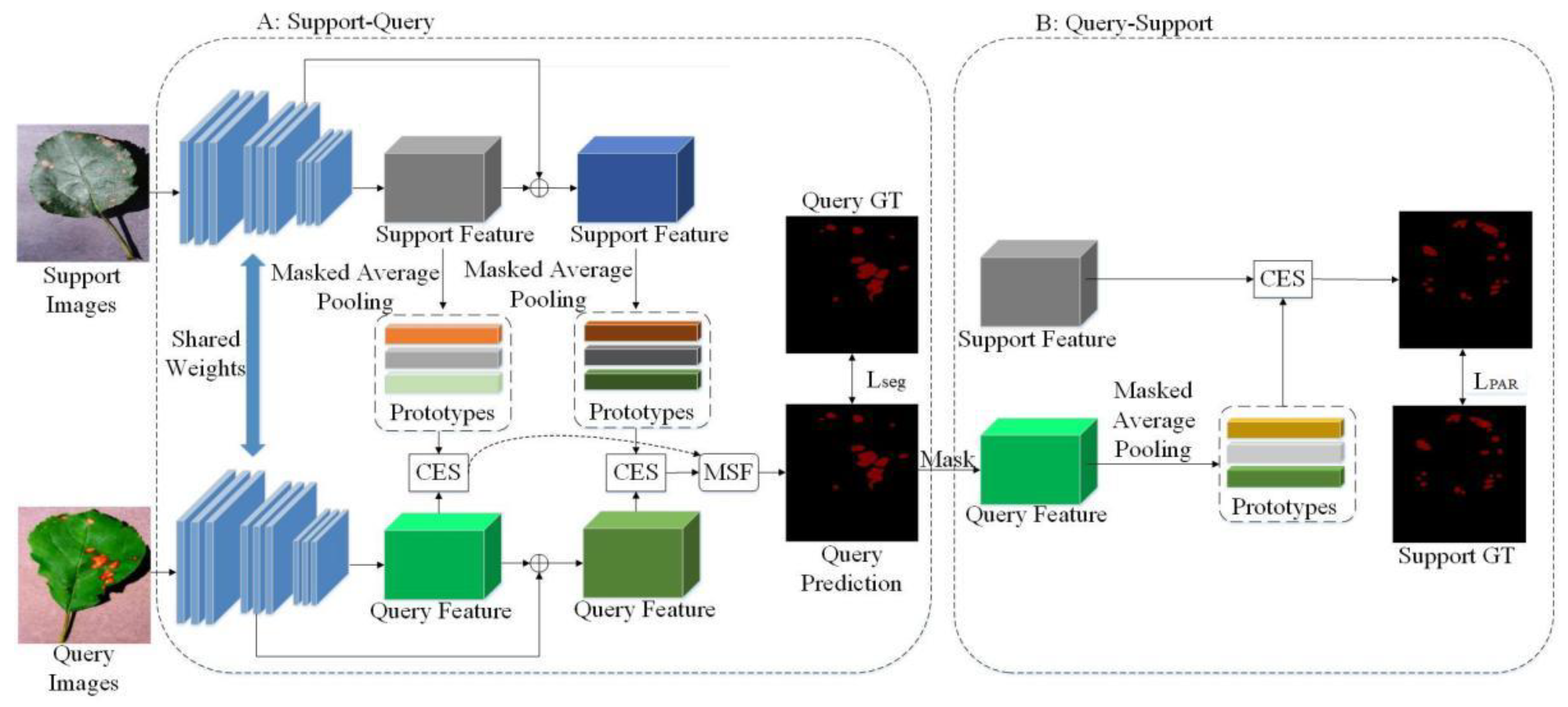
3.3. Method Overview
3.4. Multi-Scale and Multi-Prototypes Match
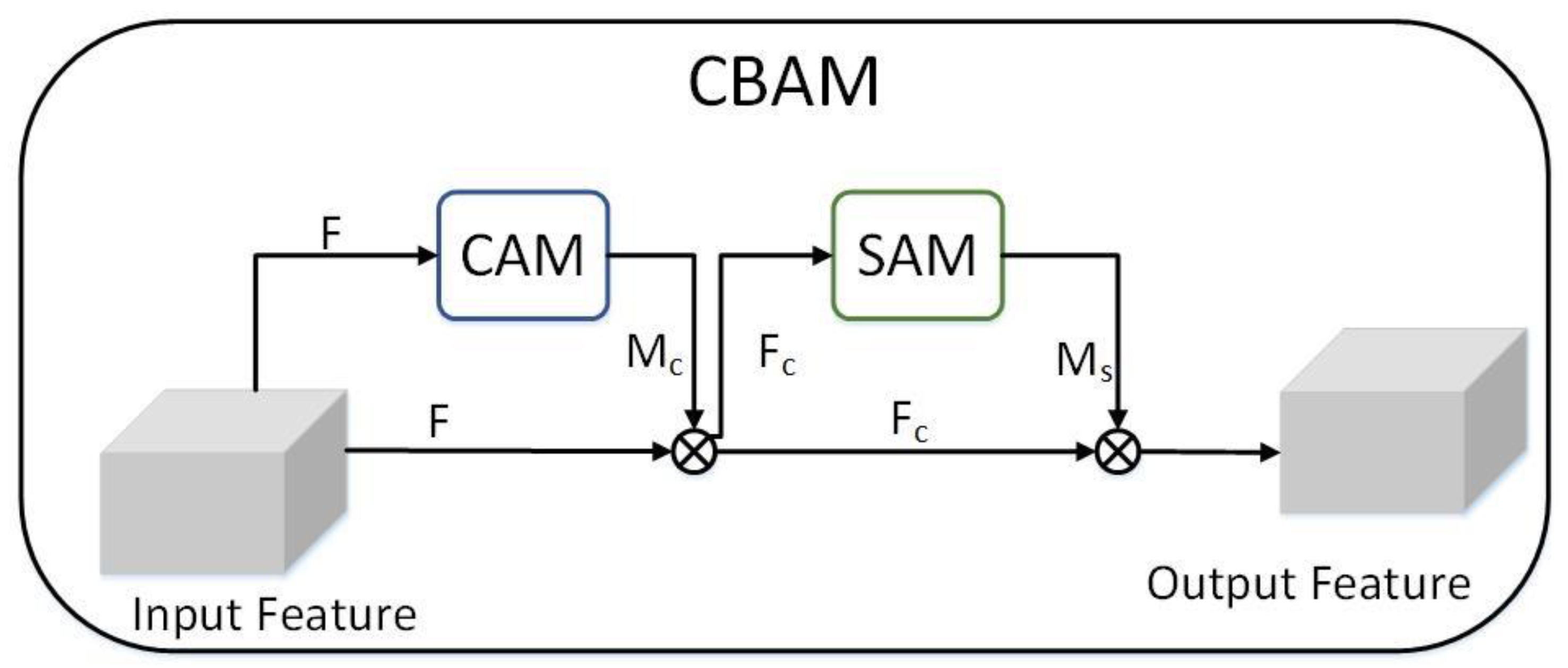
3.5. CBAM Module
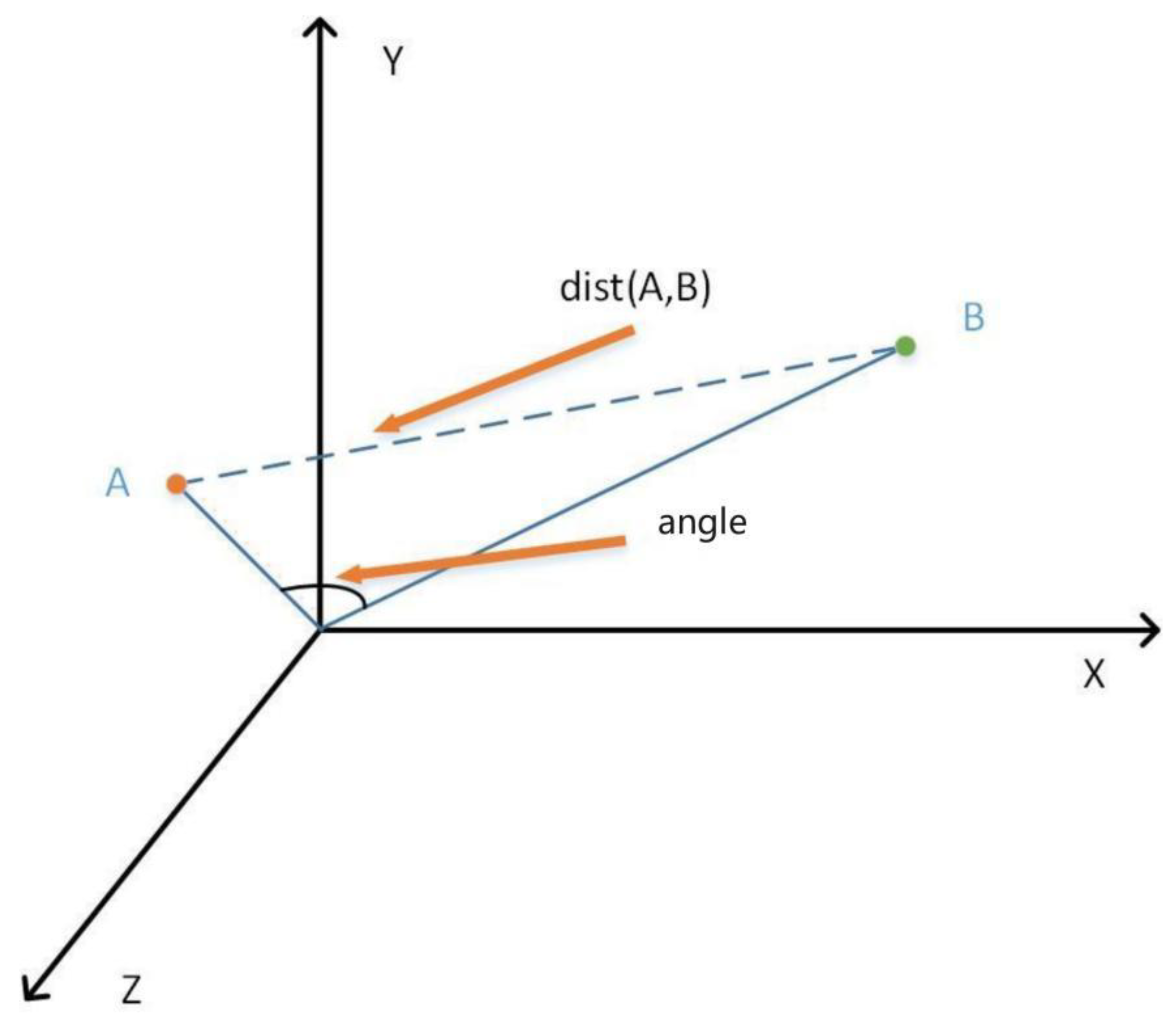
3.6. Hybrid Similarity
3.7. Loss Function
4. Experiments
4.1. Experimental Setup
4.2. Experimental Results and Discussions
4.2.1. Validation of Proposed Model
4.2.2. Performance Comparison of Different Types of Diseases
4.2.3. Qualitative Analysis
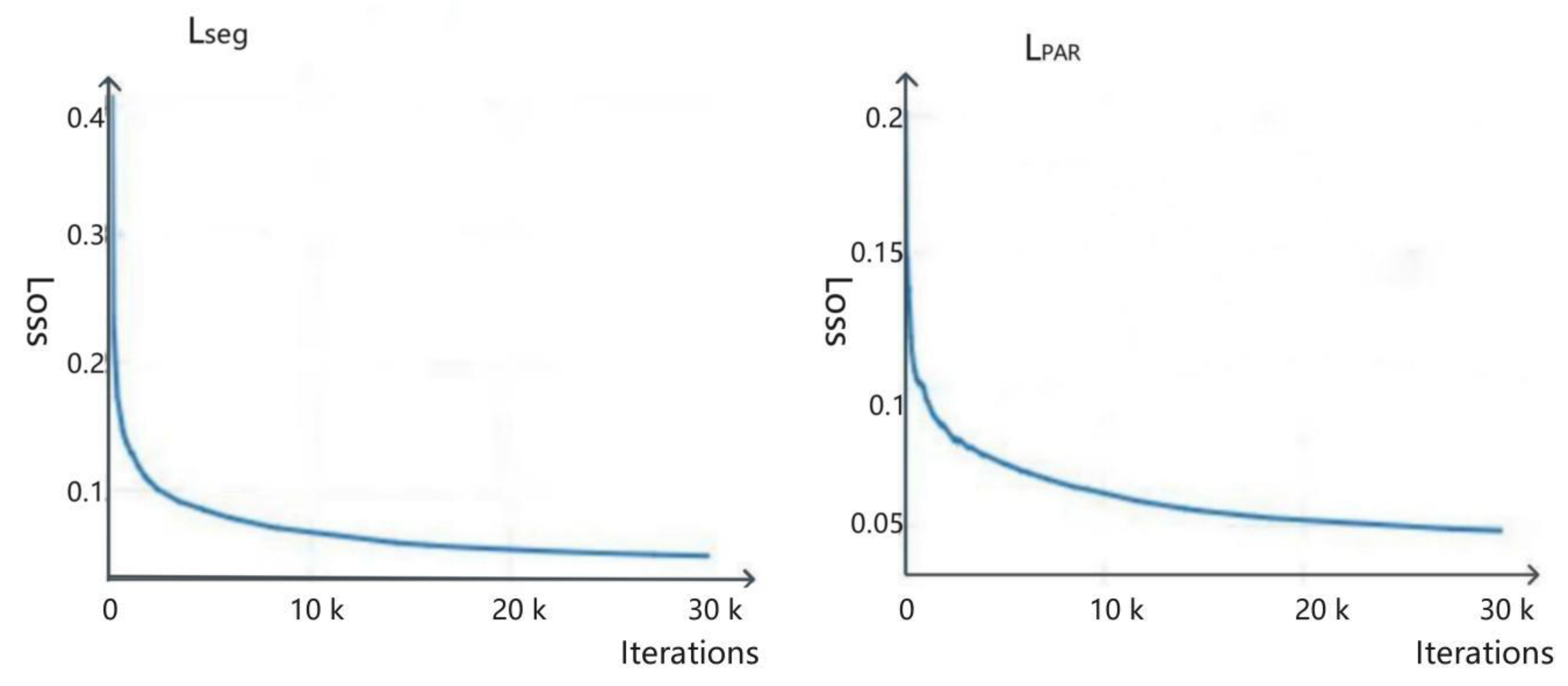
4.2.4. Training Loss Function
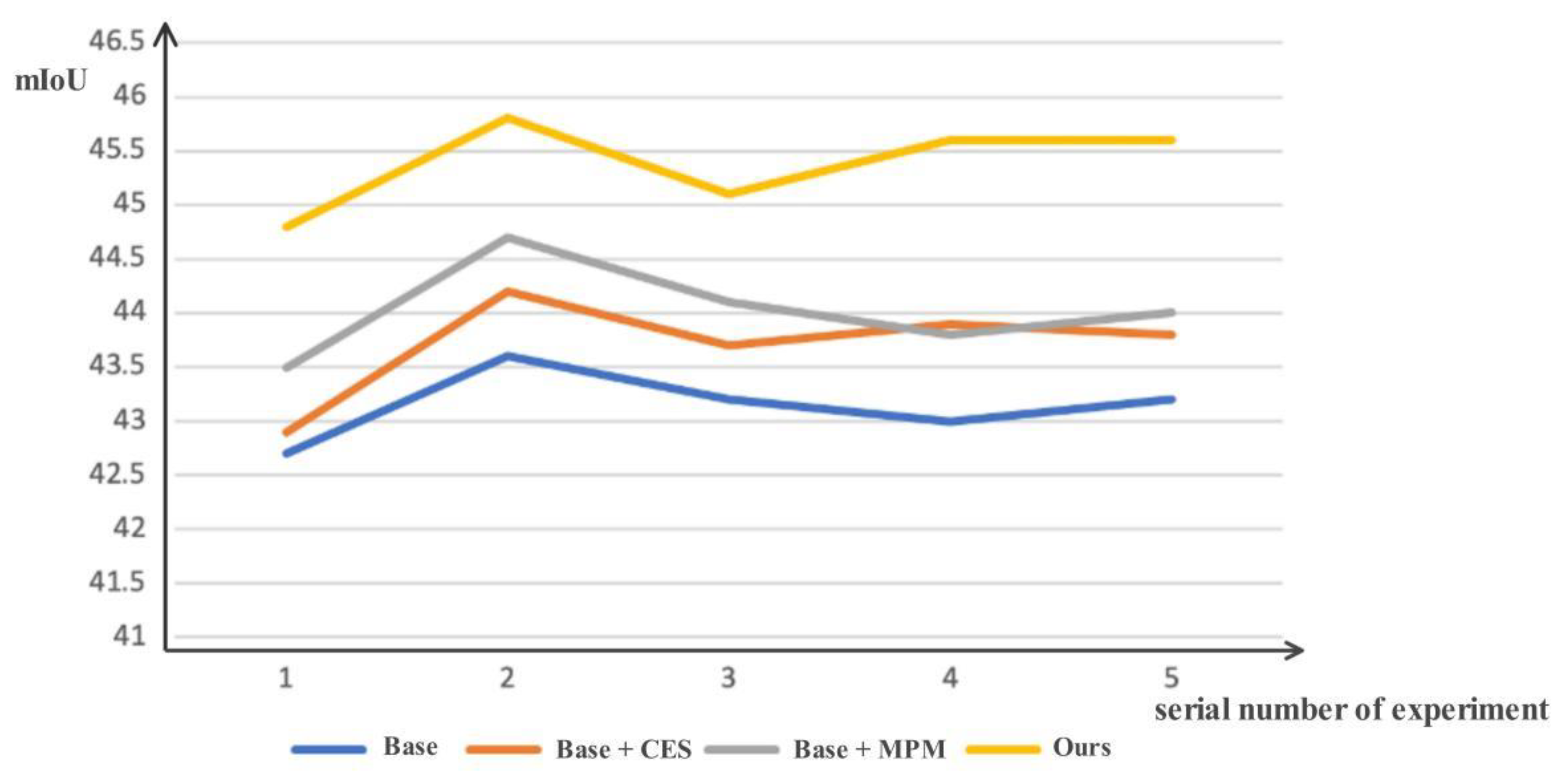
4.3. Ablation Studies
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aliane, N.; Muñoz, C.Q.G.; Sánchez-Soriano, J. Web and MATLAB-Based Platform for UAV Flight Management and Multispectral Image Processing. Sensors 2022, 22, 4243. [Google Scholar] [CrossRef] [PubMed]
- Chadalavada, K.; Anbazhagan, K.; Ndour, A.; Choudhary, S.; Palmer, W.; Flynn, J.R.; Mallayee, S.; Pothu, S.; Prasad, K.V.S.V.; Varijakshapanikar, P.; et al. NIR Instruments and Prediction Methods for Rapid Access to Grain Protein Content in Multiple Cereals. Sensors 2022, 22, 3710. [Google Scholar] [CrossRef] [PubMed]
- Chadalavada, K.; Anbazhagan, K.; Ndour, A.; Choudhary, S.; Palmer, W.; Flynn, J.R.; Mallayee, S.; Pothu, S.; Prasad, K.V.S.V.; Varijakshapanikar, P.; et al. Spectra Fusion of Mid-Infrared (MIR) and X-ray Fluorescence (XRF) Spectroscopy for Estimation of Selected Soil Fertility Attributes. Sensors 2022, 22, 3459. [Google Scholar]
- Liu, G.; Tian, S.; Mo, Y.; Chen, R.; Zhao, Q. On the Acquisition of High-Quality Digital Images and Extraction of Effective Color Information for Soil Water Content Testing. Sensors 2022, 22, 3130. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Qiao, Y.; Li, J.; Zhang, H.; Zhang, M.; Wang, M. An Improved Lightweight Network for Real-Time Detection of Apple Leaf Diseases in Natural Scenes. Agronomy 2022, 12, 2363. [Google Scholar] [CrossRef]
- Eunice, J.; Popescu, D.E.; Chowdary, M.K.; Hemanth, J. Deep Learning-Based Leaf Disease Detection in Crops Using Images for Agricultural Applications. Agronomy 2022, 12, 2395. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Snell, J.; Swersky, K.; Zemel, R. Denseaspp for semantic segmentation in street scenes. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 3684–3692. [Google Scholar]
- Snell, J.; Swersky, K.; Zemel, R. Prototypical networks for few-shot learning. Adv. Neural Inf. Process. Syst. 2017, 30, 4080–4090. [Google Scholar]
- Zhang, X.; Wei, Y.; Yang, Y.; Huang, T.S. Sg-one: Similarity guidance network for one-shot semantic segmentation. IEEE Trans. Cybern. 2020, 50, 3855–3865. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Wang, K.; Liew, J.H.; Zou, Y.; Zhou, D.; Feng, J. Panet: Few-shot image semantic segmentation with prototype alignment. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 9197–9206. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Goncalves, J.P.; Pinto, F.A.; Queiroz, D.M.; Villar, F.M.; Barbedo, J.G.; Del Ponte, E.M. Deep learning architectures for semantic segmentation and automatic estimation of severity of foliar symptoms caused by diseases or pests. Biosyst. Eng. 2021, 210, 129–142. [Google Scholar] [CrossRef]
- Tassis, L.M.; de Souza, J.E.T.; Krohling, R.A. A deep learning approach combining instance and semantic segmentation to identify diseases and pests of coffee leaves from in-field images. Comput. Electron. Agric. 2021, 186, 106191. [Google Scholar] [CrossRef]
- Yang, R.; Yu, Y. Artificial convolutional neural network in object detection and semantic segmentation for medical imaging analysis. Front. Oncol. 2021, 11, 573. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Zhang, Q.; Han, Y.; Megason, S.; Hormoz, S.; Mosaliganti, K.R.; Lam, J.C.K.; Li, V.O.K. A novel deep learning-based 3D cell segmentation framework for future image-based disease detection. Sci. Rep. 2022, 12, 342. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, F.; Yan, H.; Wang, K.; Ma, Y.; Shen, L.; Xu, M.; Alzheimer’s Disease Neuroimaging Initiative. A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in Alzheimer’s disease. Neuroimage 2020, 208, 116459. [Google Scholar] [CrossRef] [PubMed]
- Douarre, C.; Crispim-Junior, C.F.; Gelibert, A.; Tougne, L.; Rousseau, D. Novel data augmentation strategies to boost supervised segmentation of plant disease. Comput. Electron. Agric. 2019, 165, 104967. [Google Scholar] [CrossRef]
- Sodjinou, S.G.; Mohammadi, V.; Mahama, A.T.S.; Gouton, P. A deep semantic segmentation-based algorithm to segment crops and weeds in agronomic color images. Inf. Process. Agric. 2021, 9, 355–364. [Google Scholar] [CrossRef]
- Lin, K.; Gong, L.; Huang, Y.; Liu, C.; Pan, J. Deep Learning-Based Segmentation and Quantification of Cucumber Powdery Mildew Using Convolutional Neural Network. Front. Plant Sci. 2019, 10, 155. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Jiang, L.; Zhang, T.; Ji, J.; Xiong, H. A multi-part convolutional attention network for fine-grained image recognition. In Proceedings of the 2018 24th International Conference on Pattern Recognition (ICPR), Beijing, China, 18–20 August 2018; pp. 1857–1862. [Google Scholar]
- He, X.; Zhou, Y.; Zhao, J.; Zhang, M.; Yao, R.; Liu, B.; Li, H. Semantic segmentation of remote-sensing images based on multiscale feature fusion and attention refinement. IEEE Geosci. Remote Sens. Lett. 2021, 19, 1–5. [Google Scholar] [CrossRef]
- Tang, H.; Liu, X.; Sun, S.; Yan, X.; Xie, X. Recurrent mask refinement for few-shot medical image segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 10–17 October 2021; pp. 3918–3928. [Google Scholar]
- Wang, H.; Zhang, X.; Hu, Y.; Yang, Y.; Cao, X.; Zhen, X. Few-shot semantic segmentation with democratic attention networks. In Proceedings of the European Conference on Computer Vision, Glasgow, UK, 23–28 August 2020; Springer: Cham, Switzerland, 2020; pp. 730–746. [Google Scholar]
- Xie, G.S.; Liu, J.; Xiong, H.; Shao, L. Scale-aware graph neural network for few-shot semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 5475–5484. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Kwon, H.; Song, T.; Kim, S.; Sohn, K. Mask-Guided Attention and Episode Adaptive Weights for Few-Shot Segmentation. In Proceedings of the 2022 IEEE International Conference on Image Processing (ICIP), Bordeaux, France, 16–19 October 2022; pp. 2611–2615. [Google Scholar]


| C1 | C2 | C3 | C4 | C5 | |
|---|---|---|---|---|---|
| PDID-50 | Apple Frogeye Spot | Apple Scab | Grape Black Measles Fungus | Grape Black Rot Fungus | Grape Leaf Blight Fungus |
| C6 | C7 | C8 | C9 | C10 | |
| PDID-51 | Peach Bacterial Spot | Tomato Early Blight Fungus | Tomato Late Blight Water Mold | Tomato Leaf Mold Fungus | Tomato Septoria Leaf Spot Fungus |
| Method | 1-Way 1-Shot | Params | ||
|---|---|---|---|---|
| Split-0 | Split-1 | Mean | ||
| PANet | 43.1 | 34.5 | 38.8 | 14.7 M |
| Ours | 45.4 | 35.6 | 40.5 | 16.5 M |
| Method | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | C10 |
|---|---|---|---|---|---|---|---|---|---|---|
| PANet | 57.3 | 35.3 | 42 | 54.4 | 26.7 | 40.6 | 52.4 | 29.4 | 23 | 27 |
| Ours | 59.9 | 36.6 | 44.5 | 56.9 | 29.2 | 41.3 | 52.2 | 32.7 | 23.6 | 28.2 |
| Ratio | 9:1 | 8:2 | 7:3 | 6:4 | 5:5 | 4:6 | 3:7 | 2:8 | 1:9 |
|---|---|---|---|---|---|---|---|---|---|
| mIoU | 45.4 | 44.1 | 44.5 | 44.3 | 42.7 | 42.3 | 41.6 | 40.5 | 39.6 |
| binary-mIoU | 69.2 | 68.3 | 68.4 | 67.9 | 66.7 | 66.5 | 65.5 | 64.2 | 63.4 |
| Way | mIoU | Binary-mIoU |
|---|---|---|
| 1 | 45.4 | 69.2 |
| 2 | 37.7 | 64.1 |
| 3 | 42.6 | 66.5 |
| 4 | 42.9 | 67.4 |
| 5 | 44 | 68.5 |
| 6 | 36.8 | 63.8 |
| 7 | 42.1 | 66.5 |
| CES | MPM | mIoU | Binary_mIoU |
|---|---|---|---|
| 43.1 | 67.9 | ||
| √ | 43.7 | 68.5 | |
| √ | 44 | 68.8 | |
| √ | √ | 45.4 | 69.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, W.; Hu, W.; Xie, L.; Yang, Z. Multi-Scale and Multi-Match for Few-Shot Plant Disease Image Semantic Segmentation. Agronomy 2022, 12, 2847. https://doi.org/10.3390/agronomy12112847
Yang W, Hu W, Xie L, Yang Z. Multi-Scale and Multi-Match for Few-Shot Plant Disease Image Semantic Segmentation. Agronomy. 2022; 12(11):2847. https://doi.org/10.3390/agronomy12112847
Chicago/Turabian StyleYang, Wenji, Wenchao Hu, Liping Xie, and Zhenji Yang. 2022. "Multi-Scale and Multi-Match for Few-Shot Plant Disease Image Semantic Segmentation" Agronomy 12, no. 11: 2847. https://doi.org/10.3390/agronomy12112847
APA StyleYang, W., Hu, W., Xie, L., & Yang, Z. (2022). Multi-Scale and Multi-Match for Few-Shot Plant Disease Image Semantic Segmentation. Agronomy, 12(11), 2847. https://doi.org/10.3390/agronomy12112847






